Abstract
Human fibroblast growth factor (hFGF-1) is a ~17 kDa heparin binding cytokine. It lacks the conventional hydrophobic N-terminal signal sequence and is secreted through non-classical secretion routes. Under stress, hFGF-1 is released as a multiprotein complex consisting of hFGF-1, S100A13 (a calcium binding protein), and p40 synaptotagmin (Syt1). Copper (Cu2+) is shown to be required for the formation of the multiprotein hFGF-1 release complex (Landriscina et al.,2001; Di Serio et al., 2008). Syt1, containing the lipid binding C2B domain, is believed to play an important role in the eventual export of the hFGF-1 across the lipid bilayer. In this study, we characterize Cu2+ and lipid interactions of the C2B domain of Syt1 using multidimensional NMR spectroscopy. The results highlight how Cu2+ appears to stabilize the protein bound to pS vesicles. Cu2+ and lipid binding interface mapped using 2D 1H–15N heteronuclear single quantum coherence experiments reveal that residues in β-strand I contributes to the unique Cu2+ binding site in the C2B domain. In the absence of metal ions, residues located in Loop II and β-strand IV contribute to binding to unilamelar pS vesicles. In the presence of Cu2+, additional residues located in Loops I and III appear to stabilize the protein-lipid interactions. The results of this study provide valuable information towards understanding the molecular mechanism of the Cu2+-induced non-classical secretion of hFGF-1.
Keywords: Fibroblast growth factor, Secretion, Non-classical, Synaptotagmin, Lipid binding
1. Introduction
The human acidic fibroblast growth factor (hFGF-1) [1,2], a potent mitogen, is a ubiquitously expressed member of the FGF family [3–7]. hFGF-1 is a ~17-kDa all-β-sheet protein and is involved in the regulation of a wide variety of important cellular processes such as angiogenesis, morphogenesis, inflammation, tumor growth, and wound healing [8]. Interestingly, unlike most secreted proteins, hFGF-1 lacks the N-terminal signal sequence and therefore is secreted through routes that are independent of the classical endoplasmic reticulum (ER)–Golgi pathway [9].
The precise mechanism for the secretion of the signal peptide-less proteins is not completely understood. Several studies show hFGF-1 is released in response to stresses such as heat shock, hypoxia, cultivation under low serum conditions, and cell treatment with low-density lipoproteins (LDLs) [10–13]. Jackson et al. [11] demonstrated that the formation of FGF-1 homodimer is a prerequisite for its release in response to heat shock and hypoxia. Homodimer formation is facilitated by copper (Cu2+)-induced oxidation of a specific cysteine residue (Cys30) in hFGF-1. Cu2+ has been demonstrated to be required for the assembly of multiprotein hFGF-1 release complex. Prudovsky et al. [2,14], investigating the spatio-temporal characteristics of the non-classical release of hFGF-1 using real-time confocal Microscopy, showed the formation of a multiprotein hFGF-1 release complex near the inner surface of the plasma membrane [14]. The proteins assembled in the multiprotein hFGF-1 release complex include the FGF-1 homodimer, S100A13 (a calcium binding protein), and the p40 form of synaptotagmin, a protein involved in secretory vesicle docking [15,16]. More recently, the non-classically secreted enzyme sphingosine kinase 1 is also suggested to be a part of the hFGF-1 release complex [10,17–20]. However, to date, details of the molecular events leading to the formation of multiprotein FGF-1 release complex, as well as the mechanism underlying the export of this growth factor to the extracellular compartment is still an enigma.
Synaptotagmin-1 (Syt-1) is a 65 kDa synaptic vesicle protein containing a short intravesicular N-terminus, a single-pass trans-membrane domain, and a C-terminal extravesicular cytosolic portion including two calcium binding C2 (C2A and C2B) domains (Fig. 1) [21–26]. Both C2A and C2B domains bind to calcium (Ca2+) and partially penetrate into the lipid bilayer, and the interactions between C2A, C2B and Ca2+ are believed to be critical for the membrane fusion activity of Syt-1. The 40-kDa form of synaptotagmin (p40 Syt1) represents a product of the alternative in-frame initiation of synaptotagmin mRNA translation [27–29]. It lacks the intravesicular and transmembrane domains and corresponds to the extravesicular domain of p65 Syt1. p40 Syt 1 is a member of the FGF1 export complex which is critically important for stress-induced nonclassical FGF1 release [19,30]. Although, the exact role of the C2 domains of Syt-1 in the non-classical secretion of FGF-1 is still unclear, it is believed that these lipid-binding domains are important for anchoring the multi-protein hFGF-1 release complex to the cell membrane. Indeed, mutation of lysines 326, 327 and 331 in C2B domain drastically reduced the membrane destabilizing activity of p40 Syt1 and abolished its nonclassical export [31].
Fig. 1.
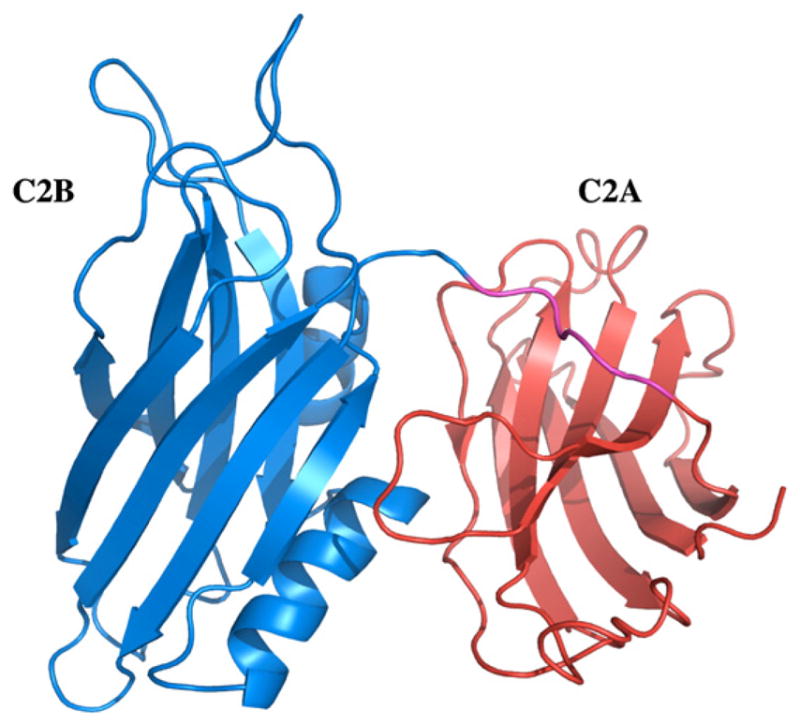
Three-dimensional structure of the C2A (orange) and C2B (blue) domains of Syt1. The linker region connecting the C2 domains is shown in magenta. The structure was generated using the pdb accession code 2R83.
The C2B domain has a β-sandwich structure formed by two layers of four antiparallel β-strands. Unlike the C2A domain, which lacks helical segments, the C2B domain contains two α-helices (Fig. 1) [32]. The helical segment located at the C-terminal end of the molecule is tightly packed against residues in β-strands III, VI, and VII [32,33]. A stretch of negatively charged residues in the C-terminal helix is in close proximity to a patch of positively charged residues that are implicated in C2B domain’s interaction with other proteins. The striking dipolar distribution of charged residues on one side of the molecule is a characteristic feature of the structure of the C2B domain. Two distinct Ca2+ binding sites have been characterized in the C2B domain [32]. However, additional calcium binding sites could not be ruled out in the presence of interacting protein or lipid components [4,15,33–35]. Although the Ca2+-dependent lipid binding affinity of the C2B domain has been well characterized, very little is known about the structural role of this domain in the Cu2+-mediated non-classical secretion of hFGF-1. In this study, we investigated the influence of metal ions (Ca2+ and Cu2+) on the structural stability and lipid binding affinity of the C2B domain. In addition, the lipid-binding interface on the C2B domain, in the absence and presence of the metal ions, has been characterized using multidimensional NMR spectroscopy. The results of this study clearly show that Cu2+ binds to distinct sites and significantly influences the lipid binding affinity of the C2B domain.
2. Materials and methods
Ingredients for Luria Broth were obtained from EMD Biosciences. Aprotinin, pepstatin, leupeptin, phenylmethylsulfonyl fluoride, triton X-100, terbium chloride, and β-mercaptoethanol were obtained from Sigma Co. (St. Louis). Heparin sepharose and glutathione sepharose were obtained from GE Healthcare. Labeled 15NH4Cl and D2O were purchased from Cambridge Isotope Laboratories. All other chemicals used were of high quality analytical grade. All experiments were performed at 25 °C. Unless specified, all solutions were made in 10 mM Tris buffer (pH 7.5) containing 100 mM NaCl.
2.1. Expression and purification of the C2B domain
cDNA encoding the C2B domain of synaptotagmin I (residues 270 to 421) and p40 Syt1 was kindly provided by Professor Thomas Sudhof. Escherichia coli expressing GST-tagged C2B were induced with isopropyl-β-D-thiogalactopyranoside (IPTG) when absorbance (at 600 nm) reached 0.5–0.6, and the cells were harvested by centrifugation at 6000 rpm after 4 h of induction. The harvested cells were resuspended, and cell walls were ruptured by sonication. The cell lysate was centrifuged at 16,000 rpm for 20 min. The supernatant was then incubated with glutathione sepharose, and the resin was then extensively washed with Tris buffer saline (TBS, 20 mM Tris 150 mM NaCl) and 20 mM Tris containing 1M NaCl to remove impurities. The column was then equilibrated with cleavage buffer (20 mM Tris, 0.2 M NaCl, 2.5 mM CaCl2, pH8.0), and an on-column thrombin cleavage was carried out (2 NIH units/mL) at 25 °C for 3 h. The cleaved protein (C2B) was eluted with TBS buffer as described by Ubach et al. [33]. The homogeneity of the protein was then assessed using SDS-PAGE, and the concentration of the protein was estimated on the [33] basis of the extinction coefficient (ε= 19940 M−1 cm−1) value calculated from the amino acid sequence of the C2B domain. Complete removal of nucleotide impurities was verified by recording UV spectrum of the protein.
2.2. Preparation of isotope-enriched C2B domain
Uniform 15N labeling of C2B was achieved using M9 minimal medium containing 15NH4Cl. To achieve maximal expression yields, the composition of the M9 medium was modified by the addition of a mixture of vitamins [36–37]. The expression host strain E. coli BL21 (DE3) is a vitamin B1-deficient host, and hence, the medium was supplemented with thiamine (vitamin B1) [36].
2.3. Preparation of the lipid vesicles
Small unilamellar vesicles (SUV) were prepared by dissolving the solid phospholipids (phosphatidyl serine (pS)) purchased from Avanti polar lipid, Inc.) in chloroform, followed by evaporation to dryness under nitrogen. The lipid film was suspended in 10 mM Tris buffer (pH 7.5) containing 100 mM NaCl and sonicated in an AQUASONIC-75D12 bath-type sonicator until optical clarity was obtained. The solution was finally centrifuged for 5 min at 14000 rpm in an Eppendorf microfuge before use and stored for a maximum of 6 h in ice.
2.4. NMR experiments
All NMR experiments were performed on a Bruker Avance-700 MHz NMR spectrometer equipped with a cryoprobe at 30 °C. 15N decoupling during data acquisition was accomplished using the globally optimized altering phase rectangular pulse sequence, and 2048 complex data points were collected in the 15N dimension. 1H–15N HSQC spectra were recorded at 32 scans. The concentration of the protein sample used was 0.1 mM in 90% H2O and 10 % D2O prepared in 50 mM MES buffer containing 150 mM NaCl and 2 mM DTT (pH 6.3). The spectra were processed on a Windows workstation using XWIN-NMR and Sparky software [38].
3. Results and discussion
S100A13 and p40 Syt1 are both Ca2+ binding proteins that chaperone the Cu2+-mediated non-classical secretion of hFGF-1 [11]. Recent evidence suggests that the C2 domains of p40 Syt1 bind to Cu2+ and facilitate the anchoring of hFGF-1 to the membrane bilayer [23,24]. In addition, C2B is known to preferentially bind to small unilamelar phosphatidylserine vesicles [26,39]. In this context, it will be interesting to map the pS binding sites on the C2B domain, in the presence and absence of Cu2+, using multidimensional NMR spectroscopy.
3.1. Conformational changes monitored by 2D NMR spectroscopy
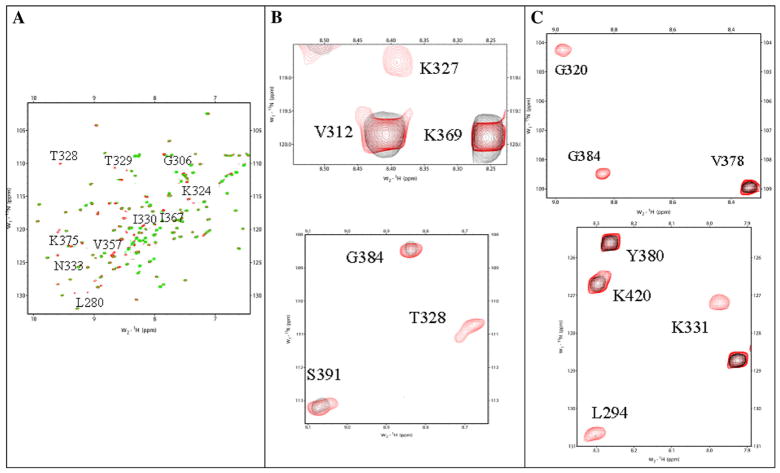
NMR spectroscopy is a powerful tool to monitor protein-ligand or protein-protein interactions [40]. Ligand binding sites on proteins are routinely mapped using 2D 1H–15N heteronuclear single quantum coherence (HSQC) spectroscopy [41]. A HSQC spectrum of a protein is a fingerprint of its backbone conformation [42]. Each 1H–15N cross-peak in the spectrum is representative of the microenvironment of a backbone amide proton in a given conformation of proteins. Therefore, backbone conformational transitions induced by metal (Cu2+)/pS vesicles can be readily monitored from the chemical shift perturbation due to decrease in intensity of the crosspeaks in the 1H–15N HSQC spectrum. The 1H–15N HSQC spectrum of the apoC2B domain is well dispersed, suggesting that most of the backbone of the protein is in an ordered conformation. Assignment of all the 1H–15N crosspeaks in the HSQC spectrum of the C2B domain was previously achieved by Fernandez et al. [32].
Copper (Cu2+) is a paramagnetic ion, and therefore, the 1H–15N crosspeaks at the vicinity of the Cu2+-binding site are expected to broaden (and consequently disappear) with the reciprocal of the sixth power of the Cu2+-nucleus distance [43]. Selected crosspeaks in the 1H–15N HSQC spectrum of the apoC2B domain, show progressively decreased intensities with increased additions of Cu2+. However, we did not observe any significant perturbation of crosspeaks in the 1H–15N HSQC spectrum when C2B domain was titrated with Cu2+. The 1H–15N crosspeaks that completely disappeared (at a 1: 4 ratio of the C2B domain to Cu2+ concentration) are Cys277, Phe278, Leu280, Gly306, Lys324, Thr 328, Thr329, Ile330, Asn333, Val357, Val358, Ile367, Gly368, and Lys375 (Figs. 2A and 3A). Most of these residues are present in β-strand I, Loop1, β-strand IV, LoopII, β-strand V1, and LoopIII of the structure of the C2B domain. Interestingly, some of the residues in Loops I, II and III (Gly306, Ile367, and Lys375) are part of the Ca2+-binding site in the C2B domain [32]. These observations suggest that Cu2+ shares common binding sites with Ca2+. It appears that of the four Cu2+ ions that bind to the C2B, two of them bind to the Ca2+ binding domain (data not shown). Two of the Cu2+ ions appear to bind to another distinct site(s) plausibly contributed by residues in β-strand II and β-strand IV. Titration of the C2B domain with pS vesicles shows selective disappearance of crosspeaks in the 1H–15N HSQC spectrum. The crosspeaks that disappear in the presence of pS vesicles include, Lys326, Lys327, Thr328, Asn333, and Thr334 (Figs. 2B and 3B). Some of these residues which bind to pS vesicles also belong to the Ca2+ binding domain. These observations are in good agreement with findings of Rufener et al. [44] who characterized the lipid binding sites on the C2B domain of Syt1 (in the presence of Ca2+) using site-directed spin labeling studies. Electron paramagnetic spin resonance spectroscopy data showed that Val304 and Gly305 in Loop1, Ile367 and Lys369 in Loop III contribute to lipid binding. Interestingly, our HSQC data on the C2B domain–pS vesicle interaction obtained in the absence of Ca2+ did not suggest the involvement of residues in Loop I (Val304 and Gly305) and Loop III (Ile367 and Lys369) in binding to pS vesicles. However, HSQC data obtained on the titration of the pS-bound C2B domain with Ca2+ (data not shown) showed excellent agreement with the C2B–lipid binding interface characterized by Rufener et al. [44].
Fig. 2.
1H–15N HSQC spectra of the C2B domain in the absence (in red) (Panel A) and in the presence of Cu2+ (in green). Portions of 1H–15N HSQC spectra of the C2B domain (in red) depicting crosspeaks of residues that show significant loss in intensity in the presence of pS (in black) (Panel B), pS and Cu2+ (in black) (Panel C). The significant decrease in the cross-peak intensity observed in the presence of Cu2+ indicates that these residues are in the vicinity of the Cu2+ binding site(s).
Fig. 3.
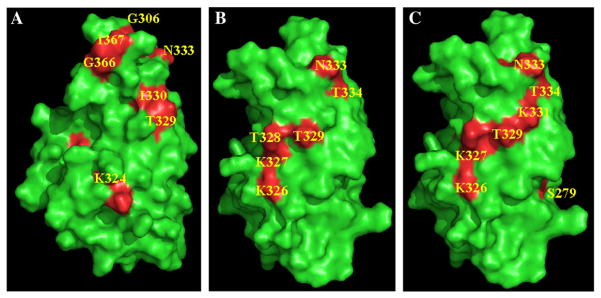
Surface diagram representing the structure of C2B domain (green). Residues that show significant decrease in intensity in presence of Cu2+ (Panel A), pS (Panel B), pS and Cu2+ (Panel C) are depicted in red. Titration of the protein (in its free and pS bound forms) with Cu2+ resulted in a significant decrease in intensity of crosspeaks of several residues which are buried in the native conformation of the C2B domain. These buried residues could not be depicted in the surface diagrams shown in this figure. For example, crosspeaks of Cys277, Phe278, Leu280 completely disappear when apoC2B domain or C2B domain bound to pS vesicles are titrated with Cu2+.
Titration of C2B domain bound to pS vesicles with Cu2+ shows significant loss (>90%) in intensity of 1H–15N crosspeaks (in the HSQC spectrum) corresponding to Cys277, F278, Ser279, Leu294, Asp309, Gly320, Lys331, Lys 324 and Gly 384 (Figs. 2C and 3C). These residues are mostly located in β-strands I and III. The 1H–15N crosspeaks that disappeared in the presence of pS vesicles remain so in the HSQC spectrum of the C2B domain obtained in the presence of both Cu2+ and the pS vesicles. Additional crosspeaks of residues which show significant loss in intensity, in the presence of both Cu2+ and pS vesicles, plausibly further stabilize the C2B-lipid interaction. These results suggest that Cu2+ increases the lipid binding affinity by stabilizing interactions in the C2B domain. In summary, the NMR data clearly suggest that there are two Cu2+ binding sites in the C2B domain. One of these Cu2+ binding site, contributed by residues in Loops I, II and III, represents the Ca2+ binding site. In absence of metal ions, the protein appears to bind to pS vesicles predominantly through residues in Loop II.
3.2. Possible sequence of molecular events in the non-classical secretion of hFGF-1
Based on the results of this study and our previous findings, we suggest a hypothetical model predicting the sequence of molecular events that plausibly occur during the Cu2+-mediated non-classical secretion of hFGF-1 (Fig. 4). The first step appears to be an interaction of hFGF-1 with the homodimer of S100A13. It appears that this interaction does not require Cu2+ because previously reported ITC data showed that these proteins interact even in the absence of the metal ion (Cu2+) [45]. As a next step, the hFGF-1/S100A13 binary complex plausibly interacts with Syt1 to form the hFGF-1/S100A13/ Syt1 ternary complex. Previous studies from our group and others suggest that C2A domain is involved in direct interactions with both hFGF-1 and S100A13 [9,39,45–47]. To-date there is no direct evidence of direct protein interactions with the C2B domain. Cu2+ binds to S100A13 [45] and the C2A [47] and C2B domains of Syt1. Cu2+ appears to be important for, (1) Specific oxidation of the thiol group of Cys30 to facilitate the formation of a homodimer of hFGF-1; (2) Conferring stability to the multiprotein complex; and (3) Increasing the lipid binding affinity of the C2B domain. Although both hFGF-1 and S100A13 have been shown to bind to lipids, the C2 domains appear to be critical for anchoring the hFGF-1/S100A13 complex [48]. It is believed that hFGF-1 is finally exported across the membrane bilayer through the ‘fiip flop’ action of annexins [9]. In the absence of solid experimental evidence, the predicted sequence of molecular events occurring in the non-classical secretion of hFGF-1 should be considered hypothetical. More detailed studies are currently underway to validate some aspects of the proposed mechanism.
Fig. 4.
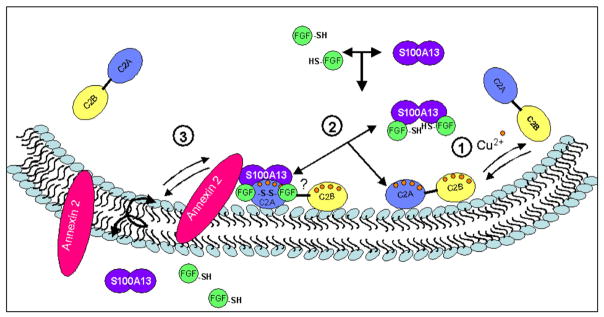
Cartoon representing the predicted sequence of molecular events in the non-classical export of hFGF-1.
Acknowledgments
We thank the National Institutes of Health (NIH NCRR COBRE Grant 1 P20 RR15569 and 1-K01-CA113753-01A2), the Department of Energy (Grant DE-FG02-01ER15161) and the Arkansas Biosciences Institute for financial support. SB was supported by the NSF sponsored REU program (NSF-REU; Grant Number: CHE-0243978), and LG is a visiting student from the State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering University, Nanjing, China. He is supported by China Scholarship Council, Grant Number: 2007U12037. IP was supported in part by NIH grants ARRA HL35627, HL 35627 and RR15555 (Project 4), and a grant of Maine Cancer Foundation.
Abbreviations
- hFGF-1
human fibroblast growth factor-1
- Syt1
Synaptotagmin 1
- HSQC
Heteronuclear Single Quantum Coherence
- NMR
nuclear magnetic resonance
References
- 1.Landriscina M, Bagala C, Mandinova A, Soldi R, Micucci I, Bellum S, Prudovsky I, Maciag T. Copper induces the assembly of a multiprotein aggregate implicated in the release of fibroblast growth factor 1 in response to stress. J Biol Chem. 2001;276:25549–25557. doi: 10.1074/jbc.M102925200. [DOI] [PubMed] [Google Scholar]
- 2.Di Serio C, Doria L, Pellerito S, Prudovsky I, Micucci I, Massi D, Landriscina M, Marchionni N, Masotti G, Tarantini F. The release of fibroblast growth factor-1 from melanoma cells requires copper ions and is mediated by phosphatidylinositol 3-kinase/Akt intracellular signaling pathway. Cancer Lett. 2008;267:67–74. doi: 10.1016/j.canlet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Chi YH, Kumar TK, Kathir KM, Lin DH, Zhu G, Chiu IM, Yu C. Investigation of the structural stability of the human acidic fibroblast growth factor by hydrogen-deuterium exchange. Biochemistry. 2002;41:15350–15359. doi: 10.1021/bi026218a. [DOI] [PubMed] [Google Scholar]
- 4.Szebenyi G, Fallon JF. Fibroblast growth factors as multifunctional signaling factors. Int Rev Cytol. 1999;185:45–106. doi: 10.1016/s0074-7696(08)60149-7. [DOI] [PubMed] [Google Scholar]
- 5.Schlessinger J. Common and distinct elements in cellular signaling via EGF and FGF receptors. Science New York, NY. 2004;306:1506–1507. doi: 10.1126/science.1105396. [DOI] [PubMed] [Google Scholar]
- 6.Korc M, Friesel RE. The role of fibroblast growth factors in tumor growth. Curr Cancer Drug Targets. 2009;9:639–651. doi: 10.2174/156800909789057006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J, Blaber M. The interaction between thermodynamic stability and buried free cysteines in regulating the functional half-life of fibroblast growth factor-1. J Mol Biol. 2009;393:113–127. doi: 10.1016/j.jmb.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 8.Friesel R, Maciag T. Fibroblast growth factor prototype release and fibroblast growth factor receptor signaling. Thromb Haemost. 1999;82:748–754. [PubMed] [Google Scholar]
- 9.Prudovsky I, Tarantini F, Landriscina M, Neivandt D, Soldi R, Kirov A, Small D, Kathir KM, Rajalingam D, Kumar TK. Secretion without Golgi. J Cell Biochem. 2008;103:1327–1343. doi: 10.1002/jcb.21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mouta Carreira C, Landriscina M, Bellum S, Prudovsky I, Maciag T. The comparative release of FGF1 by hypoxia and temperature stress. Growth factors (Chur, Switzerland) 2001;18:277–285. doi: 10.3109/08977190109029116. [DOI] [PubMed] [Google Scholar]
- 11.Jackson A, Friedman S, Zhan X, Engleka KA, Forough R, Maciag T. Heat shock induces the release of fibroblast growth factor 1 from NIH 3T3 cells. Proc Natl Acad Sci U S A. 1992;89:10691–10695. doi: 10.1073/pnas.89.22.10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin JT, Opalenik SR, Wehby JN, Mahesh VK, Jackson A, Tarantini F, Maciag T, Thompson JA. Serum-starvation induces the extracellular appearance of FGF-1. Biochim Biophys Acta. 1996;1312:27–38. doi: 10.1016/0167-4889(96)00013-4. [DOI] [PubMed] [Google Scholar]
- 13.Ananyeva NM, Tjurmin AV, Berliner JA, Chisolm GM, Liau G, Winkles JA, Haudenschild CC. Oxidized LDL mediates the release of fibroblast growth factor-1. Arterioscler Thromb Vasc Biol. 1997;17:445–453. doi: 10.1161/01.atv.17.3.445. [DOI] [PubMed] [Google Scholar]
- 14.Prudovsky I, Bagala C, Tarantini F, Mandinova A, Soldi R, Bellum S, Maciag T. The intracellular translocation of the components of the fibroblast growth factor 1 release complex precedes their assembly prior to export. J Cell Biol. 2002;158:201–208. doi: 10.1083/jcb.200203084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin OH, Xu J, Rizo J, Sudhof TC. Differential but convergent functions of Ca2+ binding to synaptotagmin-1 C2 domains mediate neurotransmitter release. Proc Natl Acad Sci U S A. 2009;106:16469–16474. doi: 10.1073/pnas.0908798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo W, Herrick DZ, Ellena JF, Cafiso DS. The calcium-dependent and calcium-independent membrane binding of synaptotagmin 1: two modes of C2B binding. J Mol Biol. 2009;387:284–294. doi: 10.1016/j.jmb.2009.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graziani I, Doyle A, Sterling S, Kirov A, Tarantini F, Landriscina M, Kumar TK, Neivandt D, Prudovsky I. Protein folding does not prevent the nonclassical export of FGF1 and S100A13. Biochem Biophys Res Commun. 2009;381:350–354. doi: 10.1016/j.bbrc.2009.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soldi R, Mandinova A, Venkataraman K, Hla T, Vadas M, Pitson S, Duarte M, Graziani I, Kolev V, Kacer D, Kirov A, Maciag T, Prudovsky I. Sphingosine kinase 1 is a critical component of the copper-dependent FGF1 export pathway. Exp Cell Res. 2007;313:3308–3318. doi: 10.1016/j.yexcr.2007.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaVallee TM, Prudovsky IA, McMahon GA, Hu X, Maciag T. Activation of the MAP kinase pathway by FGF-1 correlates with cell proliferation induction while activation of the Src pathway correlates with migration. J Cell Biol. 1998;141:1647–1658. doi: 10.1083/jcb.141.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarantini F, LaVallee T, Jackson A, Gamble S, Mouta Carreira C, Garfinkel S, Burgess WH, Maciag T. The extravesicular domain of synaptotagmin-1 is released with the latent fibroblast growth factor-1 homodimer in response to heat shock. J Biol Chem. 1998;273:22209–22216. doi: 10.1074/jbc.273.35.22209. [DOI] [PubMed] [Google Scholar]
- 21.Landriscina M, Soldi R, Bagala C, Micucci I, Bellum S, Tarantini F, Prudovsky I, Maciag T. S100A13 participates in the release of fibroblast growth factor 1 in response to heat shock in vitro. J Biol Chem. 2001;276:22544–22552. doi: 10.1074/jbc.M100546200. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez-Chacon R, Shin OH, Konigstorfer A, Matos MF, Meyer AC, Garcia J, Gerber SH, Rizo J, Sudhof TC, Rosenmund C. Structure/function analysis of Ca2+ binding to the C2A domain of synaptotagmin 1. J Neurosci. 2002;22:8438–8446. doi: 10.1523/JNEUROSCI.22-19-08438.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerber SH, Rizo J, Sudhof TC. Role of electrostatic and hydrophobic interactions in Ca(2+)-dependent phospholipid binding by the C(2)A-domain from synaptotagmin I. Diabetes. 2002;51(Suppl 1):S12–18. doi: 10.2337/diabetes.51.2007.s12. [DOI] [PubMed] [Google Scholar]
- 24.Hui E, Bai J, Wang P, Sugimori M, Llinas RR, Chapman ER. Three distinct kinetic groupings of the synaptotagmin family: candidate sensors for rapid and delayed exocytosis. Proc Natl Acad Sci U S A. 2005;102:5210–5214. doi: 10.1073/pnas.0500941102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai J, Chapman ER. The C2 domains of synaptotagmin—partners in exocytosis. Trends Biochem Sci. 2004;29:143–151. doi: 10.1016/j.tibs.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Fuson KL, Ma L, Sutton RB, Oberhauser AF. The c2 domains of human synaptotagmin 1 have distinct mechanical properties. Biophys J. 2009;96:1083–1090. doi: 10.1016/j.bpj.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajalingam D, Kumar TK, Yu C. The C2A domain of synaptotagmin exhibits a high binding affinity for copper: implications in the formation of the multiprotein FGF release complex. Biochemistry. 2005;44:14431–14442. doi: 10.1021/bi051387r. [DOI] [PubMed] [Google Scholar]
- 28.Bagala C, Kolev V, Mandinova A, Soldi R, Mouta C, Graziani I, Prudovsky I, Maciag T. The alternative translation of synaptotagmin 1 mediates the non-classical release of FGF1. Biochem Biophys Res Commun. 2003;310:1041–1047. doi: 10.1016/j.bbrc.2003.09.119. [DOI] [PubMed] [Google Scholar]
- 29.Sukumaran SS, Banerjee S, Bhasker S, Thekkuveettil A. The cytoplasmic C2A domain of synaptotagmin shows sequence specific interaction with its own mRNA. Biochem Biophys Res Commun. 2008;373:509–514. doi: 10.1016/j.bbrc.2008.06.063. [DOI] [PubMed] [Google Scholar]
- 30.Duning K, Buck F, Barnekow A, Kremerskothen J. SYNCRIP, a component of dendritically localized mRNPs, binds to the translation regulator BC200 RNA. J Neurochem. 2008;105:351–359. doi: 10.1111/j.1471-4159.2007.05138.x. [DOI] [PubMed] [Google Scholar]
- 31.LaVallee TM, Tarantini F, Gamble S, Mouta Carreira C, Jackson A, Maciag T. Synaptotagmin-1 is required for fibroblast growth factor-1 release. J Biol Chem. 1998;273:22217–22223. doi: 10.1074/jbc.273.35.22217. [DOI] [PubMed] [Google Scholar]
- 32.Graziani I, Bagala C, Duarte M, Soldi R, Kolev V, Tarantini F, Kumar TK, Doyle A, Neivandt D, Yu C, Maciag T, Prudovsky I. Release of FGF1 and p40 synaptotagmin 1 correlates with their membrane destabilizing ability. Biochem Biophys Res Commun. 2006;349:192–199. doi: 10.1016/j.bbrc.2006.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez I, Arac D, Ubach J, Gerber SH, Shin O, Gao Y, Anderson RG, Sudhof TC, Rizo J. Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine. Neuron. 2001;32:1057–1069. doi: 10.1016/s0896-6273(01)00548-7. [DOI] [PubMed] [Google Scholar]
- 34.Ubach J, Lao Y, Fernandez I, Arac D, Sudhof TC, Rizo J. The C2B domain of synaptotagmin I is a Ca2+-binding module. Biochemistry. 2001;40:5854–5860. doi: 10.1021/bi010340c. [DOI] [PubMed] [Google Scholar]
- 35.Radhakrishnan A, Stein A, Jahn R, Fasshauer D. The Ca2+ affinity of synaptotagmin 1 is markedly increased by a specific interaction of its C2B domain with phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2009;284:25749–25760. doi: 10.1074/jbc.M109.042499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montaville P, Schlicker C, Leonov A, Zweckstetter M, Sheldrick GM, Becker S. The C2A-C2B linker defines the high affinity Ca(2+) binding mode of rabphilin-3A. J Biol Chem. 2007;282:5015–5025. doi: 10.1074/jbc.M606746200. [DOI] [PubMed] [Google Scholar]
- 37.Jang JY, Krishnaswamy T, Kumar S, Jayaraman G, Yang PW, Yu C. Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid. Biochemistry. 1997;36:14635–14641. doi: 10.1021/bi971107a. [DOI] [PubMed] [Google Scholar]
- 38.Arunkumar AI, Srisailam S, Kumar TK, Kathir KM, Chi YH, Wang HM, Chang GG, Chiu I, Yu C. Structure and stability of an acidic fibroblast growth factor from Notophthalmus viridescens. J Biol Chem. 2002;277:46424–46432. doi: 10.1074/jbc.M207814200. [DOI] [PubMed] [Google Scholar]
- 39.Goddard TD, Kueller DG. SPARKY 3. University of California; San Francisco: 2006. [Google Scholar]
- 40.Hajduk PJ, Meadows RP, Fesik SW. NMR-based screening in drug discovery. Q Rev Biophys. 1999;32:211–240. doi: 10.1017/s0033583500003528. [DOI] [PubMed] [Google Scholar]
- 41.Yu L, Oost TK, Schkeryantz JM, Yang J, Janowick D, Fesik SW. Discovery of aminoglycoside mimetics by NMR-based screening of Escherichia coli A-site RNA. J Am Chem Soc. 2003;125:4444–4450. doi: 10.1021/ja021354o. [DOI] [PubMed] [Google Scholar]
- 42.Zuiderweg ER. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 43.Bertini I, Luchinat C, Piccioli M. Paramagnetic probes in metalloproteins. Methods Enzymol. 2001;339:314–340. doi: 10.1016/s0076-6879(01)39320-5. [DOI] [PubMed] [Google Scholar]
- 44.Rufener E, Frazier AA, Wieser CM, Hinderliter A, Cafiso DS. Membrane-bound orientation and position of the synaptotagmin C2B domain determined by site-directed spin labeling. Biochemistry. 2005;44:18–28. doi: 10.1021/bi048370d. [DOI] [PubMed] [Google Scholar]
- 45.Sivaraja V, Kumar TKS, Rajalingam D, Graziani I, Prudovsky I, Yu C. Copper binding affinity of S100A13, a key component of the FGF-1 nonclassical copper-dependent release complex. Biophys J. 2006;91:1832–1843. doi: 10.1529/biophysj.105.079988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prudovsky I, Mandinova A, Soldi R, Bagala C, Graziani I, Landriscina M, Tarantini F, Duarte M, Bellum S, Doherty H, Maciag T. The non-classical export routes: FGF1 and IL-1alpha point the way. J Cell Sci. 2003;116:4871–4881. doi: 10.1242/jcs.00872. [DOI] [PubMed] [Google Scholar]
- 47.Mandinova A, Soldi R, Graziani I, Bagala C, Bellum S, Landriscina M, Tarantini F, Prudovsky I, Maciag T. S100A13 mediates the copper-dependent stress-induced release of IL-1alpha from both human U937 and murine NIH 3T3 cells. J Cell Sci. 2003;116:2687–2696. doi: 10.1242/jcs.00471. [DOI] [PubMed] [Google Scholar]
- 48.Kathir KM, Ibrahim K, Rajalingam D, Prudovsky I, Yu C, Kumar TKS. S100A13-lipid interactions-role in the non-classical release of the acidic fibroblast growth factor. Biochim Biophys Acta. 2007;1768:3080–3089. doi: 10.1016/j.bbamem.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]