Abstract
G protein-coupled receptors (GPCR) are key signaling proteins that regulate how cells interact with their environments. Traditional signaling cascades involving GPCRs have been well described and are well established and very important clinical targets. With the development of more recent technologies, hints about the involvement of GPCRs in fundamental cell biological processes are beginning to emerge. In this review, we give a basic introduction to GPCR signaling and highlight some less well described roles of GPCRs, including in cell division and membrane trafficking, which may occur through canonical and non-canonical signaling pathways.
G protein-coupled receptors (GPCRs) are one of the largest protein superfamilies in mammals. Located in the cell membrane, GPCRs are a major way for cells and organisms to sense a large variety of inputs and signals from their environments, including neurotransmitters, nonsteroid hormones, biogenic amines, amino acids, ions, lipids, peptides and proteins, odorants and light. Many GPCRs are responsible for sensation, including taste, smell and light and they are often enriched in specialized sensory cells, for example in eyes, tongues, ears and the brain. Based mainly on structural predictions, it is thought that the human genome includes around 1000 GPCRs, and the biological roles of many putative GPCRs remain unexplored. Misregulation of GPCR signaling occurs in multiple diseases including psychiatric disorders, cancer, autoimmune diseases and diabetes, which makes GPCRs the most investigated targets in the pharmaceutical industry1-5. Several billion dollar selling drugs target GPCRs. For example, Eli Lilly’s Zyprexa targets serotonin receptors and is used to treat schizophrenia and bipolar disorders; GlaxoSmithKline’s Seretide/Advair targets adrenoreceptors and is used to treat asthma. It is well accepted that upon ligand binding, GPCRs trigger signaling cascades inside cells that can result in a multitude of cellular effects. Recent findings suggest that GPCRs may have additional cellular functions. This review will focus on the current understanding of GPCRs in different aspects of basic cell biology, including some unconventional functions of GPCRs in membrane trafficking and cell division.
Traditional functions of GPCRs in cell signaling
G protein-coupled receptors are also known as 7-transmembrane (TM) receptors because of their common seven cross-membrane structures, which is a characteristic commonly used to distinguish GPCRs from other receptors. GPCRs are found in all eukaryotes, including yeast, fungi, amoeba, and animals (invertebrates and vertebrates). As indicated by their name, the best known function of GPCRs is to transduce signals across cell membranes via heterotrimeric G proteins (GTP binding proteins), which are located on the cytoplasmic side of the plasma membrane (Figure 1). GPCRs’ activations produce a large variety of cellular effects mainly through interactions with multiple different G proteins, which regulate discrete signaling pathways6, 7. As these signaling events have been reviewed extensively elsewhere8-10, we will only briefly introduce them: Upon ligand binding at the cell surface, GPCRs undergo conformational alterations and change their interactions with heterotrimeric G proteins, which are composed of Gα, Gβ and Gγ subunits. Inactive GDP-bound Gα is associated with the Gβγ complex. GPCRs can be viewed as GEF (Guanine exchange factor) enzymes because GPCR activation triggers GDP to GTP exchange in Gα, which is then activated and disassociated from Gβγ11. This process is reversible because Gα’s GTPase can hydrolyze GTP into GDP, which results in re-association of Gα with Gβγ. G proteins, like free Gα, can trigger signaling cascades in cells through many different effectors, for example, adenylyl cyclases, protein kinase C (PKC) or Rho GTPases12, 13. Adenylyl cyclases are enzymes at the inner face of the plasma membrane that catalyze the conversion of ATP into the “second messenger” cyclic AMP (cAMP). “Second messengers” are cytoplasmic components that relay signals from cell surface receptors. Besides cAMP, other “second messengers” include calcium, guanosine monophosphate (GMP), inositol-1,4,5-triphosphate (IP3), diacyl glycerol (DAG) and arachidonic acid (for phospholipases). The downstream targets of the signaling cascades include enzymes, intracellular receptors and transcription factors, which eventually control gene expression14-16.
Figure 1. Schematic diagram shows a GPCR and possible ligand binding sites.
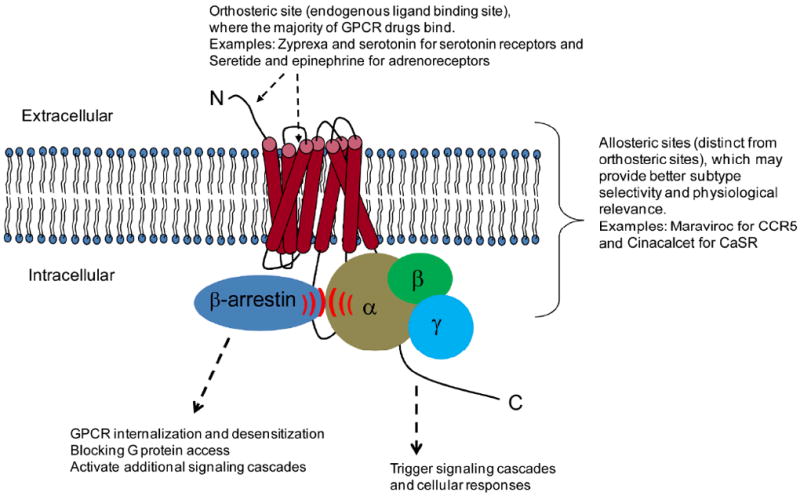
Orthosteric sites are usually at the outer surface of the receptor and sometimes in the extracellular N-terminus region. Allosteric sites are distinct from the orthosteric sites and can be located in any region of the receptor. GPCRs bound to G proteins vs. arrestins show differential signaling behaviors.
Calcium can act as both a ligand and a second messenger for GPCRs17. Calcium can bind to calcium-sensing receptor (CaSR) as an extracellular ligand and is the second messenger for many GPCRs upon activation18, 19. Since activation of many GPCRs is coupled to increased calcium concentration in cells, fluorescent reporters that monitor intracellular calcium are commonly used to measure the function of GPCRs, especially in high throughput screens. Calcium has effects on multiple cellular processes, such as vesicle fusion, autophagy, cytokinesis and apoptosis, mainly through regulating calcium-dependent kinases and phosphatases20-24.
The dogma has been that the differential functions of the large numbers of GPCRs are largely due to their specific interactions with different G proteins. G proteins form a crucial part of the GPCR signaling pathway. Humans have 23 Gα, 5 Gβ, and 12 Gγ subunits, as well as 37 RGS proteins (regulators of G protein signaling)25. Gα proteins are classified into four classes based on their functions: Gαs (“s” for stimulate), Gαi (“i” for inhibit), Gαq (or Gα11) and Gα12/13, and they signal through distinct pathways. While the specificity of GPCRs is thought to be largely due to the different G proteins they are coupled to, a given GPCR can also bind to multiple G proteins26. In the traditional model, Gα disassociates from Gβγ upon GPCR activation and invokes a signaling cascade in the cytoplasm. However, it has been shown that Gαi and Gβγ are also able to translocate into the nucleus and bind to chromatin27. This translocation is critical for Gαi to regulate mitosis but not DNA synthesis and it does not require Gαi to disassociate from Gβγ28.
In addition to the ligand-triggered G protein dependent signaling cascades, GPCRs can also trigger G protein-independent cellular processes via β-arrestin and activate a broad set of intracellular signaling molecules, including JNK, Akt, PI3 kinase, RhoA, MAPK and NF-κB29,30. The best characterized G protein-independent components downstream of GPCRs are GPCR kinases (GRKs) and β-arrestins (including β-arrestin1 and β-arrestin2), both cytosolic proteins which can bind to GPCRs. Upon ligand binding, GRKs phosphorylate GPCRs and recruit β-arrestins, which results in termination or reduction of signaling by blocking G proteins from further interaction with the receptors31. The β-arrestins are important for GPCR desensitization, sequestration and intracellular trafficking32, which prevent cells from undergoing excessive receptor stimulation. This simple model suggests that the primary role of the GRKs and arrestins is to control the feedback mechanisms that regulate what has been traditionally defined as the on/off states of GPCRs. However, given the large number and diversity of the GRKs and arrestins, it is entirely possible that they have additional roles, both in controlling non-traditional GPCR functions and independent of GPCRs.
Since GPCRs are able to trigger signaling cascades that involve many different players, they have a broad impact on several fundamental processes, including cell-cell communication, cell proliferation and regulation of gene expression33-36. In addition to these traditionally well accepted roles of GPCRs, more recent studies have suggested that they participate in other cell biological processes, both through traditional G protein mediated signaling and through non-canonical pathways. For example, some studies indicate that GPCRs can regulate actin33, 37-39, a key component for multiple cellular processes including cell division, migration and membrane trafficking, which will be discussed further below.
GPCR as drug targets
Due to the large variety of GPCR functions in sensing the environment and beyond, their ligands range from very small substances like photons to large ones like glycoproteins. However, many endogenous GPCR ligands are small molecules and small molecule binding pockets are well conserved in GPCRs. This is why GPCRs are such attractive drug targets; drug-like small molecules can bind to similar binding sites as natural ligands. In addition to these traditional binding sites (orthosteric sites), small molecule drugs can also bind to allosteric sites40-42. For example, the β1 adrenergic receptor has at least two ligand-binding sites that have differential pharmacological properties43-45. Similarly, different ligands of the same GPCRs can separately trigger G protein and β-arrestin pathways46. Such different ligands may induce different GPCR conformations and therefore distinct downstream signaling pathways, which gives rise to the concept of biased GPCR ligands and drugs47. This is probably due to differential affinities of diverse agonists and/or the existence of multiple binding sites on the receptor6.
Based on the functions of ligands that bind to GPCRs, they can be largely classified as agonists, antagonists or inverse agonists. A GPCR agonist is a chemical that binds to the GPCR and mimics the action of its endogenous ligand. Antagonist refers to chemicals that block the action of the agonist and an inverse agonist causes an action that is opposite to the agonist in an active receptor. It is usually thought that structurally similar compounds have similar biological properties, which is the rationale behind many ligand-based drug discovery programs48. However, there are some examples that agonists can switch to being antagonists and vice versa. For example, burimamide is a histamine H2 blocker that was derived from the endogenous ligand histamine49, 50.
Although GPCRs have been studied intensively for decades, the first crystal structure, of rhodopsin51, was only identified twelve years ago. The difficulty of solving GPCR structures is not only due to their membrane bound nature, but also because they are very flexible and have multiple intermediate states ranging from inactive to active. However, substantial progress in GPCR structural biology has now been made, including solving the structures of β1 and β2 adrenergic receptors, as well as adenosine A2a receptor, which facilitated GPCR based drug design to a large extent52. Thanks largely to the structural information, our understanding of the architecture of orthosteric sites is now sufficiently sophisticated to allow computer-based predictions of potential ligands, as was recently shown in the case of the dopamine receptor D353.
Drugs that target GPCRs comprise the largest family of currently available medicines in the market and they are intensely studied for drug development. Although most of the currently available GPCR drugs are agonists or antagonists that target orthosteric sites of GPCRs, the discovery of allosteric sites on GPCRs is a promising development in GPCR drug discovery. Because they are not competitive with orthosteric ligands, allosteric inhibitors have to be found in functional assays instead of traditional ligand replacement assay-based screens, which makes their discovery challenging. However, the presence of allosteric sites allows many more potential ligand-GPCR interactions to occur, which opens a new field for GPCR drug development. Moreover, the allosteric binding sites can provide much better subtype selectivity because the orthosteric sites are often highly conserved across all receptor subtypes40, 54, 55. For example, there are two allosteric modulators of GPCRs in the pharmaceutical market: Cinacalcet (targets the calcium-sensing receptor and is used to treat hyperparathyroidism) and maraviroc (targets CCR5 and is used to treat HIV infection).
GPCRs and effectors in cell division
Many GPCRs are important for cell proliferation but which stages of the cell cycle they participate in has been less studied. DNA synthesis is often used as a standard to measure proliferation, but this assay does not report on which stage of the cell cycle has been affected if proliferation is inhibited. For example, arrests at G1, G2 or mitosis will eventually affect DNA synthesis. For this reason, it is likely that there are still unexplored roles of GPCR effectors/regulators in cell cycle regulation, and especially in cell division, which occupies a very short time late in the cell cycle. Some hints for the participation of GPCR-mediated pathways in the regulation of cell division have emerged from the literature and are described below.
G proteins are the most studied components of GPCR signaling in the cell cycle. For example, roles for heterotrimeric G proteins in cell division have been proposed in some model organisms56, 57. As early as 1996, heterotrimeric G proteins (Gβ) were indicated to function in mitotic spindle orientation in C. elegans embryos58 and Gα was also shown to affect the spindle asymmetric division in C. elegans59-61. Then multiple groups found that AGS (Activators of G protein signaling) can regulate G proteins in mitotic spindle positioning in C. elegans embryos and Drosophila62-66. Similarly, investigations of the subunits in plants show that they have differential roles in cell division67. The mechanism of G proteins in cell division regulation is beginning to be illustrated. Although G proteins are the major effectors of GPCRs at the cell membrane, they also localize to other subcellular locations, such as endosomes, mitochondria and the Golgi, and function in multiple cellular processes that may be independent of GPCR activation68. The subcellular localizations away from the plasma membrane for G proteins likely contribute to their function in cell division. For example, Giα proteins localize to centrosomes, the spindle midzone, and midbodies and are involved in cell division69. In addition, G proteins were also shown to have direct regulatory functions on cytoskeletal actin and microtubules. For example, a few G proteins, such as Gαi, Gαs, Gαq and Gβγ directly interact with and regulate microtubules70. Gα12 proteins (including Gα12 and Gα13) can also regulate actin as well as the dynamic turnover of growth factor-induced dorsal ruffles71-73. In addition, knockdown of Gβ2 can cause microtubule and mitotic phenotypes74. All these studies suggest that G proteins function in cell division and cytoskeleton regulation, either independently or via GPCR signaling.
Besides the G proteins themselves, some regulators of G proteins are also involved in cell division, for example some RGS proteins, which are important regulators of GPCR signaling. They bind directly to GTP-bound Gα and activate its intrinsic GTPase activity (acting as GTPase-activating proteins (GAPs)), which decreases the lifetime of Gα-GTP and terminates signaling75. However, some (e.g. Pins, GPR-1/GPR-2, LGN, and RGS14) have additional functions as guanine nucleotide dissociation inhibitors (GDIs) of GDP bound Gαi subunits, which slows down GDP release and inhibits the reassociation of Gα and Gβγ76, 77. For example, the Gα regulator RGS14 is localized to the nucleus in interphase, and to the spindle, centrioles and midbody during mitosis78. It is critical for the first cell division of the fertilized zygote in mice and plays an essential role in mitosis. RGS14 knockdown induces multinucleated cell formation, a consequence of cell division failure where the daughter cells fail to separate from each other78. In addition to the involvement of some G proteins and G protein regulators, other proteins associated with GPCRs have also been shown to function in cell division. For example, both the β-arrestins and GRKs can regulate the cytoskeleton and cell division32, 79, 80. β-arrestins can interact with and control the actin cytoskeleton, including cofilin, an F-actin severing protein32, 79. GRK5 knockdown induces increased mitotic index, chromosome misalignment and apoptotic cells80. These data support a role for GPCR-associated proteins in cell division regulation, but have not yet led to a comprehensive understanding of the signaling cascades involved.
All these studies provide indirect evidence for GPCR involvement in cell division, but we have little information about whether the most upstream players in this whole team, the GPCRs themselves, actually participate. A systematic examination of GPCRs and their functions in cell division will help to clear up this point. A recent study from our group took a first step in this direction. We used RNAi to knock down different GPCRs in different cancer cell lines and showed that several GPCRs that were traditionally thought to be involved in sensory signal transduction also cause cell division defects, suggesting that GPCRs may play broader roles than previously assumed81. We further discuss these findings below.
GPCRs and effectors in membrane trafficking
As membrane proteins, GPCRs mostly localize to the plasma membrane. However, GPCRs not only sit on the membrane and transmit extracellular signals into the cytosol through effectors and secondary messengers, they also enter the cytoplasm themselves. The internalization of GPCRs is through endocytosis, mostly through the clathrin-dependent pathway82. Internalized GPCRs can either be delivered to the lysosome for degradation if they are to be inactivated permanently or recycled back to the cell membrane through recycling endosomes for re-activation (Figure 2). Some GPCRs, such as TSHR (thyroid stimulating hormone receptor), PTHR (parathyroid hormone receptor) and S1P1 (sphingosine1-phosphate receptor 1), can continue signaling by producing cAMP when they are located inside cells83. In some cases, this continued signaling from the internalized GPCRs is crucial for their cellular functions. For example, TSH stimulation alone induces actin depolymerization but it can be antagonized by the endocytosis inhibitor dynasore. This indicates that the internalization of TSHR is required for its regulation of the actin cytoskeleton84.
Figure 2. GPCR localizations and trafficking pathways in interphase cells.
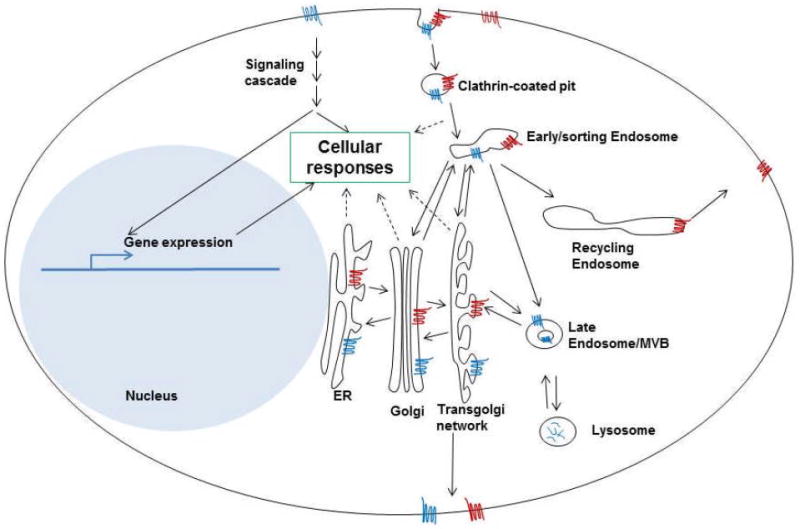
Two different GPCRs (blue and red) are shown in this cartoon to illustrate the different membrane trafficking pathways that they can go through. The blue GPCR goes through the lysosomal degradation pathway and the red GPCR goes through the recycling pathway. The two pathways can coexist for a given GPCR. Cells’ responses to GPCRs can happen through external ligand-induced signaling cascades, as well as through internalized GPCRs.
GPCRs were recently found to take part in a process related to membrane trafficking, autophagy. Autophagy is a cytoprotective response to cellular stress, such as nutrient deprivation85. Its malfunction leads to numerous diseases including cancer, cardiovascular disease and neuronal disorders86-88. Autophagosome formation is associated with membrane trafficking. Interestingly, the taste receptor T1R1/T1R3 complex, originally found in taste receptor cells in the mouth as an umami flavor detector89, is able to sense amino acid availability and regulate autophagy90. This regulation of autophagy by the taste receptors occurs through a well-studied component in autophagy pathway, mTOR1 (mammalian target of rapamycin complex 1).
Although the desensitization of GPCRs by endocytosis has been well studied, not much is known about the regulation of endocytosis by GPCRs, similarly to their potential involvement in other basic cell biological processes. A recent study from our group found that a GPCR, the dopamine receptor D3, is involved in the regulation of endocytic sorting and cell division91. After internalization, endocytic cargoes can be delivered to late endosomes/lysosomes for degradation or recycling endosomes for recycling. Dopamine receptor D3 knockdown causes endocytic cargoes to be trapped in early/sorting endosomes.
Both studies discussed in this section showed that GPCRs can have much more general roles than previously predicted based on specific expression in specialized tissues such as the brain (for dopamine receptor D3) or the tongue (for T1R1/T1R3). For T1R1/T1R3, it is likely that its involvement in autophagy is related to its original function of sensing tastes/amino acids. This is less clear for the dopamine receptor D3’s participation in endocytosis. Our work showed that small molecule dopamine receptor agonists and antagonists do not seem to affect D3’s role in endocytosis. We used Prazosin, a different small molecule, to show that D3’s role in endocytic sorting might be mediated by a transient interaction with the coatomer complex COPI, which is stabilized by Prazosin91. The detailed mechanism of how this receptor participates in the regulation of endocytic sorting requires further investigation.
Other non-canonical roles of GPCRs
Growing data, including some presented above, suggest that GPCRs are likely to be more complex and diverse than anticipated. For example, not all of the seven transmembrane domains are always necessary for their functions92. In addition, the two-state model of GPCRs being in either active or inactive states is outdated. Different GPCR structures that have been determined as well as functional experimental data suggest a continuum of conformations rather than specific “on” and “off” conformations93-95. Further supporting a more nuanced view of GPCR activation states, different agonists for the same GPCR can cause different cellular responses. For example, different agonists for the human δ-opioid receptor can differentially trigger different outcomes such as endosomal recycling or degradation in the lysosomal pathway96, 97. GPCR responses can be dependent on agonist concentration, for example by neurokinin 1 receptor and δ-opioid receptor trafficking into different intracellular compartments98, 99. It was also shown that aromatic residues in an allosteric site on M2 muscarinic acetylcholine receptor (mAChR) regulate the activation state of the receptor100. A recent report showed that GPCRs not only stimulate cell proliferation by signaling cascades that relay into the nucleus and regulate gene expression, but they can also activate purisomes, a cytosolic nucleotide factory101.
Many GPCRs exist as dimers or oligomers, which makes both functional studies and drug design more complex. For example, it is possible for an allosteric ligand that works on one monomer to affect the binding and/or function of the orthosteric ligand of the other monomer within the same complex102. For some GPCRs, monomers are sufficient for G protein coupling, but it appears that for other receptors dimerization or oligomerization may be a prerequisite for many of their biological functions103, 104. Oligomerization states of GPCRs and their functional consequences are an emerging area of research that still needs to be further investigated.
A key to understanding non-traditional roles of GPCRs is to evaluate their expression in different tissues. One would expect GPCRs that govern more general biological processes rather than specialized sensory events, for example, to be expressed in a variety of different tissues. While GPCRs compose one of the largest protein families in the human genome, their expression is usually low81, 105, 106. GPCRs can be classified as odorant/olfactory and non-odorant. To provide systematic information about non-odorant GPCRs, Regard et al. analyzed the expression level of 353 GPCRs in mouse tissues and found that the GPCRs’ expression levels can vary dramatically among different tissues. As expected, they found that many receptors are highly expressed in the tissues in which their activities were initially defined, but also noted that many of these GPCRs are expressed in additional, less expected, tissues106. Similarly, emerging evidence shows that odorant GPCRs are also expressed in other cells and tissues, sometimes at comparable and sometimes at low levels compared to the specialized sensory organs81, 91, 107, 108. For example, dozens of olfactory receptors are ectopically expressed in human and chimpanzee liver, heart, testis and lung108. They are also found in myocardial and erythroid cells109, 110, as well as in spleen, brainstem, colon and prostate111-114. Olfactory receptors expressed in testis may play a role in sperm chemotaxis115, 116, however, the detailed mechanism is not clear. The potential functions of other ectopically expressed olfactory receptors in non-olfactory tissues remain largely unexplored, suggesting these GPCRs may play broader roles than originally thought.
In agreement with these emerging ideas, our recent study found that several GPCRs are unexpectedly expressed in HeLa cells (human cervical cancer cell line) and function in cytokinesis, the last step of cell division81. These include the sensory GPCRs OPN1MW (opsin1, medium-wave-sensitive), OR2A4 (olfactory receptor family 2, subfamily A, member 4) and TAS2R13 (taste receptor, type2, member 13) as well as dopamine receptors D2 and D3. The putative odorant receptor OR2A4 localizes to cell division compartments such as centrosomes, spindle midzone and midbodies (Figure 3). OR2A4 knockdown caused cytokinesis failure at an early stage and causes defects in the actin cytoskeleton, which is the likely reason for cytokinesis failure81. Other GPCRs have been reported to participate in the regulation of actin dynamics, mostly through canonical signaling cascades33, 38, suggesting that OR2A4 might follow a similar pattern. Unlike OR2A4, knockdown of the other GPCRs identified in this study cause late cytokinesis defects and the mechanisms by which these occur are not clear. We showed that dopamine receptor D3 participates in endocytic sorting91 and its effects on cytokinesis might be a consequence of inhibiting membrane trafficking. Given that they are membrane proteins, it is possible that other GPCRs are also involved in the regulation of membrane trafficking through as yet unstudied mechanisms. They may exert their functions through nontraditional G protein signaling cascades and if so it is likely that additional intermediate proteins are involved. It is also possible that traditional signaling cascades are activated that have yet not been studied because it was thought that the expression of some of these interesting GPCRs was limited to specialized tissues. There are still many outstanding questions to be addressed in the future about the expression and function of GPCRs and their effectors in different tissues and their involvement in basic processes such as cell division.
Figure 3. Putative odorant receptor OR2A4 localizes to cell division compartments such as centrosomes and midbodies.
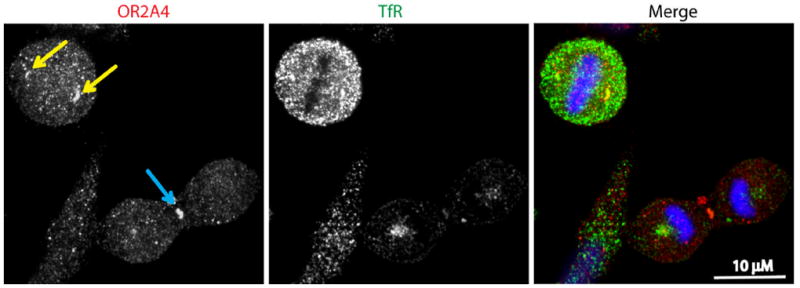
Yellow arrows show centrosomes and the blue arrow shows a midbody. Transferrin receptor (TfR) positive-endosomes have overlapping localizations with OR2A4 at centrosomes and midbody (adapted from reference 81).
Conclusions
As the largest gene family and the most targeted protein class in drug development, understanding different aspects of GPCRs is of great interest both to basic scientists and to clinicians. Elegant research using, amongst others, structural biology, medicinal chemistry and cell biology has allowed the field to progress to a point where we have a good working knowledge of the traditional signaling cascades that GPCRs trigger and participate in and we understand the basic wiring of these cascades. Major contributions to this field were recognized by the award of the Nobel Prize in Chemistry to Robert Lefkowitz and Brian Kobilka in 2012. While defining these basic parameters, we are beginning to get some hints that GPCRs are even more complex than predicted. For example, GPCRs do not just exist in two opposite on/off states and antagonists that bind to different sites on the same GPCR can induce differential effects. GPCRs and their associated proteins such as G proteins are not just located at the plasma membrane, but also internalize to intracellular locations where they can regulate different cellular processes.
New data created by technological advances including bioinformatics and expression profiling suggest that GPCR biology is more comprehensive than previously thought. For example, many GPCRs are ectopically expressed in tissues other than the specialized tissues where they were initially discovered. The field of GPCR signaling research is at an exciting juncture. Many GPCRs are still orphans, which means that their functions have not been determined and new functions are being discovered for established receptors. Expression profiling is beginning to allow us to look for GPCR function more broadly, in cells or tissues that we would not have previously considered. The wealth of drugs that target GPCRs, both orthosterically and especially allosterically, provides an arsenal of chemical biology tools to perturb different GPCR functions. We expect that the hints of non-canonical GPCR function discussed in this review are just the beginning in our understanding of this very important, and diverse, protein family.
Acknowledgments
Research in the Eggert lab is funded by NIH grant R01 GM082834, a Human Frontier Science Program Young Investigator Award, Marie Curie CIG grant 304137 and ERC Starting Grant 306659.
References
- 1.Filmore D. It’s a GPCR world. Modern Drug Discovery (American Chemical Society) 2004;7:24–28. [Google Scholar]
- 2.Gilchrist A. GPCR molecular pharmacology and drug targeting: shifting paradigms and new directions. Wiley; 2010. [Google Scholar]
- 3.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5(12):993–6. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 4.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291(5507):1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 5.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7(2):79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 6.Baker JG, Hill SJ. Multiple GPCR conformations and signalling pathways: implications for antagonist affinity estimates. Trends Pharmacol Sci. 2007;28(8):374–81. doi: 10.1016/j.tips.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maudsley S, Martin B, Luttrell LM. The origins of diversity and specificity in G protein-coupled receptor signaling. J Pharmacol Exp Ther. 2005;314(2):485–94. doi: 10.1124/jpet.105.083121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wess J. G-protein-coupled receptors: molecular mechanisms involved in receptor activation and selectivity of G-protein recognition. FASEB J. 1997;11(5):346–54. [PubMed] [Google Scholar]
- 9.Brady AE, Limbird LE. G protein-coupled receptor interacting proteins: emerging roles in localization and signal transduction. Cell Signal. 2002;14(4):297–309. doi: 10.1016/s0898-6568(01)00239-x. [DOI] [PubMed] [Google Scholar]
- 10.Ostrom RS, Post SR, Insel PA. Stoichiometry and compartmentation in G protein-coupled receptor signaling: implications for therapeutic interventions involving G(s) J Pharmacol Exp Ther. 2000;294(2):407–12. [PubMed] [Google Scholar]
- 11.Jacobson ED, Bunnett NW. G protein-coupled receptor signaling: implications for the digestive system. Dig Dis. 1997;15(4-5):207–42. doi: 10.1159/000171600. [DOI] [PubMed] [Google Scholar]
- 12.McCudden CR, Hains MD, Kimple RJ, Siderovski DP, Willard FS. G-protein signaling: back to the future. Cell Mol Life Sci. 2005;62(5):551–77. doi: 10.1007/s00018-004-4462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris AJ, Malbon CC. Physiological regulation of G protein-linked signaling. Physiol Rev. 1999;79(4):1373–430. doi: 10.1152/physrev.1999.79.4.1373. [DOI] [PubMed] [Google Scholar]
- 14.Nathanson NM. An array of details on G-protein coupled receptor signaling: differential effects of alpha1-adrenergic receptor subtypes on gene expression and cytokine receptor signaling. Mol Pharmacol. 2003;63(5):959–60. doi: 10.1124/mol.63.5.959. [DOI] [PubMed] [Google Scholar]
- 15.Naor Z. Signaling by G-protein-coupled receptor (GPCR): studies on the GnRH receptor. Front Neuroendocrinol. 2009;30(1):10–29. doi: 10.1016/j.yfrne.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Naor Z, Benard O, Seger R. Activation of MAPK cascades by G-protein-coupled receptors: the case of gonadotropin-releasing hormone receptor. Trends Endocrinol Metab. 2000;11(3):91–9. doi: 10.1016/s1043-2760(99)00232-5. [DOI] [PubMed] [Google Scholar]
- 17.Brown EM. G protein-coupled, extracellular Ca2+ (Ca2+(o))-sensing receptor enables Ca2+(o) to function as a versatile extracellular first messenger. Cell Biochem Biophys. 2000;33(1):63–95. doi: 10.1385/cbb:33:1:63. [DOI] [PubMed] [Google Scholar]
- 18.Bai M, Quinn S, Trivedi S, Kifor O, Pearce SH, Pollak MR, Krapcho K, Hebert SC, Brown EM. Expression and characterization of inactivating and activating mutations in the human Ca2+o-sensing receptor. J Biol Chem. 1996;271(32):19537–45. doi: 10.1074/jbc.271.32.19537. [DOI] [PubMed] [Google Scholar]
- 19.Chakravarti B, Chattopadhyay N, Brown EM. Signaling through the extracellular calcium-sensing receptor (CaSR) Adv Exp Med Biol. 2012;740:103–42. doi: 10.1007/978-94-007-2888-2_5. [DOI] [PubMed] [Google Scholar]
- 20.Atilla-Gokcumen GE, Castoreno AB, Sasse S, Eggert US. Making the cut: the chemical biology of cytokinesis. ACS Chem Biol. 2010;5(1):79–90. doi: 10.1021/cb900256m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aballay A, Arenas GN, Mayorga LS. Calcium- and zinc-binding proteins in intracellular transport. Biocell. 1996;20(3):339–42. [PubMed] [Google Scholar]
- 22.Jackson MB, Chapman ER. The fusion pores of Ca2+ -triggered exocytosis. Nat Struct Mol Biol. 2008;15(7):684–9. doi: 10.1038/nsmb.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harr MW, Distelhorst CW. Apoptosis and autophagy: decoding calcium signals that mediate life or death. Cold Spring Harb Perspect Biol. 2010;2(10):a005579. doi: 10.1101/cshperspect.a005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinton P, Giorgi C, Siviero R, Zecchini E, Rizzuto R. Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene. 2008;27(50):6407–18. doi: 10.1038/onc.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones AM, Assmann SM. Plants: the latest model system for G-protein research. EMBO Rep. 2004;5(6):572–8. doi: 10.1038/sj.embor.7400174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Gilchrist A, Stern PH. Antagonist minigenes identify genes regulated by parathyroid hormone through G protein-selective and G protein co-regulated mechanisms in osteoblastic cells. Cell Signal. 2011;23(2):380–8. doi: 10.1016/j.cellsig.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crouch MF. Growth factor-induced cell division is paralleled by translocation of Gi alpha to the nucleus. FASEB J. 1991;5(2):200–6. doi: 10.1096/fasebj.5.2.1900794. [DOI] [PubMed] [Google Scholar]
- 28.Crouch MF, Simson L. The G-protein G(i) regulates mitosis but not DNA synthesis in growth factor-activated fibroblasts: a role for the nuclear translocation of G(i) FASEB J. 1997;11(2):189–98. doi: 10.1096/fasebj.11.2.9039962. [DOI] [PubMed] [Google Scholar]
- 29.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Beta-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 30.Defea K. Beta-arrestins and heterotrimeric G-proteins: collaborators and competitors in signal transduction. Br J Pharmacol. 2008;153(Suppl 1):S298–309. doi: 10.1038/sj.bjp.0707508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hupfeld CJ, Olefsky JM. Regulation of receptor tyrosine kinase signaling by GRKs and beta-arrestins. Annu Rev Physiol. 2007;69:561–77. doi: 10.1146/annurev.physiol.69.022405.154626. [DOI] [PubMed] [Google Scholar]
- 32.Min J, Defea K. beta-arrestin-dependent actin reorganization: bringing the right players together at the leading edge. Mol Pharmacol. 2011;80(5):760–8. doi: 10.1124/mol.111.072470. [DOI] [PubMed] [Google Scholar]
- 33.Kabarowski JH, Feramisco JD, Le LQ, Gu JL, Luoh SW, Simon MI, Witte ON. Direct genetic demonstration of G alpha 13 coupling to the orphan G protein-coupled receptor G2A leading to RhoA-dependent actin rearrangement. Proc Natl Acad Sci U S A. 2000;97(22):12109–14. doi: 10.1073/pnas.97.22.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kehrl JH. G-protein-coupled receptor signaling, RGS proteins, and lymphocyte function. Crit Rev Immunol. 2004;24(6):409–23. doi: 10.1615/critrevimmunol.v24.i6.20. [DOI] [PubMed] [Google Scholar]
- 35.Rohrer DK, Kobilka BK. G protein-coupled receptors: functional and mechanistic insights through altered gene expression. Physiol Rev. 1998;78(1):35–52. doi: 10.1152/physrev.1998.78.1.35. [DOI] [PubMed] [Google Scholar]
- 36.Marinissen MJ, Gutkind JS. G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol Sci. 2001;22(7):368–76. doi: 10.1016/s0165-6147(00)01678-3. [DOI] [PubMed] [Google Scholar]
- 37.Cant SH, Pitcher JA. G protein-coupled receptor kinase 2-mediated phosphorylation of ezrin is required for G protein-coupled receptor-dependent reorganization of the actin cytoskeleton. Mol Biol Cell. 2005;16(7):3088–99. doi: 10.1091/mbc.E04-10-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies SL, Gibbons CE, Vizard T, Ward DT. Ca2+-sensing receptor induces Rho kinase-mediated actin stress fiber assembly and altered cell morphology, but not in response to aromatic amino acids. Am J Physiol Cell Physiol. 2006;290(6):C1543–51. doi: 10.1152/ajpcell.00482.2005. [DOI] [PubMed] [Google Scholar]
- 39.Cotton M, Claing A. G protein-coupled receptors stimulation and the control of cell migration. Cell Signal. 2009;21(7):1045–53. doi: 10.1016/j.cellsig.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Christopoulos A, Kenakin T. G protein-coupled receptor allosterism and complexing. Pharmacol Rev. 2002;54(2):323–74. doi: 10.1124/pr.54.2.323. [DOI] [PubMed] [Google Scholar]
- 41.Lazareno S, Dolezal V, Popham A, Birdsall NJ. Thiochrome enhances acetylcholine affinity at muscarinic M4 receptors: receptor subtype selectivity via cooperativity rather than affinity. Mol Pharmacol. 2004;65(1):257–66. doi: 10.1124/mol.65.1.257. [DOI] [PubMed] [Google Scholar]
- 42.Birdsall NJ, Lazareno S. Allosterism at muscarinic receptors: ligands and mechanisms. Mini Rev Med Chem. 2005;5(6):523–43. doi: 10.2174/1389557054023251. [DOI] [PubMed] [Google Scholar]
- 43.Granneman JG. The putative beta4-adrenergic receptor is a novel state of the beta1-adrenergic receptor. Am J Physiol Endocrinol Metab. 2001;280(2):E199–202. doi: 10.1152/ajpendo.2001.280.2.E199. [DOI] [PubMed] [Google Scholar]
- 44.Molenaar P. The ‘state’ of beta-adrenoceptors. Br J Pharmacol. 2003;140(1):1–2. doi: 10.1038/sj.bjp.0705420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arch JR. Do low-affinity states of beta-adrenoceptors have roles in physiology and medicine? Br J Pharmacol. 2004;143(5):517–8. doi: 10.1038/sj.bjp.0705991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei H, Ahn S, Shenoy SK, Karnik SS, Hunyady L, Luttrell LM, Lefkowitz RJ. Independent beta-arrestin 2 and G protein-mediated pathways for angiotensin II activation of extracellular signal-regulated kinases 1 and 2. Proc Natl Acad Sci U S A. 2003;100(19):10782–7. doi: 10.1073/pnas.1834556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Violin JD, DeWire SM, Yamashita D, Rominger DH, Nguyen L, Schiller K, Whalen EJ, Gowen M, Lark MW. Selectively engaging beta-arrestins at the angiotensin II type 1 receptor reduces blood pressure and increases cardiac performance. J Pharmacol Exp Ther. 2010;335(3):572–9. doi: 10.1124/jpet.110.173005. [DOI] [PubMed] [Google Scholar]
- 48.Martin YC, Kofron JL, Traphagen LM. Do structurally similar molecules have similar biological activity? J Med Chem. 2002;45(19):4350–8. doi: 10.1021/jm020155c. [DOI] [PubMed] [Google Scholar]
- 49.Eyre P. Histamine H2-receptors in the sheep bronchus and cat trachea: the action of burimamide. Br J Pharmacol. 1973;48(2):321–3. doi: 10.1111/j.1476-5381.1973.tb06920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyllie JH, Hesselbo T, Black JW. Effects in man of histamine H 2 -receptor blockade by burimamide. Lancet. 1972;2(7787):1117–20. doi: 10.1016/s0140-6736(72)92719-5. [DOI] [PubMed] [Google Scholar]
- 51.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289(5480):739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 52.Congreve M, Marshall F. The impact of GPCR structures on pharmacology and structure-based drug design. Br J Pharmacol. 2010;159(5):986–96. doi: 10.1111/j.1476-5381.2009.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carlsson J, Coleman RG, Setola V, Irwin JJ, Fan H, Schlessinger A, Sali A, Roth BL, Shoichet BK. Ligand discovery from a dopamine D3 receptor homology model and crystal structure. Nat Chem Biol. 2011;7(11):769–78. doi: 10.1038/nchembio.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Christopoulos A. Allosteric binding sites on cell-surface receptors: novel targets for drug discovery. Nat Rev Drug Discov. 2002;1(3):198–210. doi: 10.1038/nrd746. [DOI] [PubMed] [Google Scholar]
- 55.Bridges TM, Lindsley CW. G-protein-coupled receptors: from classical modes of modulation to allosteric mechanisms. ACS Chem Biol. 2008;3(9):530–41. doi: 10.1021/cb800116f. [DOI] [PubMed] [Google Scholar]
- 56.Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107(2):183–94. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- 57.Bringmann H. Mechanical and genetic separation of aster- and midzone-positioned cytokinesis. Biochem Soc Trans. 2008;36(Pt 3):381–3. doi: 10.1042/BST0360381. [DOI] [PubMed] [Google Scholar]
- 58.Zwaal RR, Ahringer J, van Luenen HG, Rushforth A, Anderson P, Plasterk RH. G proteins are required for spatial orientation of early cell cleavages in C elegans embryos. Cell. 1996;86(4):619–29. doi: 10.1016/s0092-8674(00)80135-x. [DOI] [PubMed] [Google Scholar]
- 59.Miller KG, Rand JB. A role for RIC-8 (Synembryn) and GOA-1 (G(o)alpha) in regulating a subset of centrosome movements during early embryogenesis in Caenorhabditis elegans. Genetics. 2000;156(4):1649–60. doi: 10.1093/genetics/156.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gotta M, Ahringer J. Distinct roles for Galpha and Gbetagamma in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat Cell Biol. 2001;3(3):297–300. doi: 10.1038/35060092. [DOI] [PubMed] [Google Scholar]
- 61.Gotta M, Ahringer J. Axis determination in C elegans: initiating and transducing polarity. Curr Opin Genet Dev. 2001;11(4):367–73. doi: 10.1016/s0959-437x(00)00206-9. [DOI] [PubMed] [Google Scholar]
- 62.Gotta M, Dong Y, Peterson YK, Lanier SM, Ahringer J. Asymmetrically distributed C elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr Biol. 2003;13(12):1029–37. doi: 10.1016/s0960-9822(03)00371-3. [DOI] [PubMed] [Google Scholar]
- 63.Srinivasan DG, Fisk RM, Xu H, van den Heuvel S. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev. 2003;17(10):1225–39. doi: 10.1101/gad.1081203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Colombo K, Grill SW, Kimple RJ, Willard FS, Siderovski DP, Gonczy P. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 2003;300(5627):1957–61. doi: 10.1126/science.1084146. [DOI] [PubMed] [Google Scholar]
- 65.Schwabe T, Bainton RJ, Fetter RD, Heberlein U, Gaul U. GPCR signaling is required for blood-brain barrier formation in Drosophila. Cell. 2005;123(1):133–44. doi: 10.1016/j.cell.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 66.Wang H, Ng KH, Qian H, Siderovski DP, Chia W, Yu F. Ric-8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G proteins. Nat Cell Biol. 2005;7(11):1091–8. doi: 10.1038/ncb1317. [DOI] [PubMed] [Google Scholar]
- 67.Chen JG, Gao Y, Jones AM. Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots. Plant Physiol. 2006;141(3):887–97. doi: 10.1104/pp.106.079202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hewavitharana T, Wedegaertner PB. Non-canonical signaling and localizations of heterotrimeric G proteins. Cell Signal. 2012;24(1):25–34. doi: 10.1016/j.cellsig.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cho H, Kehrl JH. Localization of Gi alpha proteins in the centrosomes and at the midbody: implication for their role in cell division. J Cell Biol. 2007;178(2):245–55. doi: 10.1083/jcb.200604114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dave RH, Saengsawang W, Yu JZ, Donati R, Rasenick MM. Heterotrimeric G-proteins interact directly with cytoskeletal components to modify microtubule-dependent cellular processes. Neurosignals. 2009;17(1):100–8. doi: 10.1159/000186693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buhl AM, Johnson NL, Dhanasekaran N, Johnson GL. G alpha 12 and G alpha 13 stimulate Rho-dependent stress fiber formation and focal adhesion assembly. J Biol Chem. 1995;270(42):24631–4. doi: 10.1074/jbc.270.42.24631. [DOI] [PubMed] [Google Scholar]
- 72.Lowry WE, Huang J, Ma YC, Ali S, Wang D, Williams DM, Okada M, Cole PA, Huang XY. Csk, a critical link of g protein signals to actin cytoskeletal reorganization. Dev Cell. 2002;2(6):733–44. doi: 10.1016/s1534-5807(02)00175-2. [DOI] [PubMed] [Google Scholar]
- 73.Wang D, Tan YC, Kreitzer GE, Nakai Y, Shan D, Zheng Y, Huang XY. G proteins G12 and G13 control the dynamic turnover of growth factor-induced dorsal ruffles. J Biol Chem. 2006;281(43):32660–7. doi: 10.1074/jbc.M604588200. [DOI] [PubMed] [Google Scholar]
- 74.Wu HC, Huang PH, Chiu CY, Lin CT. G protein beta2 subunit antisense oligonucleotides inhibit cell proliferation and disorganize microtubule and mitotic spindle organization. J Cell Biochem. 2001;83(1):136–46. doi: 10.1002/jcb.1210. [DOI] [PubMed] [Google Scholar]
- 75.Neubig RR, Siderovski DP. Regulators of G-protein signalling as new central nervous system drug targets. Nat Rev Drug Discov. 2002;1(3):187–97. doi: 10.1038/nrd747. [DOI] [PubMed] [Google Scholar]
- 76.Willard FS, Kimple RJ, Siderovski DP. Return of the GDI: the GoLoco motif in cell division. Annu Rev Biochem. 2004;73:925–51. doi: 10.1146/annurev.biochem.73.011303.073756. [DOI] [PubMed] [Google Scholar]
- 77.Willars GB. Mammalian RGS proteins: multifunctional regulators of cellular signalling Semin. Cell Dev Biol. 2006;17(3):363–76. doi: 10.1016/j.semcdb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 78.Martin-McCaffrey L, Willard FS, Oliveira-dos-Santos AJ, Natale DR, Snow BE, Kimple RJ, Pajak A, Watson AJ, Dagnino L, Penninger JM, et al. RGS14 is a mitotic spindle protein essential from the first division of the mammalian zygote. Dev Cell. 2004;7(5):763–9. doi: 10.1016/j.devcel.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 79.Pontrello CG, Sun MY, Lin A, Fiacco TA, DeFea KA, Ethell IM. Cofilin under control of beta-arrestin-2 in NMDA-dependent dendritic spine plasticity, long-term depression (LTD), and learning. Proc Natl Acad Sci U S A. 2012;109(7):E442–51. doi: 10.1073/pnas.1118803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang F LH, Chen Y*, Ma L. The Role of GRK5 in Mitosis. Chinese Journal of Cell Biology. 2012;34(3):234–239. [Google Scholar]
- 81.Zhang X, Bedigian AV, Wang W, Eggert US. G protein-coupled receptors participate in cytokinesis. Cytoskeleton (Hoboken) 2012;69(10):810–818. doi: 10.1002/cm.21055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wolfe BL, Trejo J. Clathrin-dependent mechanisms of G protein-coupled receptor endocytosis. Traffic. 2007;8(5):462–70. doi: 10.1111/j.1600-0854.2007.00551.x. [DOI] [PubMed] [Google Scholar]
- 83.Calebiro D, Nikolaev VO, Persani L, Lohse MJ. Signaling by internalized G-protein-coupled receptors. Trends Pharmacol Sci. 2010;31(5):221–8. doi: 10.1016/j.tips.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 84.Calebiro D, Nikolaev VO, Gagliani MC, de Filippis T, Dees C, Tacchetti C, Persani L, Lohse MJ. Persistent cAMP-signals triggered by internalized G-protein-coupled receptors. PLoS Biol. 2009;7(8):e1000172. doi: 10.1371/journal.pbio.1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marino G, Madeo F, Kroemer G. Autophagy for tissue homeostasis and neuroprotection. Curr Opin Cell Biol. 2011;23(2):198–206. doi: 10.1016/j.ceb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 87.Moreau K, Luo S, Rubinsztein DC. Cytoprotective roles for autophagy. Curr Opin Cell Biol. 2010;22(2):206–11. doi: 10.1016/j.ceb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martinet W, Knaapen MW, Kockx MM, De Meyer GR. Autophagy in cardiovascular disease. Trends Mol Med. 2007;13(11):482–91. doi: 10.1016/j.molmed.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 89.Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, Zuker CS. An amino-acid taste receptor. Nature. 2002;416(6877):199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- 90.Wauson EM, Zaganjor E, Lee AY, Guerra ML, Ghosh AB, Bookout AL, Chambers CP, Jivan A, McGlynn K, Hutchison MR, et al. The G Protein-Coupled Taste Receptor T1R1/T1R3 Regulates mTORC1 and Autophagy. Mol Cell. 2012;47(6):851–862. doi: 10.1016/j.molcel.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang X, Wang W, Bedigian AV, Coughlin ML, Mitchison TJ, Eggert US. Dopamine receptor D3 regulates endocytic sorting by a Prazosin-sensitive interaction with the coatomer COPI. Proc Natl Acad Sci U S A. 2012;109(31):12485–90. doi: 10.1073/pnas.1207821109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ling K, Wang P, Zhao J, Wu YL, Cheng ZJ, Wu GX, Hu W, Ma L, Pei G. Five-transmembrane domains appear sufficient for a G protein-coupled receptor: functional five-transmembrane domain chemokine receptors. Proc Natl Acad Sci U S A. 1999;96(14):7922–7. doi: 10.1073/pnas.96.14.7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gether U, Kobilka BK. G protein-coupled receptors. II. Mechanism of agonist activation. J Biol Chem. 1998;273(29):17979–82. doi: 10.1074/jbc.273.29.17979. [DOI] [PubMed] [Google Scholar]
- 94.Kenakin T. Ligand-selective receptor conformations revisited: the promise and the problem. Trends Pharmacol Sci. 2003;24(7):346–54. doi: 10.1016/S0165-6147(03)00167-6. [DOI] [PubMed] [Google Scholar]
- 95.Kobilka BK. G protein coupled receptor structure and activation. Biochim Biophys Acta. 2007;1768(4):794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marie N, Lecoq I, Jauzac P, Allouche S. Differential sorting of human delta-opioid receptors after internalization by peptide and alkaloid agonists. J Biol Chem. 2003;278(25):22795–804. doi: 10.1074/jbc.M300084200. [DOI] [PubMed] [Google Scholar]
- 97.Lecoq I, Marie N, Jauzac P, Allouche S. Different regulation of human delta-opioid receptors by SNC-80 [(+)-4-[(alphaR)-alpha-((2S,5R)-4-allyl-2,5-dimethyl-1-piperazinyl)-3-methoxybenz yl]-N,N-diethylbenzamide] and endogenous enkephalins. J Pharmacol Exp Ther. 2004;310(2):666–77. doi: 10.1124/jpet.103.063958. [DOI] [PubMed] [Google Scholar]
- 98.Roosterman D, Cottrell GS, Schmidlin F, Steinhoff M, Bunnett NW. Recycling and resensitization of the neurokinin 1 receptor Influence of agonist concentration and Rab GTPases. J Biol Chem. 2004;279(29):30670–9. doi: 10.1074/jbc.M402479200. [DOI] [PubMed] [Google Scholar]
- 99.Trapaidze N, Gomes I, Bansinath M, Devi LA. Recycling and resensitization of delta opioid receptors. DNA Cell Biol. 2000;19(4):195–204. doi: 10.1089/104454900314465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gregory KJ, Sexton PM, Tobin AB, Christopoulos A. Stimulus bias provides evidence for conformational constraints in the structure of a G protein-coupled receptor. J Biol Chem. 2012;287(44):37066–37077. doi: 10.1074/jbc.M112.408534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Verrier F, An S, Ferrie AM, Sun H, Kyoung M, Deng H, Fang Y, Benkovic SJ. GPCRs regulate the assembly of a multienzyme complex for purine biosynthesis. Nat Chem Biol. 2011;7(12):909–15. doi: 10.1038/nchembio.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Binet V, Brajon C, Le Corre L, Acher F, Pin JP, Prezeau L. The heptahelical domain of GABA(B2) is activated directly by CGP7930, a positive allosteric modulator of the GABA(B) receptor. J Biol Chem. 2004;279(28):29085–91. doi: 10.1074/jbc.M400930200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gurevich VV, Gurevich EV. GPCR monomers and oligomers: it takes all kinds. Trends Neurosci. 2008;31(2):74–81. doi: 10.1016/j.tins.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Milligan G. G protein-coupled receptor dimerization: function and ligand pharmacology. Mol Pharmacol. 2004;66(1):1–7. doi: 10.1124/mol.104.000497.. [DOI] [PubMed] [Google Scholar]
- 105.Brattelid T, Levy FO. Quantification of GPCR mRNA using real-time RT-PCR. Methods Mol Biol. 2011;746:165–93. doi: 10.1007/978-1-61779-126-0_9. [DOI] [PubMed] [Google Scholar]
- 106.Regard JB, Sato IT, Coughlin SR. Anatomical profiling of G protein-coupled receptor expression. Cell. 2008;135(3):561–71. doi: 10.1016/j.cell.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Feldmesser E, Olender T, Khen M, Yanai I, Ophir R, Lancet D. Widespread ectopic expression of olfactory receptor genes. BMC Genomics. 2006;7:121. doi: 10.1186/1471-2164-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.De la Cruz O, Blekhman R, Zhang X, Nicolae D, Firestein S, Gilad Y. A signature of evolutionary constraint on a subset of ectopically expressed olfactory receptor genes. Mol Biol Evol. 2009;26(3):491–4. doi: 10.1093/molbev/msn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Drutel G, Arrang JM, Diaz J, Wisnewsky C, Schwartz K, Schwartz JC. Cloning of OL1, a putative olfactory receptor and its expression in the developing rat heart. Receptors Channels. 1995;3(1):33–40. [PubMed] [Google Scholar]
- 110.Feingold EA, Penny LA, Nienhuis AW, Forget BG. An olfactory receptor gene is located in the extended human beta-globin gene cluster and is expressed in erythroid cells. Genomics. 1999;61(1):15–23. doi: 10.1006/geno.1999.5935. [DOI] [PubMed] [Google Scholar]
- 111.Blache P, Gros L, Salazar G, Bataille D. Cloning and tissue distribution of a new rat olfactory receptor-like (OL2) Biochem Biophys Res Commun. 1998;242(3):669–72. doi: 10.1006/bbrc.1997.8041. [DOI] [PubMed] [Google Scholar]
- 112.Raming K, Konzelmann S, Breer H. Identification of a novel G-protein coupled receptor expressed in distinct brain regions and a defined olfactory zone. Receptors Channels. 1998;6(2):141–51. [PubMed] [Google Scholar]
- 113.Conzelmann S, Levai O, Bode B, Eisel U, Raming K, Breer H, Strotmann J. A novel brain receptor is expressed in a distinct population of olfactory sensory neurons. Eur J Neurosci. 2000;12(11):3926–34. doi: 10.1046/j.1460-9568.2000.00286.x. [DOI] [PubMed] [Google Scholar]
- 114.Yuan TT, Toy P, McClary JA, Lin RJ, Miyamoto NG, Kretschmer PJ. Cloning and genetic characterization of an evolutionarily conserved human olfactory receptor that is differentially expressed across species. Gene. 2001;278(1-2):41–51. doi: 10.1016/s0378-1119(01)00709-0. [DOI] [PubMed] [Google Scholar]
- 115.Spehr M, Gisselmann G, Poplawski A, Riffell JA, Wetzel CH, Zimmer RK, Hatt H. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 2003;299(5615):2054–8. doi: 10.1126/science.1080376. [DOI] [PubMed] [Google Scholar]
- 116.Spehr M, Schwane K, Riffell JA, Zimmer RK, Hatt H. Odorant receptors and olfactory-like signaling mechanisms in mammalian sperm. Mol Cell Endocrinol. 2006;250(1-2):128–36. doi: 10.1016/j.mce.2005.12.035. [DOI] [PubMed] [Google Scholar]


