Abstract
Since its introduction to the market in the 1970s, the synthetic biocide triclosan has had widespread use in household and medical products. Although decreased triclosan susceptibility has been observed for several bacterial species, when exposed under laboratory settings, no in vivo studies have associated triclosan use with decreased triclosan susceptibility or cross-resistance to antibiotics. One major challenge of such studies is the lack of strains that with certainty have not been exposed to triclosan. Here we have overcome this challenge by comparing current isolates of the human opportunistic pathogen Staphylococcus epidermidis with isolates collected in the 1960s prior to introduction of triclosan to the market. Of 64 current S. epidermidis isolates 12.5% were found to have tolerance towards triclosan defined as MIC≥0.25 mg/l compared to none of 34 isolates obtained in the 1960s. When passaged in the laboratory in the presence of triclosan, old and current susceptible isolates could be adapted to the same triclosan MIC level as found in current tolerant isolates. DNA sequence analysis revealed that laboratory-adapted strains carried mutations in fabI encoding the enoyl-acyl carrier protein reductase isoform, FabI, that is the target of triclosan, and the expression of fabI was also increased. However, the majority of the tolerant current isolates carried no mutations in fabI or the putative promoter region. Thus, this study indicates that the widespread use of triclosan has resulted in the occurrence of S. epidermidis with tolerance towards triclosan and that the adaptation involves FabI as well as other factors. We suggest increased caution in the general application of triclosan as triclosan has not shown efficacy in reducing infections and is toxic to aquatic organisms.
Introduction
Triclosan is a synthetic broad spectrum biocide that was introduced to the market in the early 1970s [1]. In low concentrations triclosan inhibits the growth of many bacteria and higher concentrations can be bactericidal [2], [3]. It is now widely used as an antiseptic, disinfectant and preservative in clinical settings and in various consumer products including cosmetics, plastic materials, toys and textiles [4]. Environmental exposure, toxicity and mechanisms of action have recently been reviewed by Dann et Hontela [5]. Triclosan is excreted in the urine and has been found in human urine (2.4–3790 µg/l), plasma (0.01–38 µg/l) and breast milk (0.018–0.95 µg/l). The extensive use in human products has led to accumulation in the environment with concentrations of 20–133,000 µg/kg dry-weight in biosolids from wastewater treatment plants. Very low concentrations, in the ng/l range, have been found in lakes, rivers, seawater and drinking water. Toxicological studies indicate that triclosan is not toxic for mammals, however, triclosan has an estimated bioaccumulation factor of more than 1000 in algae and can be highly toxic to green algae. [4]–[6]
It has been wondered whether the extensive use of triclosan could select for antimicrobial resistance. For staphylococci only low-level triclosan resistance, tolerance, has been detected and the few exposure studies that have been conducted, have not shown any dependency between triclosan usage and increased tolerance towards triclosan or co/cross-resistance to antibiotics [7]–[10]. A confounder for exposure studies is the omnipresence of triclosan in the environment, which affects both the user groups and the controls [7]. In vitro studies have shown that bacteria can be stably adapted to increasing concentrations of triclosan and in some cases develop cross-resistance towards antibiotics. This has mostly been found for Gram-negative rods, caused by multidrug efflux pumps [11]–[14].
Staphylococcus epidermidis is an abundant human skin commensal as well as an important opportunistic pathogen responsible for a significant number of severe foreign body related infections. S. epidermidis isolated from blood cultures span the clinical spectrum from skin contaminants to severe infections [15]. S. epidermidis are exposed to triclosan through personal care products and the incidence of triclosan tolerance have been found to be higher in clinical S. epidermidis than in clinical Staphylococcus aureus isolates and it is speculated if the mechanisms are similar [16]. In S. aureus the mechanism for tolerance has been recognized as mutations in or increased expression of the gene fabI. The FabI enzyme, enoyl-acyl carrier protein reductase, catalyses the final step in bacterial type II fatty acid biosynthesis. Triclosan functions as a slow binding inhibitor that inactivates the enzyme through the formation of a stable, non-covalent, FabI-NAD+−triclosan ternary complex. [17]
In this study we describe, for the first time, the susceptibility towards triclosan in S. epidermidis that have never been exposed to triclosan, namely S. epidermidis isolated from blood in 1965–66, well before the introduction of triclosan to the market. We compare these old isolates with current isolates of S. epidermidis. The old S. epidermidis were exposed to triclosan, leading to development of tolerance to triclosan. The fabI gene and its expression was characterized in laboratory developed triclosan tolerant S. epidermidis and current triclosan tolerant S. epidermidis.
Methods
Ethics
Scientific ethical committee approval is not required for this project. We worked with bacteria that were already isolated from blood cultures as part of the standard operating procedures with samples referred to the Department of Clinical Microbiology at Hvidovre Hospital, Denmark. No material relating to humans was used or stored in this experiment and all person related data were blinded for the researchers. This studies results or the researchers had no influence on any aspects concerning patient care.
Bacterial isolates
A collection of coagulase negative staphylococci isolated from blood cultures of Danish patients during 1965 and 66 has been stored in agar sticks at Statens Serum Institut, Copenhagen, Denmark and have stayed untouched since then. From 149 of those old agar sticks isolates could be cultured from 51, and 34 of these isolates were identified as S. epidermidis. These 34 isolates were used as pre-triclosan era controls in relation to triclosan exposure. As comparable isolates, which have been exposed to “the uncontrolled” modern usage of triclosan, 64 S. epidermidis isolates were collected from blood cultures of patients hospitalized in Copenhagen, Denmark, 2010–11. We collected these 64 isolates consecutively, until we had 40 methicillin resistant S. epidermidis. After that we continued to collect methicillin susceptible isolates for a total of 24 to increase the diversity of the collection.
All isolates were species-identified by MALDI-TOF MS (Bruker Daltonics), S. epidermidis ATCC 12228 (NCBI Reference Sequence: NC_004461.1) was included in the susceptibility tests as control. Isolates were onward stored at −80°C. Tryptic soy broth/agar (TSB/TSA) (Oxoid, Roskilde, Denmark) was used as growth medium unless otherwise stated. All incubations were performed at 37°C atmospheric air.
Antimicrobial and biocide susceptibility testing
The MIC of triclosan was determined by following the recommendations of the British Society of Antimicrobial Chemotherapy using broth micro dilutions [18]. A stock solution of triclosan (Irgasan, SIGMA-ALDRICH®, Germany) of 1 mg/ml dissolved in 96% ethanol (KEMETYL A/S, Denmark) was prepared in advance and a doubling dilution range from 0.0156–16 mg/l triclosan in Mueller Hinton bouillon (MHB) (Oxoid, Roskilde, Denmark) was made for each experiment. 100 µl of the triclosan dilution+100 µl of an overnight bacterial suspension adjusted to 106 CFU/ml was mixed in each well. The MIC was determined as the lowest concentration that inhibited visible growth after 24 hours. Positive (bacterial suspension+MHB) and negative (MHB and triclosan dilutions without bacterial suspension) controls were included in each measurement. Bacterial growth in the highest used dilution of 96% ethanol (0.77%) was also tested and did not differ visually from growth in MHB alone. 10 µl from each well (0.0625 mg/l and above) was, after 24 hours incubation spotted on to Muller-Hinton agar (MHA) plates containing no triclosan. The MBC was read as the lowest concentration with no growth after 48 hours. All MIC experiments were repeated on two separate occasions each with two technical replicates and MIC is given as the mean. The MBC was repeated for all isolates with a high MBC defined as 8 mg/l and for one third of the rest (below 8 mg/l). All of the repeat MBCs stayed at 8 mg/l or below 8 mg/l, respectively. Antibiotic susceptibility was determined using the disc diffusion method (Oxoid, Roskilde, Denmark) following the EUCAST guidelines and breakpoints [19], [20].
Adaptation experiments
Six S. epidermidis isolates were adapted to triclosan using the gradient plate approach [21]. Initial gradients consisted of zero mg/l at one end of the plate up to 0.025 mg/l or 4 mg/l at the other end depending on the initial MIC of each isolate. After 48 h of incubation, colonies from the leading edge of growth were passed to a new gradient plate with the same concentrations until growth occurred across the entire plate. This process was repeated using doubling increases in triclosan gradients until no further adaptation was possible. The current S. epidermidis isolates, with a high triclosan MIC, were passed repeatedly on plates going up to 4 mg/l as many times as the other isolates were passed. Each isolate was passed in duplicate and with a control passed along on TSA plates without triclosan. Colonies from the leading edges from the final gradient plates as well as controls were frozen at −80°C. Furthermore, colonies from the leading edges of each of the adapted isolates were passed on TSA without triclosan for 5 days and then frozen. Adapted isolates, controls and adapted isolates passed for 5 days without triclosan were subjected to triclosan susceptibility testing and antibiotic susceptibility testing.
FabI sequencing and MLST
Template DNA was prepared by suspending colonies from an overnight culture in Milli-Q water, heating at 100°C for 10 minutes, centrifugation at 8000 rpm for to minutes and then the supernatant was used immediately or frozen at −20°C.
The fabI gene including the putative promoter region in S. epidermidis was amplified using the primers 5′-GGTGTTGTTGAAGATCAAATATAC-3′ and 5′-GTCCTCTTATTAAACTCCG-3′.
Multilocus sequence typing (MLST) was conducted following the mlst.net guidelines and the same thermocycler program was used for fabI as for the MLST reactions [22]. PCR products were purified using 1 µl exonuclease1 and 2 µl alkaline phosphatase (Fermentas, Roskilde, Denmark) for 10 µl PCR product, activated at 37° C for 15 minutes and terminated at 85°C for 15 minutes. The purified products were sequenced at both strands using the same primer set as amplification and Macrogenservice (Macrogen Europe, Netherlands). The results were analyzed using CLC Main Workbench 6.2.
RNA extraction and northern hybridization
Cells of S. epidermidis were grown to mid logarithmic growth phase (OD600 = 0.6–0.8) in MH broth and samples were immediately cooled in ice-water bath. The bacterial cells were lysed using the Fast Prep FP120 instrument (BIO101, ThermoSavent) for 45 s at speed 6.0. Total RNA was extracted from the cells using the RNeasy mini kit (Qiagen, Denmark) according to the manufacturer's directions. Analysis of transcripts was done as previously described [23]. Hybridization probes were generated by PCR from chromosomal DNA of S. aureus 8325–4 using specific primers for the fabI gene (fab2F: atgttaaatcttgaaaacaaaac and fab2R: ttatttaattgcgtggaatccgc), (TAG Copenhagen A/S, Denmark). RNA extracted from at least two independent experiments was analyzed.
Statistics
Based on a pilot study (data not shown) showing that none of 22 old S. epidermidis isolates were triclosan tolerant while 15% of current isolates were, a power analyses was performed using G*Power 3.1.5 (freely distributed at http://www.psycho.uni-duesseldorf.de/abteilungen/aap/gpower3/download-and-register). Given a power of 0.80 and α = 0.05 a required sample size of minimum 32 old isolates and 59 current isolates was calculated.
Categorical variables were compared using Fisher's exact test.
Results
Triclosan susceptibility in old and in current S. epidermidis isolates
The 34 old S. epidermidis isolates were all methicillin susceptible. The results of the triclosan MIC/MBC determinations are summarized in Table 1. Old and current S. epidermidis isolates had the same MIC50 (MIC required to inhibit the growth of 50% of isolates) of 0.0625 mg/l and for the old isolates the MIC90 (MIC required to inhibit the growth of 90% of isolates) was also 0.0625 mg/l. This was in contrast to the current isolates that had a MIC90 that was 8 fold higher than the MIC50. The highest MIC value among the current isolates was 4 mg/l; 32-fold higher than the highest MIC value of 0.125 mg/l, among the old isolates. The same pattern was true for the MBC values though on a narrower scale.
Table 1. Susceptibility to triclosan among S. epidermidis isolates from 1965–66 (old) and from 2010–11 (current) as determined by their minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC).
| Number | MIC50 | MIC90 | MIC range | MBC50 | MBC90 | MBC range | |
| Old | 34 | 0.0625 | 0.0625 | ≤0.0156–0.125 | 1 | 2 | ≤0.0625–4 |
| Current | 64 | 0.0625 | 0.5 | ≤0.0156–4 | 2 | 8 | ≤0.0625–8 |
MIC/MBC50 = Minimum Inhibitory/Bactericidal Concentration required to inhibit the growth/kill of 50% of isolates. MIC/MBC90 = Minimum Inhibitory/Bactericidal Concentration required to inhibit the growth/kill of 90% of isolates
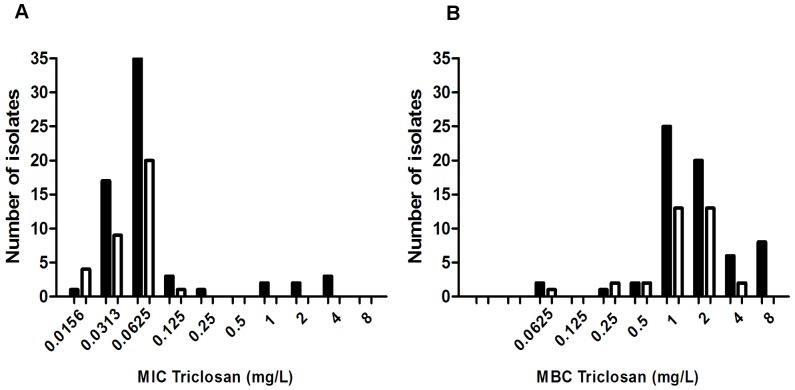
The distributions of the MIC and MBC values are shown in Figure 1. A group of 8 isolates were identified amongst the current isolates having a MIC≥0.25 mg/l and a MBC of 8 mg/l not seen among the old isolates. This suggests a wild-type population cut-off MIC of 0.25 mg/l and MBC of 8 mg/l. By this definition the isolates fell in two natural groups (Figure 1, Table 2) and based on their MIC values they could be divided into triclosan susceptible isolates (MIC<0.25 mg/l) and triclosan tolerant (MIC≥0.25 mg/l). The current S. epidermidis isolates had significantly more triclosan tolerant isolates compared to the group of old isolates; 12.5% versus 0%. This was even more evident when comparing current methicillin resistant S. epidermidis isolates to the old isolates, but triclosan tolerance was also seen in the current methicillin susceptible S. epidermidis isolates (Table 2).
Figure 1. Triclosan MIC and MBC distributions for S. epidermidis isolates from 1965-66 and from 2010-11.
A: Triclosan MIC (Mininmum inhibitory concentration) distributions. B: Triclosan MBC (Mininmum bactericidal concentration) distributions. Black bars = S. epidermidis blood isolates from 2010-11. White bars = S. epidermidis blood isolates from 1965-66. The lowest dilutions represent a MIC of≤0.0156 mg/l and a MBC of≤0.0652 mg/l. Eight isolates in the current group have a MIC ≥0.25 mg/l and the same eight isolates are the ones with a MBC = 8 mg/l.
Table 2. Distribution of triclosan tolerance among S. epidermidis isolates from 1965-66 (old) and 2010-11 (current).
| N | MIC≥0.25 mg/l | Fishers exact testa | |
| Old | 34 | 0 (0%) | |
| Current | 64 | 8 (12.5%) | p≤0.048 |
| Current, cefoxitin S | 24 | 2 (8.3%) | Not significant |
| Current, cefoxitin R | 40 | 6 (15%) | p≤0.028 |
MIC <0.25 mg/l is defined as susceptible and MIC ≥0.25 mg/l is defined as tolerant.
Old isolates versus current isolates.
Triclosan tolerant isolates are not clonal
MLST results (supplementary material, Table S1) revealed that the triclosan tolerant isolates were not due to the expansion of a clone. Out of 15 current isolates (seven triclosan susceptible and the eight triclosan tolerant) nine different ST types were found with six different ST types in the group of triclosan tolerant isolates and three ST types were represented in both the susceptible and the tolerant isolates.
Old S. epidermidis isolates could be adapted to triclosan tolerance
Two old (65–13, 66–1) and two current (BD-62, Van-1) isolates, all triclosan susceptible, and two current isolates with a high triclosan MIC (BD-12, BD-24) were attempted adapted to triclosan. The four triclosan susceptible isolates all acquired stable tolerance to triclosan. Three isolates maintained a stabile triclosan MIC of 4 mg/l, while one old isolate peaked at MIC of 2 mg/l and following 5 passages without triclosan dropped to 0.375 mg/l, while maintaining a MBC of 8 mg/l. It was not possible to increase the MIC of the two isolates that were already triclosan tolerant with an MIC of 4 mg/l. Results are presented in Table 3 left side columns.
Table 3. Triclosan and antibiotic susceptibility of parental strains from 1965-66 (65–13 and 66–01) and from 2010–11 (BD-62, Van-1, BD-12 and BD-24) and their triclosan laboratory exposed descendents.
| Isolate ID (ST) | MIC_0 | MBC_0 | MIC_5 | MBC_5 | PEN | FOX | FA | GEN | ERY | CLIN | RIF | LIN | NOF |
| 65-13 (410) | 0.0625 | 0.25 | R | S | S | S | S | S | S | S | S | ||
| 65-13a | 2 | 8 | 0.25 | 8 | R | S | S | S | S | S | S | S | S |
| 65-13b | 2 | 8 | R | S | S | S | S | S | S | S | S | ||
| 65-13Ka | 0.0313 | 0.125 | R | S | S | S | S | S | S | S | S | ||
| 65-13Kb | 0.0313 | 0.25 | |||||||||||
| 66-1 (190) | 0.125 | 4 | R | S | S | S | S | S | S | S | S | ||
| 66-1a | 4 | 8 | 4 | 8 | R | S | S | S | S | S | S | S | S |
| 66-1b | 4 | 8 | R | S | S | S | S | S | S | S | S | ||
| 66-1Ka | 0.0625 | 4 | R | S | S | S | S | S | S | S | S | ||
| 66-1Kb | 0.0625 | 4 | |||||||||||
| BD-62 (327) | 0.0313 | 1 | S | S | S | S | S | S | S | S | S | ||
| BD-62a | 4 | 4 | 4 | 4 | S | S | S | S | S | S | S | S | S |
| BD-62b | 4 | 4 | S | S | S | S | S | S | S | S | S | ||
| BD-62Ka | 0.0313 | 1 | S | S | S | S | S | S | S | S | S | ||
| BD-62Kb | 0.0313 | 1 | |||||||||||
| Van-1 (2) | 0.0625 | 1 | R | R | R | R | R | R | S | S | R | ||
| Van-1a | 4 | 8 | 4 | 8 | R | R | R | R | S | S | S | S | R |
| Van-1b | 4 | 8 | R | R | R | R | S | S | S | S | R | ||
| Van-1Ka | 0.0313 | R | R | R | R | S | S | S | S | R | |||
| Van-1Kb | 0.0313 | 2 | |||||||||||
| BD-12 (88) | 2 | 8 | R | R | S | S | S | S | S | S | S | ||
| BD-12a | 4 | 8 | 4 | 8 | R | R | S | S | S | S | S | S | S |
| BD-12b | 4 | 8 | R | R | S | S | S | S | S | S | S | ||
| BD-12Ka | 4 | 8 | R | R | R | S | R | S | S | S | S | ||
| BD-12Kb | 4 | 8 | |||||||||||
| BD-24 (ND) | 4 | 8 | R | R | R | S | R | S | S | S | S | ||
| BD-24a | 4 | 8 | 4 | 8 | R | R | R | S | R | S | S | S | S |
| BD-24b | 4 | 8 | R | R | R | S | R | S | S | S | S | ||
| BD-24Ka | 4 | 8 | R | R | R | S | R | S | S | S | S | ||
| BD-24Kb | 4 | 8 |
Adapted strains are named with the parent name and the suffix a and b. The control strains passed along without triclosan have the suffix Ka and Kb. MIC_0/MBC_0, measured after ended adaptation. MIC_5/MBC_5, measured after 5 passages on triclosan free media. PEN = penicillin, FOX = cefoxitin, FA = fucidic acid, GEN = gentamicin, ERY = erythromycin, CLIN = clindamycin, RIF = rifampicin, LIN = linezolid, NOF = norfloxacin. ST = Sequence Type, determined by MLST.
No antibiotic cross-resistance in S. epidermidis
There was no significant association between antibiotic resistance and triclosan tolerance in the current isolates (supporting information, Table S2). Furthermore, none of the isolates, old or current, that were adapted to triclosan tolerance developed any resistance towards the tested antibiotics. Indeed one of the adapted isolates, Van-1a and b (see Table 3 for nomenclature), changed from being resistant to become susceptible towards erythromycin and clindamycin but that was also seen in the control strain, Van-1Ka, only passed without triclosan in the media and with no change in triclosan MIC.
FabI and triclosan susceptibility in S. epidermidis
The fabI gene of S. epidermidis has 82–84% nucleotide similarity to the fabI gene of S. aureus when blasting published sequences at NCBI. Five old and seven current triclosan susceptible as well as the eight current triclosan tolerant and the six triclosan adapted S. epidermidis isolates were fabI sequenced (Table 4). We did not get a PCR product from one old (as well as its triclosan adapted isogenic descendent) and two current susceptible isolates and they are excluded from interpretation. The current triclosan susceptible isolates all had the same fabI amino acid sequence, identical to the sequence predicted for the fabI of the triclosan susceptible S. epidermidis ATCC12228 (MIC 0.0625 mg/l and MBC 2 mg/l in our assay). Five of the eight clinical S. epidermidis isolates with tolerance towards triclosan also had identical fabI amino acid sequence to ATCC 12228 and none of these isolates had mutations in the putative promoter region of fabI. The three remaining tolerant S. epidermidis isolates had between one and three non synonymous mutations causing changes in the predicted amino acid sequence: Isolate BD-18 had a substitution of a phenylalanine for a valine, at position 204 (F204V), isolate BD-38 had a A198G substitution. Mutations at those two positions have previously been described in fabI of S. aureus with triclosan tolerance [24]–[27]. BD-24 had three amino acid substitutions a H60Q, a H61Y and a S78A as well as several nucleotide changes in the putative promoter region. None of these substitutions have been described previously and interestingly H61Y and S78A were also found in two old, triclosan susceptible, S. epidermidis isolates (65–02, 65–12) while the other two old isolates from which we obtained a fabI sequence were identical to the ATCC 12228 fabI.
Table 4. Analysis of mutations in the fabI gene of S. epidermidis and the upstream promoter region.
| Isolate (year of isolation) | ST | TriclosanMIC mg/l | fabI non synonymous mutations | Up-stream (putative promoter region) mutations in relation to current susceptible isolates |
| 65-02 (1965) | 414 | <0.25 | H61Y, S78A, G85S | |
| 65-06 (1965) | 86 | <0.25 | ( = ATCC 12228) | |
| 65-12 (1965) | 409 | <0.25 | H61Y, S78A | |
| 65-13 (1965) | 410 | <0.25 | no data | no data |
| 66-01 (1966) | 190 | <0.25 | ( = ATCC 12228) | |
| BD-09 (2010) | 5 | <0.25 | ( = ATCC 12228) | Current susceptible |
| BD-10 (2010) | 32 | <0.25 | ( = ATCC 12228) | Current susceptible |
| BD-26 (2011) | ND | <0.25 | no data | no data |
| BD-44 (2011) | 5 | <0.25 | ( = ATCC 12228) | Current susceptible |
| BD-50 (2011) | 73 | <0.25 | no data | no data |
| BD-62 (2011) | 327 | <0.25 | ( = ATCC 12228) | Current susceptible |
| Van-1 (2011) | 2 | <0.25 | ( = ATCC 12228) | Current susceptible |
| BD-06 (2010) | 88 | ≥0.25 | ( = ATCC 12228) | ( = current susceptible) |
| BD-12 (2011) | 88 | ≥0.25 | ( = ATCC 12228) | ( = current susceptible) |
| BD-18 (2011) | 73 | ≥0.25 | F204V | Two NT changes |
| BD-19 (2011) | 88 | ≥0.25 | ( = ATCC 12228) | ( = current susceptible) |
| BD-24 (2011) | ND | ≥0.25 | H60Q, H61Y, S78A | Many NT changes |
| BD-32 (2011) | 2 | ≥0.25 | ( = ATCC 12228) | ( = current susceptible) |
| BD-38 (2011) | New | ≥0.25 | A198G | ( = current susceptible) |
| BD-53 (2011) | 5 | ≥0.25 | ( = ATCC 12228) | ( = current susceptible) |
| 65-13a* | ≥0.25 | no data | no data | |
| 66-01a* | ≥0.25 | A95V | One NT change compared to 66-01 | |
| BD-62a* | ≥0.25 | A95V | Deletion in −69 to −77 compared to the current susceptible | |
| Van-1a* | ≥0.25 | A95V | ( = current susceptible) | |
| BD-12a* | ≥0.25 | ( = ATCC 12228) | ( = current susceptible) | |
| BD-24a* | ≥0.25 | ( = BD-24) | ( = BD-24) |
The suffix a* indicates adapted strains that have been passed for 5 days on triclosan free media. The fabI primers did not work on three isolates (65-13, BD-26 and BD-50). S. epidermidis ATCC 12228 is published on NCBI with its full sequence. It has a triclosan MIC <0.25 mg/l and its fabI gene is identical to all the current triclosan susceptible isolates we have sequenced. The putative promoter region of ATCC 12228 is different from the current triclosan susceptible isolates we have sequenced but those are all identical. ST, Sequence type determined by MLST. ND = not determined. Abbreviations for amino acids: H = histidine, Y = tyrosine, S = serine, A = alanine, G = glycine, F = phenylalanine, V = valine, Q = glutamine.
Three of the triclosan-adapted isolates, one old and two current, developed an identical mutation that predicted a substitution in amino acid residue 95 from alanine to valine (A95V). This change was not found in any of the current triclosan tolerant S. epidermidis isolates, but it has also been identified as the most frequent mutation in in vitro selected S. aureus mutants with triclosan tolerance [26], [27]. The two triclosan tolerant isolates (that were not possible to adapt further to triclosan in measure of MIC/MBC) did also not develop further mutations in fabI following the adaptation experiments with the BD-12 descendent, BD-12a*, keeping the fabI sequence identical to ATCC12228, and BD-24a* just keeping the mutations from its parent BD-24 (Table 4).
The mRNA expression of fabI of the six parent and six descendant isolates from the adaptation experiment was measured (Figure 2). It was thereby visualized that all the triclosan-exposed descendants had a variable increase in fabI expression compared to their own parental isolate including the already tolerant isolates that did not change their MIC/MBC further. When comparing the different isolates, it is seen that the parent isolate BD-12 that had a high MIC/MBC (4 mg/l/8 mg/l) and no mutations in fabI or the putative promoter region also had a relatively low mRNA expression of fabI.
Figure 2. Northern blot analysis of fabI transcripts in parental strains and their triclosan adapted descendant.
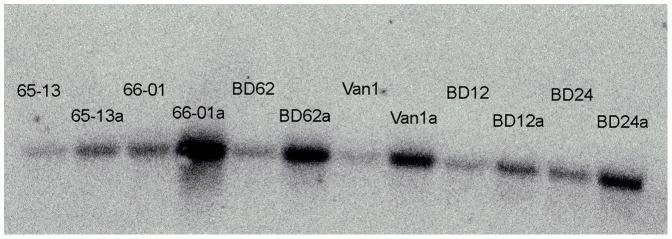
Parental strains from 1965-66 (65-13 and 66-01) and from 2010-11 (BD-62, Van-1, BD-12 and BD-24) and the descendant strains isolated after triclosan adaptation. Adapted strains are named with the parent name and the suffix a.
Discussion
This study indicates that wild-type S. epidermidis has changed their population structure to adapt to the widespread use of triclosan. We now have a triclosan tolerant subpopulation of S. epidermidis, so far identified in Denmark and USA [7], [16]. While no Danish isolates from 1965-66 where tolerant, 12.5% of the current isolates studied have decreased triclosan susceptibility with MIC values that were up to 32 fold higher, than the highest value found in the old isolates. We suggest that this change is caused by the general large-scale use of triclosan in society and to a lesser degree in the healthcare system.
We could reproduce this evolution through laboratory experiments exposing triclosan susceptible isolates to increasing concentrations of triclosan. Old, current, antibiotic susceptible, multiresistant and isolates with different ST types could be mutated to increased triclosan MICs. Interestingly it was not possible to adapt S. epidermidis isolates to a higher MIC than 4 mg/l and a MBC of 8 mg/l that correlated with the maximum values found in the clinical isolates. A number of studies have examined triclosan tolerance and its mechanisms in S. aureus [7], [16], [17], [24]–[26], [28]–[32]. Only two other studies have reported the distribution of triclosan MICs for S. epidermidis [7], [16]. In the first study, households were randomized to using or not using liquid soaps containing 0.2% triclosan (2000 mg/l). After one year triclosan MIC values remained in the range ≤0.0312–4 mg/l with no decrease in triclosan susceptibility. The other study investigated clinical S. epidermidis collected between 2001 and 2002 from all over USA [16]. They found a triclosan MIC range of ≤0.03–8 mg/l. This is similar to what we have found in the current S. epidermidis isolates, and we believe that the prior use of triclosan-containing products may have influenced the overall susceptibility which also may explain why intervention studies have not shown increased MIC levels [7]–[10].
Schmid and Kaplan found that clinical S. epidermidis isolates have a higher prevalence of reduced triclosan susceptibility (22%>0.5 mg/l) than clinical S. aureus isolates have (5%>0.5 mg/l) and that this was associated with the methicillin resistant (32%) versus the methicillin susceptible phenotype (12%) [16]. We found a non-significant higher prevalence of triclosan tolerant, methicillin resistant S. epidermidis isolates (15%) compared to methicillin susceptible S. epidermidis isolates (8.3%) (Table 2). Schmidt et al speculated whether the higher incidence of triclosan tolerance in S. epidermidis compared to S. aureus could be caused by the more frequent contact of S. epidermidis with triclosan -containing products or due to an alternative mechanism in S. epidermidis than in S. aureus.
Different mechanisms of decreased susceptibility towards triclosan have been described for different bacterial species. Moderation of outer membrane permeability barriers and upregulation and adaption of pre-existing effux pumps, is seen in Gram-negative rods for example P. aeruginosa and E. coli [33]–[35]. In Gram-negative rods also increased expression of the target, the FabI enoyl-ACP reductase, is seen in e.g. E. coli and Salmonella and mutations in the fabI gene, in E.coli, Salmonella, P. aeruginosa and Mycobacterium have been described [33]. Some bacterial species harbor an alternative to the FabI enoyl-ACP reductase, e.g. FabK, FabV and FabL, that are not or less affected by triclosan. These bacteria are intrinsically less susceptible to triclosan [33], [36]–[38]. Streptococcus pneumoniae, Enterococcus faecalis, Vibrio cholerae, Bacillus subtilis, E. coli and P. aeruginosa are examples [36], [37], [39]. Production of triclosan degradative enzymes, has been identified in soil isolates of the bacteria Pseudomonas putida and Alcaligenes xylosoxidans with intrinsicly reduced susceptibility towards triclosan [40].
In staphylococci, efflux pumps have not been identified as the cause for decreased susceptible to triclosan and only increased expression of the target FabI, and mutations in the fabI gene, has been identified. It has been shown that when wild-type fabI is introduced to S. aureus on a multicopy plasmid the MIC is raised 30 fold from 0.06 mg/l to 2 mg/l [30]. In clinical S. aureus isolates, it has been found that increased MIC levels towards triclosan is correlated with both increased expression of fabI as well as a single-point mutations in fabI [24].
By DNA sequencing of S. epidermidis fabI and the putative promoter region we found that mutations in fabI are partially involved in the development of triclosan tolerance in this organism, as has been found for S. aureus. However, still undefined mechanisms seem to be involved as maximum levels of triclosan tolerance was seen in isolates without mutations in or over expression of fabI. Ciusa, Furi and Knight et al [26] recently showed that an additional fabI allele most likely achieved by horizontal transfer from S. haemolyticus could explain reduced triclosan susceptibility of clinical S. aureus isolates with a wild-type fabI allele. The homology of the nucleotide sequence between the fabI alleles of Staphylococci is in the order of 82–84% (published sequences at NCBI) and we expect that our hybridization probe used in the mRNA expression assay would bind equally well to the fabI alleles of S. epidermidis and eg S. haemolyticus. Therefore yet other mechanisms seem to be involved.
One isolate had a predicted change at amino acid residue 204. An alteration of fabI in this position is well known from clinical as well as in vitro triclosan selected mutants of S. aureus with triclosan tolerance [24], [26]. The mutation has been shown, in S. aureus fabI, to correlate with elevated triclosan MIC through the formation of a stable triclosan −NAD+−FabI complex not seen in the wild-type fabI [24]. The amino acid residue at position 95 has, just as that at position 204, been thought to lie in the cofactor binding region of S. aureus fabI [27]. It is interesting though that the most frequently in vitro selected mutation, A95V, that is also identified as the mutation with the greatest impact on inhibition of S. aureus fabI [27] has never, as far as we are aware, been detected in clinical staphylococci isolates. An explanation could be that this mutation has a relatively larger fitness cost than the other mutations.
As other in situ studies [7]–[10] we did not find a significant correlation between triclosan tolerant isolates and antibiotic resistance in staphylococci. Only the previous discussed study [16] has found such an association, where triclosan tolerance was associated to methicillin resistance in S. epidermidis. Importantly we did not see increased antibiotic resistance in our laboratory adapted isolates either. It seems at least that in S. epidermidis there is no cross-resistance to antibiotics but the possibility of co-selection in vivo with for example methicillin and maybe other, not yet identified traits is still not fully elucidated. We believe, based on others and our findings, that the targeted use of triclosan is safe in regard to development of antibiotic cross-resistance and in working concentrations should be effective against S. epidermidis. A claim has been made for using triclosan in medical devices such as sutures with the aim to lower rates of nosocomial infections. Recently a multicenter, prospective, double-blinded, parallel group study was conducted in Finland [41] comparing triclosan-coated suture material with noncoated sutures in the control group. Among the 276 patients undergoing lower limb revascularization surgery, similar surgical wound infection rates of 21.9 to 22.3% were found.
The widespread (and often unregulated) use of triclosan in textiles, chopping boards and in antibacterial soaps in the domestic setting might need to be reconsidered. Aiello et al. [42] reviewed the literature on commonly used soaps containing triclosan in the community setting. In working concentrations 0.1%–0.45% wt/vol (1000–4500 mg/l) triclosan containing soaps were no more effective than plain soap in preventing infectious illness symptoms and reducing bacterial levels on the hands. Finally, taking the environmental concerns about aquatic organisms and toxic by-products into consideration together with the still not fully understood consequences of an increased tolerance in some bacterial species, we think the use of triclosan should be more regulated and restricted for purposes that have been proven beneficial.
Supporting Information
Multilocus sequence typing (MLST) was performed for 15 S. epidermidis isolated from blood in 2010-11, 7 triclosan susceptible (MIC<0.25 mg/l) and 8 triclosan tolerant (MIC≥0.25).
(DOC)
Antibiotic susceptibility, of the 64 S. epidiermidis isolated from blood in 2010-11, given for the triclosan tolerant isolates (MIC≥0.25 mg/l, n = 8) and for the triclosan susceptible (n = 56).
(DOC)
Acknowledgments
We thank Susanne Mie Rohde for technical help and Kit Boye and Wilhelm Paulander for academic advice and laboratory support.
Funding Statement
Funded by the Danish Council for Strategic Research, 09-0030. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Russell AD (2002) Introduction of biocides into clinical practice and the impact on antibiotic-resistant bacteria. J Appl Microbiol 92 Suppl: 121S–135S [PubMed] [Google Scholar]
- 2. Escalada MG, Russell AD, Maillard JY, Ochs D (2005) Triclosan-bacteria interactions: single or multiple target sites? Lett Appl Microbiol 41: 476–481. [DOI] [PubMed] [Google Scholar]
- 3. Jones RD, Jampani HB, Newman JL, Lee AS (2000) Triclosan: a review of effectiveness and safety in health care settings. Am J Infect Control 28: 184–196. [PubMed] [Google Scholar]
- 4.SCCS (Scientific committee on Consumer Safety) (2010) Opinion on triclosan (antimicrobial resistance). Available: http://ec.europa.eu/health/scientific_committees/consumer_safety/docs/sccs_o_023.pdf. Accessed 27 December 2012
- 5. Dann AB, Hontela A (2011) Triclosan: environmental exposure, toxicity and mechanisms of action. J Appl Toxicol 31: 285–311. [DOI] [PubMed] [Google Scholar]
- 6.Tamura I, Kagota KI, Yasuda Y, Yoneda S, Morita J, et al.. (2012) Ecotoxicity and screening level ecotoxicological risk assessment of five antimicrobial agents: triclosan, triclocarban, resorcinol, phenoxyethanol and p-thymol. J Appl Toxicol. DOI: 10.1002/jat.2771. [DOI] [PubMed]
- 7. Aiello AE, Marshall B, Levy SB, la-Latta P, Larson E (2004) Relationship between triclosan and susceptibilities of bacteria isolated from hands in the community. Antimicrob Agents Chemother 48: 2973–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cole EC, Addison RM, Rubino JR, Leese KE, Dulaney PD, et al. (2003) Investigation of antibiotic and antibacterial agent cross-resistance in target bacteria from homes of antibacterial product users and nonusers. J Appl Microbiol 95: 664–676. [DOI] [PubMed] [Google Scholar]
- 9. Sullivan A, Wretlind B, Nord CE (2003) Will triclosan in toothpaste select for resistant oral streptococci? Clin Microbiol Infect 9: 306–309. [DOI] [PubMed] [Google Scholar]
- 10. Jones CL, Ritchie JA, Marsh PD, Van der OF (1988) The effect of long-term use of a dentifrice containing zinc citrate and a non-ionic agent on the oral flora. J Dent Res 67: 46–50. [DOI] [PubMed] [Google Scholar]
- 11. Tkachenko O, Shepard J, Aris VM, Joy A, Bello A, et al. (2007) A triclosan-ciprofloxacin cross-resistant mutant strain of Staphylococcus aureus displays an alteration in the expression of several cell membrane structural and functional genes. Res Microbiol 158: 651–658. [DOI] [PubMed] [Google Scholar]
- 12. Sanchez P, Moreno E, Martinez JL (2005) The biocide triclosan selects Stenotrophomonas maltophilia mutants that overproduce the SmeDEF multidrug efflux pump. Antimicrob Agents Chemother 49(2): 781–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Braoudaki M, Hilton AC (2004) Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross-resistance to antimicrobial agents. J Clin Microbiol 42: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chuanchuen R, Beinlich K, Hoang TT, Becher A, Karkhoff-Schweizer RR, et al. (2001) Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob Agents Chemother 45: 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rogers KL, Fey PD, Rupp ME (2009) Coagulase-negative staphylococcal infections. Infect Dis Clin North Am 23: 73–98. [DOI] [PubMed] [Google Scholar]
- 16. Schmid MB, Kaplan N (2004) Reduced triclosan susceptibility in methicillin-resistant Staphylococcus epidermidis . Antimicrob Agents Chemother 48: 1397–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heath RJ, Li J, Roland GE, Rock CO (2000) Inhibition of the Staphylococcus aureus NADPH-dependent enoyl-acyl carrier protein reductase by triclosan and hexachlorophene. J Biol Chem 275: 4654–4659. [DOI] [PubMed] [Google Scholar]
- 18. Andrews JM (2001) Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48 Suppl 15–16. [DOI] [PubMed] [Google Scholar]
- 19.European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Available: http://www.eucast.org/clinical_breakpoints/. Accessed 27 December 2012.
- 20.European Committee on Antimicrobial Susceptibility Testing. Antimicrobial susceptibility testing EUCAST disk diffusion method. Available: http://www.eucast.org/antimicrobial_susceptibility_testing/disk_diffusion_methodology/. Accessed 27 December 2012.
- 21. Bryson V, Szybalski W (1952) Microbial Selection. Science 116: 45–51. [PubMed] [Google Scholar]
- 22.Multilocus sequence typing. Available: http://sepidermidis.mlst.net/. Accessed 27 December 2012.
- 23. Frees D, Chastanet A, Qazi S, Sorensen K, Hill P, et al. (2004) Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus . Mol Microbiol 54: 1445–1462. [DOI] [PubMed] [Google Scholar]
- 24. Fan F, Yan K, Wallis NG, Reed S, Moore TD, et al. (2002) Defining and combating the mechanisms of triclosan resistance in clinical isolates of Staphylococcus aureus . Antimicrob Agents Chemother 46: 3343–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brenwald NP, Fraise AP (2003) Triclosan resistance in methicillin-resistant Staphylococcus aureus (MRSA). J Hosp Infect 55: 141–144. [DOI] [PubMed] [Google Scholar]
- 26. Ciusa ML, Furi L, Knight D, Decorosi F, Fondi M, et al. (2012) A novel resistance mechanism to triclosan that suggests horizontal gene transfer and demonstrates a potential selective pressure for reduced biocide susceptibility in clinical strains of Staphylococcus aureus . Int J Antimicrob Agents 40: 210–220. [DOI] [PubMed] [Google Scholar]
- 27. Xu H, Sullivan TJ, Sekiguchi J, Kirikae T, Ojima I, et al. (2008) Mechanism and inhibition of saFabI, the enoyl reductase from Staphylococcus aureus . Biochemistry 47: 4228–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bayston R, Ashraf W, Smith T (2007) Triclosan resistance in methicillin-resistant Staphylococcus aureus expressed as small colony variants: a novel mode of evasion of susceptibility to antiseptics. J Antimicrob Chemother 59: 848–853. [DOI] [PubMed] [Google Scholar]
- 29. Seaman PF, Ochs D, Day MJ (2007) Small-colony variants: a novel mechanism for triclosan resistance in methicillin-resistant Staphylococcus aureus . J Antimicrob Chemother 59: 43–50. [DOI] [PubMed] [Google Scholar]
- 30. Slater-Radosti C, Van AG, Greenwood R, Nicholas R, Keller PM, et al. (2001) Biochemical and genetic characterization of the action of triclosan on Staphylococcus aureus . J Antimicrob Chemother 48: 1–6. [DOI] [PubMed] [Google Scholar]
- 31. Suller MT, Russell AD (2000) Triclosan and antibiotic resistance in Staphylococcus aureus . J Antimicrob Chemother 46: 11–18. [DOI] [PubMed] [Google Scholar]
- 32. Wootton M, Walsh TR, Davies EM, Howe RA (2009) Evaluation of the effectiveness of common hospital hand disinfectants against methicillin-resistant Staphylococcus aureus, glycopeptide-intermediate S. aureus, and heterogeneous glycopeptide-intermediate S. aureus . Infect Control Hosp Epidemiol 30: 226–232. [DOI] [PubMed] [Google Scholar]
- 33. Saleh S, Haddadin RN, Baillie S, Collier PJ (2011) Triclosan-an update. Lett Appl Microbiol 52: 87–95. [DOI] [PubMed] [Google Scholar]
- 34. Russell AD (2004) Whither triclosan? J Antimicrob Chemother 53: 693–695. [DOI] [PubMed] [Google Scholar]
- 35. McMurry LM, Oethinger M, Levy SB (1998) Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli . FEMS Microbiol Lett 166: 305–309. [DOI] [PubMed] [Google Scholar]
- 36. Massengo-Tiasse RP, Cronan JE (2008) Vibrio cholerae FabV defines a new class of enoyl-acyl carrier protein reductase. J Biol Chem 283: 1308–1316. [DOI] [PubMed] [Google Scholar]
- 37. Massengo-Tiasse RP, Cronan JE (2009) Diversity in enoyl-acyl carrier protein reductases. Cell Mol Life Sci 66: 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heath RJ, Rock CO (2000) A triclosan-resistant bacterial enzyme. Nature 406: 145–146. [DOI] [PubMed] [Google Scholar]
- 39. Yazdankhah SP, Scheie AA, Hoiby EA, Lunestad BT, Heir E, et al. (2006) Triclosan and antimicrobial resistance in bacteria: an overview. Microb Drug Resist 12: 83–90. [DOI] [PubMed] [Google Scholar]
- 40. Meade MJ, Waddell RL, Callahan TM (2001) Soil bacteria Pseudomonas putida and Alcaligenes xylosoxidans subsp. denitrificans inactivate triclosan in liquid and solid substrates. FEMS Microbiol Lett 204: 45–48. [DOI] [PubMed] [Google Scholar]
- 41. Turtiainen J, Saimanen EI, Makinen KT, Nykanen AI, Venermo MA, et al. (2012) Effect of triclosan-coated sutures on the incidence of surgical wound infection after lower limb revascularization surgery: a randomized controlled trial. World J Surg 36: 2528–2534. [DOI] [PubMed] [Google Scholar]
- 42. Aiello AE, Larson EL, Levy SB (2007) Consumer antibacterial soaps: effective or just risky? Clin Infect Dis 45 Suppl 2S137–S147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multilocus sequence typing (MLST) was performed for 15 S. epidermidis isolated from blood in 2010-11, 7 triclosan susceptible (MIC<0.25 mg/l) and 8 triclosan tolerant (MIC≥0.25).
(DOC)
Antibiotic susceptibility, of the 64 S. epidiermidis isolated from blood in 2010-11, given for the triclosan tolerant isolates (MIC≥0.25 mg/l, n = 8) and for the triclosan susceptible (n = 56).
(DOC)