Abstract
We provide an NMRPipe macro to meet an increasing need in membrane biophysics for facile de-Pake-ing of axially symmetric deuterium, and to an extent phosphorous, static lineshapes. The macro implements the development of McCabe & Wassall (1997), and is run as a simple replacement for the usual Fourier transform step in an NMRPipe processing procedure.
Keywords: dePake, NMRPipe, Membrane perturbation, Lipid biophysics, Deuterium NMR
There has been a resurgence of interest in solid-state 2H and 31P NMR, particularly in the burgeoning area of antimicrobial peptides (Pinheiro & Watts, 1994; Tremouilhac et al., 2006; Ouellet et al., 2007; Wi & Kim, 2008; Gehman et al., 2008a; Pabst et al., 2008; Fernandez, Gehman & Separovic, 2009; Cheng et al., 2009), but also in many other research programs for which membrane-protein/peptide interactions are integral (e.g. Dufourc, Bonmatin & Dufourcq, 1989; Gehman et al., 2008b; van den Brink-van der Laan, Killian & de Kruijff, 2004; Vogel et al., 2007). Unoriented lipid vesicles are typically the most convenient sample systems used in these studies, for which broad static lineshapes are analyzed to assess perturbation by peptide. Static 2H NMR of enriched fluid phase acyl chain and/or head group CHn sites produce the canonical Pake pattern (Pake, 1948), where reduction or increases in the quadrupole splitting corresponds to an increase or decrease in fluctuation in the frequency regime of 105 Hz (Seelig & Seelig, 1974). Similarly, reduction or increases in the width of the 31P chemical shift anisotropy corresponds to an increase or decrease, respectively, in orientational order in the 103 Hz regime and/or changes in the average orientation of the phospholipid headgroup (Kohler & Klein, 1977; Gehman et al., 2008b).
De-Pake-ing is one method which aids in the analysis of these static spectra, for fluid phase membranes. The procedure is a numerical transform which converts the unoriented axially symmetric static spectrum into a 0°-oriented spectrum. The de-Paked spectrum mimics that which would have been obtained if the lipids had been uniformly oriented in an aligned bilayer with the surface perpendicular to the static magnetic field (i.e. with the membrane normal oriented parallel to the static magnetic field), rather than in spherical (or spheroidal Schäfer, Mädler & Sternin, 1998) lipid vesicles. De-Pake-ing was introduced first as a computationally intensive iterative procedure (Bloom, Davis & Mackay, 1981; Sternin, Bloom & Mackay, 1983), then treated as an “ill-posed problem” using inverse theory (Whittall et al., 1989), and regularization (Schäfer, Mädler & Volke, 1995) and finally using Fourier transform (FT) of modified signal (McCabe & Wassall, 1997).
In our experience, de-Pake-ing is something of a black art within individual laboratories, including our own, owing in part to the difficulty in propagating the expertise required to control the nuanced behaviors of the procedure with freely available documentation alone. To address this problem, we offer here a simple NMRPipe (Delaglio et al., 1995) macro that can be substituted for the Fourier transform step in an otherwise identical NMRPipe script, which implements the method of de-Pake-ing by McCabe & Wassall (1997). The procedure is therefore easy to implement, and should share the same future stability as NMRPipe itself.
The theory behind “de-Paking” of axially symmetric powder patterns using weighted FT culminates in the expression (McCabe & Wassall, 1997)
| (1) |
This relates the intensity of the frequency in the oriented spectrum (F0) to the intensity of the Fourier transform of a weighted time domain signal at half the frequency on the opposite side of the spectrum. The apodization is simply multiplication by the square root of time t. This windowing function, unfortunately, decreases signal-to-noise in the de-Paked spectrum relative to the unoriented spectrum. The ± refers to positive and negative frequencies, i.e. the left and right halves of the spectrum, respectively. The (1 ± i) means the left and right halves of the spectrum come out 90° out of phase with each other. Thus the time domain can be converted to a purely absorptive de-Paked F0 spectrum by resetting spectral width and referencing parameters, reversing the spectrum, exchanging the real and inverse-imaginary channels in the right half of the spectrum, and zero-order phase correcting by −45°. The new NMRPipe macro listed and described in Fig. 1 accomplishes these steps in a straightforward fashion, and should be invoked where one would normally invoke the Fourier transform, for example:
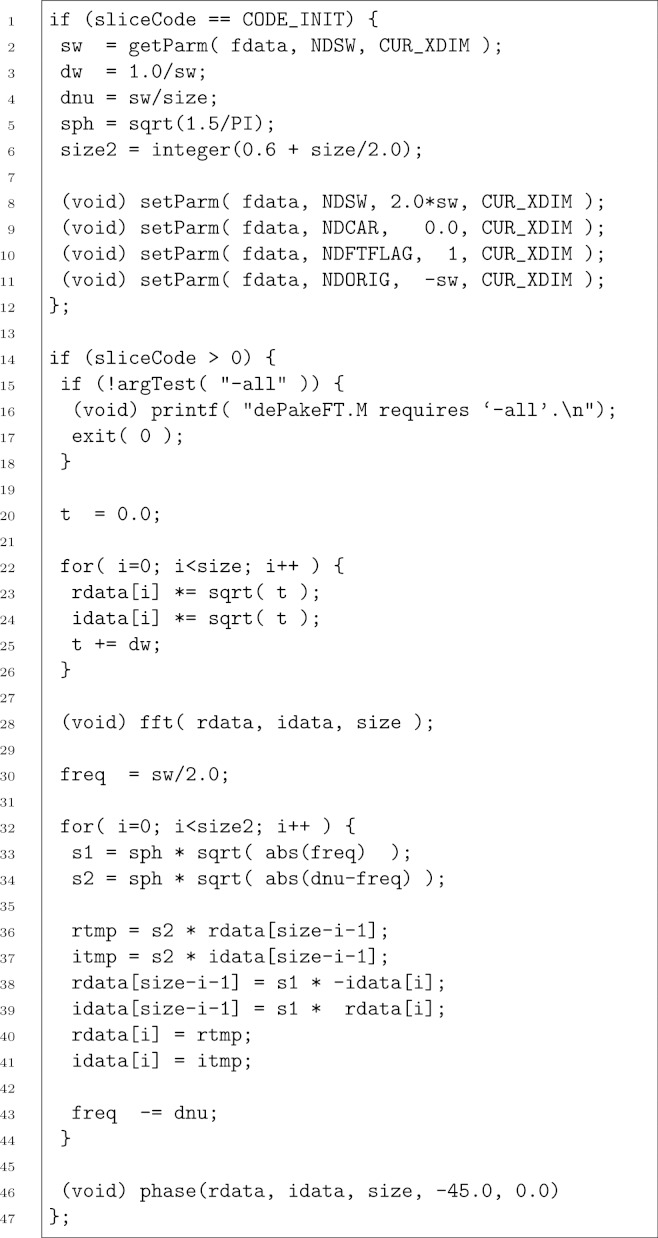
Figure 1.
The dePakeFT macro. The first block (lines 1–12) reads the acquisition spectral width from the data header, sets a collection of constants, and resets necessary header parameters. The loop in lines 22–26 performs the apodization, followed by the Fourier transformation in line 28. Lines 32–44 apply the frequency-dependent intensity scaling and differential phase corrections, followed by the uniform phase correction (line 46).
| nmrPipe -in test.fid | ∖ |
| | nmrPipe -fn LS -ls 6 -sw | ∖ |
| | nmrPipe -fn GM -g2 200.0 -c 0.5 | ∖ |
| | nmrPipe -fn MAC -macro $NMRTXT/dePakeFT.M -all | ∖ |
| | nmrPipe -fn PS -p0 259.0 -p1 0.0 -di | ∖ |
| | nmrPipe -ov -out depake.ft |
where LS compensates for having begun acquisition prior to the top of the solid echo, and effectively discards the early points which suffer from probe coil and (especially analog) audio filter ringing; GM is a usual apodization function; and PS applies the same frequency-independent (-p0) phase shift as required by the regular, unoriented FT spectrum. Note that the LS step requires an integral number of left-shifts, and it is too difficult to reliably adjust the frequency-dependent (-p1) phase correction in the PS step. Both of these constraints require careful optimization of the preacquisition delay in the solid echo pulse sequence (Davis, 1983).
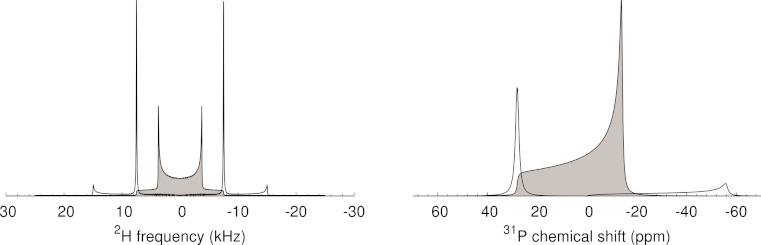
Figure 2 shows the Pake pattern of a regular Fourier transform, and a de-Paked spectrum using the macro in Fig. 1, for data simulated using Simpson (Bak, Rasmussen & Nielsen, 2000). The NMRPipe macro works as expected: oriented spectral intensity appears in 2H spectra at positions corresponding to the 0° frequencies for the simulated lineshape, consistent with a splitting of 3/2 × the 10 kHz quadrupole coupling constant used for simulation, and in 31P spectra at the δ = 28 ppm chemical shift anisotropy value used for simulation (equivalent to the 0° edge, and corresponding to more typically quoted Δδ = −42 ppm).
Figure 2.
De-Pake-ing of data representative of a single CDn site from phospholipid bilayers, and simulated with Simpson (Bak, Rasmussen & Nielsen, 2000). Regular FT static lineshapes are shown in gray, and de-Paked spectra are shown as black curves. Isotropic chemical shifts were set to zero, and the zcw4180 crystal file was used. (Left) A quadrupole splitting of 10 kHz was used in a solid echo pulse sequence simulation for 2H, and (Right) a chemical shift anisotropy (δ under the Haeberlen convention) of 28 ppm (Δδ = −42 ppm) was used in a simple Bloch decay simulation for 31P.
An artifact appears with weak intensity on the opposite side of the peak intensity in the de-Paked spectrum. This has been noted previously (McCabe & Wassall, 1997), and is a consequence of using an approximation of the asymptotic value of the underlying integral. The infinite signal-to-noise of 31P simulations in Figs. 2 and 3 indicate that the artifacts are attenuated approximations of the full static lineshape of each component, shifted and scaled along the frequency axis such that it spans from the center of the spectrum to ∼ 2 × δ, with opposite sign to the peak de-Paked intensity.
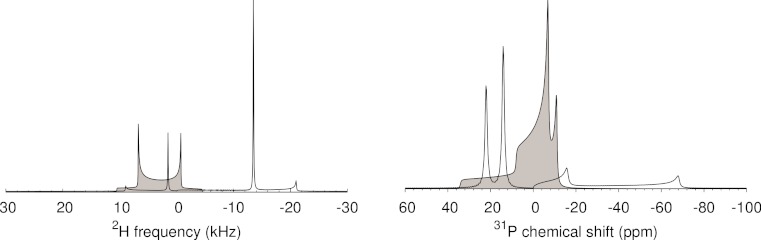
Figure 3.
De-Pake-ing of data simulated with Simpson, using non-zero isotropic offsets. Simulations and display parameters are used as in Fig. 2, except (Left) An isotropic shift offset of 3000 Hz was used for 2H, and (Right) two 31P components are included, as may be seen under some circumstances, e.g. for a mixed phospholipid bilayer, one with a chemical shift anisotropy δ = 30 ppm (Δδ = −45 ppm) and isotropic frequency offset δ0 = 4 ppm, and another with δ = 10 ppm (Δδ = −15 ppm) and isotropic shift offset of δ0 = −2 ppm.
One of the tedious aspects of most of the de-Pake-ing methods is the need to center the first moment of the static spectrum within the spectral window, at a point with frequency of exactly zero. This requirement is less stringent for the FT method. If the carrier frequency is not centered at the isotropic chemical shift, the negation and doubling of the frequency axis involved in the de-Paking means that the oriented intensity for each side will appear at −2 × the offset compared to where it would have appeared if the carrier frequency had been centered in the Pake pattern. For example, in Fig. 2, the de-Paked (oriented) 2H intensity appears at ± 7.5 kHz, but the carrier offset of +3 kHz in Fig. 3 causes the de-Paked peaks to shift −6 kHz to 1.5 and −13.5 kHz. The scaling of intensities proportional to the square root of distance from the center of the spectrum (Fig. 1 lines 36–45) also causes an imbalance between the two theoretically symmetric halves of the doublet. While the frequency axis can be adjusted to center the lineshape, this is unnecessary for small shifts as the quadrupole splitting is the same with or without the offset. This is of particular benefit, as the lipid acyl CD2 and CD3 isotropic chemical shifts are slightly different. Larger offsets are, of course, a more serious concern, from an experimental set-up perspective, due to finite excitation profiles that would likely impact upon ideally uniform excitation of the very broad line.
The same rule applies for the position of 31P oriented intensity when the carrier is not placed exactly on the isotropic frequency of a given phospholipid species. This is beneficial, insofar as spectra with multiple components that may differ in isotropic chemical shift can still be de-Paked. However, interpretation in this case will be more difficult, particularly for the more complicated mixtures being used to better approximate natural bilayer environments (Pinheiro & Watts, 1994; Sani, Dufourc & Gröbner, 2009). For the sake of illustration, two clearly distinct 31P species are shown in Fig. 3, as seen in some cases (Pukala et al., 2007). Where isotropic shift offsets are different for each species, no one frequency axis shift will satisfy all species. Consequently, some form of deconvolution is necessary to interpret the relationship between the positions of oriented intensities and the chemical shift parameters of each component line. While this may be possible, in practice, de-Paked 31P spectra may not always give sufficient resolution (e.g. Fig. 4). We find the maximum entropy-based analysis of slow-spinning MAS spectra (Sani, Separovic & Gehman, 2011) to be a more general solution to this problem.
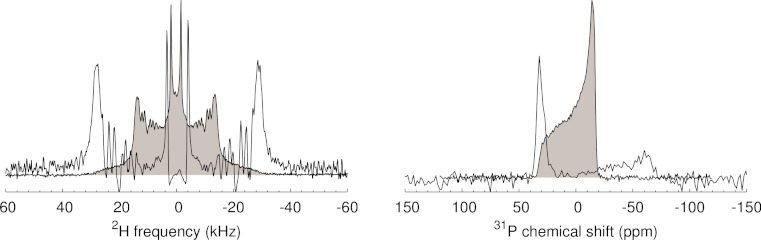
Figure 4.
FT and de-Paked spectra of d54-dimyristoylphosphatidylcholine (dDMPC) multilamellar vesicles at 30 °C for: (Left) a 2H solid-echo pulse sequence, and (Right) Hahn-echo pulse sequence using EXORCYCLE phase cycling (Rance & Byrd, 1983).
Processing of real data (Fig. 4) indicates that the NMRPipe macro works well, and is as comparable to the Single Value Decomposition (SVD) in our experience as initially reported (McCabe & Wassall, 1997). In contrast to the SVD approach, as well as a nonlinear-least squares approach (Whittall et al., 1989), which often took an hour or two of processing and iterative optimization of parameters, processing with this macro is essentially instantaneous. For existing NMRPipe installations, the macro uses numeric parameter codes, and can be downloaded from the Gehman webpage at http://www.chemistry.unimelb.edu.au, or as a Supplementary File to this note, and placed in $NMRTXT. NMRPipe distributions of version 6.1 or greater use parameter names as in Fig. 1, and include the dePakeFT.M macro by default.
Supplemental Information
This macro applies to many current versions of NMRPipe in circulation. It should be edited to output a reference to this article, if it is accepted.
Funding Statement
JD Gehman is funded under an Australian Research Council Future Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
Frances Separovic is an Academic Editor for PeerJ. There are no other competing interests.
Author Contributions
Marc-Antoine Sani performed the experiments, analyzed the data, wrote the paper.
Daniel K. Weber performed the experiments, analyzed the data.
Frank Delaglio contributed reagents/materials/analysis tools, wrote the paper, expertise with NMRPipe internals.
Frances Separovic contributed reagents/materials/analysis tools, Manuscript advice.
John D. Gehman conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper.
References
- Bak, Rasmussen & Nielsen (2000).Bak M, Rasmussen JT, Nielsen NC. SIMPSON: a general simulation program for solid-state NMR spectroscopy. Journal of Magnetic Resonance. 2000;147:296–300. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
- Bloom, Davis & Mackay (1981).Bloom M, Davis JH, Mackay A. Direct determination of the oriented sample NMR spectrum from the powder spectrum for systems with local axial symmetry. Chemical Physics Letters. 1981;80:198–202. doi: 10.1016/0009-2614(81)80089-9. [DOI] [Google Scholar]
- Cheng et al. (2009).Cheng J, Hale J, Elliot M, Hancock R, Straus S. Effect of membrane composition on antimicrobial peptides aurein 2.2 and 2.3 from Australian southern bell frogs. Biophysical Journal. 2009;96:552–565. doi: 10.1016/j.bpj.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis (1983).Davis J. The description of membrane lipid conformation, order and dynamics by 2H-NMR. Biochimica et Biophysica Acta. 1983;737:117–171. doi: 10.1016/0304-4157(83)90015-1. [DOI] [PubMed] [Google Scholar]
- Delaglio et al. (1995).Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of Biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dufourc, Bonmatin & Dufourcq (1989).Dufourc E, Bonmatin J, Dufourcq J. Membrane structure and dynamics by 2H- and 31P-NMR. Effects of amphipatic peptidic toxins on phospholipid and biological membranes. Biochimie. 1989;71:117–23. doi: 10.1016/0300-9084(89)90141-7. [DOI] [PubMed] [Google Scholar]
- Fernandez, Gehman & Separovic (2009).Fernandez DI, Gehman JD, Separovic F. Membrane interactions of antimicrobial peptides from Australian frogs. Biochimica et Biophysica Acta. 2009;1788:1630–1638. doi: 10.1016/j.bbamem.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Gehman et al. (2008a).Gehman JD, Luc F, Hall K, Lee T-H, Boland MP, Pukala TL, Bowie JH, Aguilar M-I, Separovic F. Effect of antimicrobial peptides from Australian tree frogs on anionic phospholipid membranes. Biochemistry. 2008a;47:8557–8565. doi: 10.1021/bi800320v. [DOI] [PubMed] [Google Scholar]
- Gehman et al. (2008b).Gehman JD, O’Brien CC, Shabanpoor F, Wade JD, Separovic F. Metal effects on the membrane interactions of amyloid-β peptides. European Biophysics Journal. 2008b;37:333–344. doi: 10.1007/s00249-007-0251-2. [DOI] [PubMed] [Google Scholar]
- Kohler & Klein (1977).Kohler SJ, Klein MP. Orientation and dynamics of phospholipid head groups in bilayers and membranes determined from 31P nuclear magnetic resonance chemical shielding tensors. Biochemistry. 1977;16:519–526. doi: 10.1021/bi00622a028. [DOI] [PubMed] [Google Scholar]
- McCabe & Wassall (1997).McCabe MA, Wassall SR. Rapid deconvolution of NMR powder spectra by weighted fast Fourier tranformation. Solid State Nuclear Magnetic Resonance. 1997;10:63–61. doi: 10.1016/S0926-2040(97)00024-6. [DOI] [PubMed] [Google Scholar]
- Ouellet et al. (2007).Ouellet M, Doucet J-D, Voyer N, Auger M. Membrane topology of a 14-mer model amphipathic peptide: a solid-state NMR spectroscopy study. Biochemistry. 2007;46:6597–6606. doi: 10.1021/bi0620151. [DOI] [PubMed] [Google Scholar]
- Pabst et al. (2008).Pabst G, Grage SL, Danner-Pongratz S, Jing W, Ulrich AS, Watts A, Lohner K, Hickel A. Membrane thickening by the antimicrobial peptide PGLa. Biophysical Journal. 2008;95:5779–5788. doi: 10.1529/biophysj.108.141630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pake (1948).Pake GE. Nuclear resonance absorption in hydrated crystals: fine structure of the proton line. Journal of Chemical Physics. 1948;16:327–336. doi: 10.1063/1.1746878. [DOI] [Google Scholar]
- Pinheiro & Watts (1994).Pinheiro T, Watts A. Resolution of individual lipids in mixed phospholipid membranes and specific lipid-cytochrome c interactions by magic-angle spinning solid-state phosphorus-31 NMR. Biochemistry. 1994;33:2459–2467. doi: 10.1021/bi00175a014. [DOI] [PubMed] [Google Scholar]
- Pukala et al. (2007).Pukala TL, Boland MP, Gehman JD, Kuhn-Nentwig L, Separovic F, Bowie JH. Solution structure and interaction of Cupiennin 1a, a spider venom peptide with phospholipid bilayers. Biochemistry. 2007;46:3576–3585. doi: 10.1021/bi062306+. [DOI] [PubMed] [Google Scholar]
- Rance & Byrd (1983).Rance M, Byrd RA. Obtaining high-fidelity spin-1/2 powder spectra in anisotropic media: phase-cycled Hahn echo spectroscopy. Journal of Magnetic Resonance. 1983;52:221–240. [Google Scholar]
- Sani, Dufourc & Gröbner (2009).Sani M, Dufourc E, Gröbner G. How does the Bax-α1 targeting sequence interact with mitochondrial membranes? The role of cardiolipin. Biochimica et Biophysica Acta. 2009;1788:623–631. doi: 10.1016/j.bbamem.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Sani, Separovic & Gehman (2011).Sani M-A, Separovic F, Gehman JD. Disentanglement of heterogenous dynamics in mixed lipid systems. Biophysical Journal. 2011;100:L40–L42. doi: 10.1016/j.bpj.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer, Mädler & Sternin (1998).Schäfer H, Mädler B, Sternin E. Determination of orientational order parameters from 2H NMR spectra of magnetically partially oriented lipid bilayers. Biophyical Journal. 1998;74:1007–1014. doi: 10.1016/S0006-3495(98)74025-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer, Mädler & Volke (1995).Schäfer H, Mädler B, Volke F. De-Pake-ing of NMR powder spectra by nonnegative least-squares analysis with Tikhonov regularization. Journal of Magnetic Resonance A. 1995;116:145–149. doi: 10.1006/jmra.1995.0002. [DOI] [Google Scholar]
- Seelig & Seelig (1974).Seelig A, Seelig J. The dynamic structure of fatty acyl chains in a phospholipid bilayer measured by deuterium magnetic resonance. Biochemistry. 1974;13:4839–4845. doi: 10.1021/bi00720a024. [DOI] [PubMed] [Google Scholar]
- Sternin, Bloom & Mackay (1983).Sternin E, Bloom M, Mackay AL. De-Pake-ing of NMR-spectra. Journal of Magnetic Resonance. 1983;55:274–282. [Google Scholar]
- Tremouilhac et al. (2006).Tremouilhac P, Strandberg E, Wadhwani P, Ulrich A. Synergistic transmembrane alignment of the antimicrobial heterodimer pgla/magainin. Journal of Biological Chemistry. 2006;281:32089–94. doi: 10.1074/jbc.M604759200. [DOI] [PubMed] [Google Scholar]
- van den Brink-van der Laan, Killian & de Kruijff (2004).van den Brink-van der Laan E, Killian JA, de Kruijff B. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochimica et Biophysica Acta. 2004;1666:275–288. doi: 10.1016/j.bbamem.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Vogel et al. (2007).Vogel A, Tan K-T, Waldmann H, Feller SE, Brown MF. Flexibility of ras lipid modifications studied by 2H solid state NMR and molecular dynamics simulations. Biophysical Journal. 2007;93:2697–2712. doi: 10.1529/biophysj.107.104562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittall et al. (1989).Whittall KP, Sternin E, Bloom M, Mackay AL. Time- and frequency-domain “dePakeing” using inverse theory. Journal of Magnetic Resonance. 1989;84:64–71. [Google Scholar]
- Wi & Kim (2008).Wi S, Kim C. Pore structure, thinning effect, and lateral diffusive dynamics of oriented lipid membranes interacting with antimicrobial peptide protegrin-1: 31P and 2H solid-state NMR study. Journal of Physical Chemistry B. 2008;112:11402–11414. doi: 10.1021/jp801825k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This macro applies to many current versions of NMRPipe in circulation. It should be edited to output a reference to this article, if it is accepted.