Abstract
“It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.” (Watson and Crick, 1953)
In the years since this remarkable understatement, we have come to realize the enormous complexity of the cellular machinery devoted to replicating DNA with the accuracy needed to maintain genetic information over many generations, balanced by the emergence of mutations on which selection can act. This complexity is partly based on the need to remove or tolerate cytotoxic and mutagenic lesions in DNA generated by environmental stress. Considered here is the fidelity with which undamaged and damaged DNA is replicated by the many DNA polymerases now known to exist. Some of these seriously violate Watson-Crick base pairing rules such that, depending on the polymerase, the composition and location of the error and the ability to correct errors (or not), DNA synthesis error rates can vary by more than a million-fold. This offers the potential to modulate rates of point mutations over a wide range, with consequences that can be either deleterious or beneficial.
In organisms from viruses to man, the fidelity with which genetic information is replicated depends on the ability of polymerases to select correct rather than incorrect and/or damaged nucleotides for incorporation without adding or deleting nucleotides. Polymerase selectivity is the prime determinant of fidelity both at the replication fork and during synthesis to repair DNA damage generated by endogenous cellular metabolism or exposure to the environment (Friedberg et al., 2006). In many organisms, fidelity can be increased by exonucleolytic proofreading of mismatches during replication and by DNA mismatch repair (MMR, reviewed in (Hsieh and Yamane, 2008; Iyer et al., 2006; Kunkel and Erie, 2005). Certain proteins involved in MMR also can also signal for DNA damage responses, prevent homeologous recombination, promote meiotic recombination, modulate somatic hypermutation of immunoglobulin genes, or even stabilize certain misaligned repetitive DNA sequences. When DNA damage is not removed prior to replication, helix-distorting lesions can impede replication fork progression. In such circumstances, cell survival can be enhanced by specialized DNA transactions, some of which can be mutagenic via translesion DNA synthesis (Chang and Cimprich, 2009; Jansen et al., 2007; Yang and Woodgate, 2007). Considered here is the amazing diversity of evolutionarily conserved DNA polymerases involved in these transactions, many of which have been discovered relatively recently. Emphasis is on their fidelity, and on the contributions of proofreading and MMR to replication fidelity, which can vary over a much wider range than was appreciated even a decade ago.
Multiple polymerases with multiple, overlapping functions
DNA polymerases were first discovered using assays for polymerization activity (Bessman et al., 1956). This approach revealed that bacteria and eukaryotes harbor multiple polymerases (Kornberg and Baker, 1992). However, just how many only came to light more recently when sequence alignments and recombinant DNA technology were used to find low activity, low abundance polymerases. Sequence alignments now permit classification of DNA polymerases into several different families, with most organisms encoding more than one (Bebenek and Kunkel, 2004; Loeb and Monnat, 2008; Shcherbakova et al., 2003a). For example, E. coli encodes five polymerases (Friedberg et al., 2005), one each from families A, B and C and two from different subfamilies of family Y, each with important but somewhat different functions. The human genome encodes even more (Table 1) from families A (3 pols), B (4 pols), X (4 pols), Y (4 pols) and RT (telomerase). Because polymerases can have multiple functions (Table 1) and can sometimes compensate one for another, it is a continuing challenge to understand exactly where and when each polymerase operates in vivo.
Table 1.
Human DNA-Template-Dependent DNA Polymerases
| Polymerase | Family | Mass (kDa) | Gene (alias) | Associated activities | Proposed functions |
|---|---|---|---|---|---|
| α (alpha) | B | 165 | POLA | RNA primase | nuclear genome replication, S-phase checkpoint |
| β (beta) | X | 39 | POLB | dRP lyase AP lyase |
BER MMR |
| γ (gamma) | A | 140 | POLG | 3′→5′ exonuclease dRP lyase |
mitochondrial genome replication and BER |
| δ (delta) | B | 125 | POLD1 | 3′→5′ exonuclease | nuclear genome replication NER, BER, MMR, DSB repair |
| ε (epsilon) | B | 255 | POLE | 3′→5′ exonuclease | nuclear genome replication NER, BER, MMR, DSB repair S-phase checkpoint |
| ζ (zeta) | B | 353 | POLZ (REV3) | – | TLS, DSB repair, ICL repair, SHM |
| η (eta) | Y | 78 | POLH (RAD30, RAD30A, XPV) | – | TLS, SHM, BER? recombination repair |
| θ (theta) | A | 198 | POLQ | helicase motifs | ICL repair? TLS, SHM? |
| ι (iota) | Y | 80 | POLI (RAD30B) | dRP lyase | TLS, BER SHM? |
| κ (kappa) | Y | 76 | POLK (DINB1) | – | TLS NER |
| λ (lambda) | X | 66 | POLL | dRP lyase | V(D)J recombination NHEJ, BER |
| μ(mu) | X | 55 | POLM | terminal transferase | V(D)J recombination NHEJ |
| ν (nu) | A | 100 | POLN | – | TLS? |
| σ (sigma) | X | 60 | POLS (TRF4-1) | 3′→5′ exonuclease | sister chromatid cohesion |
| TdT | X | 58 | TdT | – | V(D)J recombination |
| REV1 | Y | 138 | REV1 | dCTP incorporation | TLS |
BER, Base Excision Repair, NER, Nucleotide Excision Repair, MMR, Mismatch Repair, DSB repair, double strand break repair, TLS, Translesion Synthesis, SHM, somatic hypermutation, ICL repair, Interstrand Crosslink repair, NHEJ, Non-Homologous End Joining of double strand breaks.
Despite differences in primary sequence, DNA polymerases in different families (four examples in Fig. 1A) share a common general structure for the polymerase domain (Ollis et al., 1985), which is comprised of fingers, thumb and palm subdomains (colored blue, green and red, respectively). The palm contains three highly conserved carboxylates that bind two divalent metal ions required for catalysis via an in-line nucleophilic attack of the 3′-OH on the α-phosphate of the incoming dNTP. This mechanism is thought to be common to all DNA polymerases (Steitz, 1993), yet it appears to have resulted from convergent evolution, because some polymerase families have the active site carboxylates in a “right-handed” configuration while others (families X and C) have in a “left-handed” configuration (e.g., see (Wing et al., 2008) and references therein).
Figure 1. X ray crystal structures of DNA polymerases.

A. Shown is the structure of a representative replicative DNA polymerase from bacteriophage RB69 (family B). Polymerase domains share three common sub-domains, designated fingers (blue), palm (red) and thumb (green). Other domains for specialized functions are shown in purple and yellow. (B) The active site of human DNA polymerase β. The surface of Arg283 is highlighted in pink to emphasize the importance to fidelity of polymerase interactions with the DNA minor groove. (C) The more open and solvent-accessible active site of low-fidelity Sulfolobus sulfataricus Dpo4. See text for further descriptions. Panel (A) was prepared by Miguel Garcia-Diaz, using the structure in (Franklin et al., 2001). Panel (B) and (C) are reproduced from (Kunkel et al., 2003), with permission.
The polymerase domains are usually attached to other domains needed for the variable functions of these proteins. For example, polymerases that perform the bulk of genome replication often have a domain harboring 3′ exonuclease activity that proofreads replication errors (Fig. 1A). Nonetheless, most DNA polymerases lack an intrinsic 3′ exonuclease activity (Table 1), which is interesting given the importance of proofreading to genome stability (see below). Other specialized domains include a “little finger” domain (Yang and Woodgate, 2007) unique to family Y members involved in translesion DNA synthesis, and an 8 kDa domain unique to family X pols (Moon et al., 2007) that assists in filling small gaps during DNA repair and that, in pols β and λ, harbors a dRP lyase activity needed for repair (Table 1). Still other domains include the BRCT domains of family X pols involved in non-homologous end joining of double strand DNA breaks and amino- or carboxyl-terminal regions of polymerase catalytic subunits that are involved in cellular responses to DNA damage, including via partnerships with other proteins. In fact, DNA polymerases typically operate in DNA transactions that require coordinated interactions with many other proteins (e.g., non-catalytic accessory subunits, processivity clamps, single stranded DNA binding proteins), whose properties and functions are subjects of continuing interest (e.g., see (Bebenek and Kunkel, 2004; Burgers, 2009; Chang and Cimprich, 2009; Friedberg et al., 2005; Jansen et al., 2007; Loeb and Monnat, 2008; Shcherbakova et al., 2003a) and references therein.)
The fidelity of DNA synthesis
Measurements of the fidelity of DNA synthesis in vitro by purified DNA polymerases reveal a remarkable variation in error rates for the two major types of errors that polymerases generate, single base pair substitutions and single base deletions (Fig. 2). These error rates reflect the contribution of nucleotide selectivity at the polymerase active site and proofreading by those polymerases harboring an associated 3′ exonuclease.
Figure 2. Polymerase error rates and the contributions of each fidelity process to mutation rate.
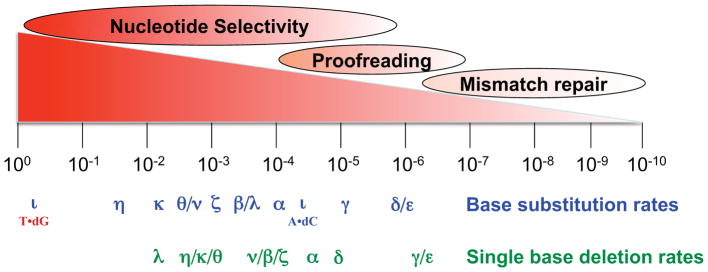
The image illustrates the wide ranges over which polymerase nucleotide selectivity, exonucleolytic proofreading and mismatch repair contribute to spontaneous mutation rates of organisms. Also depicted are the average rates at which purified eukaryotic DNA polymerases generate single base substitution and single base deletion errors when performing gap-filing DNA synthesis in vitro. See text for further descriptions. Details on the source and composition of the polymerases used, and on their error specificity, can be found in (McCulloch and Kunkel, 2008) and references therein.
Major replicative pols
In order to maintain species identity, the accuracy of genomic replication is expected to be high. Consistent with this expectation, the major replicative polymerases nearly always insert correct dNTPs onto properly aligned primer-templates (exemplified in Figure 2 by pols α, δ, ε and γ, but also true for replicative pols from other organisms). High nucleotide selectivity at the polymerase active site is illustrated by the relatively low base substitution and indel error rates (about 10−4) of pol α, which naturally lacks proofreading activity. Similarly low error rates are seen for pols δ, ε and γ when their intrinsic proofreading exonucleases are inactivated (Fortune et al., 2005; Longley et al., 2001; Shcherbakova et al., 2003b).
Nucleotide selectivity
What determines the high nucleotide selectivity of accurate DNA polymerases? Hydrogen bonding between template bases and incoming dNTPs is clearly important for replication fidelity. However, this alone is unlikely to explain high selectivity because the free energy difference between correct and incorrect base pairs in solution accounts for error rates of ~1:100 (Loeb and Kunkel, 1982). Thus other ideas have been put forth to account for the higher selectivity of accurate polymerases. For example, in order for the incoming dNTP to hydrogen bond to a template base, water molecules that are hydrogen bonded to the base of the incoming dNTP must be removed, thereby decreasing the entropy of the system. This magnifies the contribution of enthalpy to the free energy difference (Petruska and Goodman, 1995), thereby increasing nucleotide selectivity. Another idea supported by substantial evidence (reviewed in (Beard and Wilson, 2003; Kim et al., 2005; Kool, 2002; Kunkel and Bebenek, 2000)) is that high nucleotide selectivity partly results from the shape complementarity in the nascent base pair binding pocket. The four canonical Watson-Crick base pairs are nearly identical in size and shape. Structural studies reveal that correct base pairs fit within the nascent base pair binding pocket without steric clashes. Particularly important to fidelity are amino acid side chains (e.g., Fig. 1B, Arg283 (purple) in pol β) that interact with the O2 atom of pyrimidines and the N3 atom of purines, which are isosteric in the four correct Watson-Crick base pairs. This is illustrated in Figure 1B, which shows the active site of DNA polymerase β, a relatively accurate repair enzyme, with a correct base pair poised for catalysis. The correct pair fits snugly, while mismatches with different and variable geometries are predicted to have steric clashes that would reduce incorrect dNTP binding affinity, affect subsequent conformational changes needed to set up the proper geometry for catalysis, and/or reduce the rate of phosphodiester bond formation.
Insertion-deletion errors (indels)
DNA polymerases also insert and delete nucleotides during DNA synthesis. These errors result from strand misalignments that generate unpaired bases in the primer strand, leading to additions, or in the template strand, leading to deletions. Ideas to account for how these misalignments initiate and are stabilized for continued synthesis include classical DNA strand slippage, misinsertion followed by primer relocation, and misalignment of a nucleotide at the polymerase active site. Biochemical and structural support exists for all three models (reviewed in (Bebenek and Kunkel, 2000; Garcia-Diaz and Kunkel, 2006). Replicative DNA polymerases generate single base deletions at rates that are similar to those for single base substitutions (Fig. 2). Single base deletion error rates are usually higher than single base addition error rates or rates for indels involving large numbers of nucleotides, with possible explanations considered elsewhere (Bebenek and Kunkel, 2000; Garcia-Diaz and Kunkel, 2006). Importantly, the single base substitution and deletion error rates in Figure 2 are average values, with wide variations observed depending on the type of mismatch and the sequence context in which the mismatch is located (Kunkel and Bebenek, 2000). Prime examples of such variability among lower fidelity polymerases involved in DNA repair and translesion synthesis are considered below.
Proofreading by replicative DNA polymerases
Average base substitution error rates of proofreading-proficient replicative DNA polymerases are typically ≥ 10−6. Their exonuclease-deficient derivatives are considerably less accurate, indicating that on average, proofreading improves replication fidelity by about 10- to 100-fold (Fig. 2). The energetic cost of improving fidelity by more than this could be unacceptable due to excessive excision of correctly paired bases (Fersht et al., 1982). The biological importance of proofreading is illustrated by studies showing that when highly conserved residues near the active sites of S. cerevisiae replicative pols are replaced with non-conservative amino acids, the mutant enzymes have decreased DNA synthesis fidelity in vitro (Fortune et al., 2005; Longley et al., 2001; Shcherbakova et al., 2003b) and generate mutator phenotypes in vivo (Morrison and Sugino, 1994). Moreover, mice with homologous replacements in pol δ have decreased genomic stability and accelerated tumorigenesis (Goldsby et al., 2001).
The key to proofreading efficiency is the balance between polymerization and excision at a growing primer terminus (Fig. 3A). Under normal circumstances, correct incorporation allows subsequent incorporations to occur rapidly with little opportunity for proofreading (line 1). However, misinsertion generates a mismatched primer terminus that is more difficult to extend. This slows polymerization, allowing the primer terminus to fray and move single stranded DNA into the exonuclease active site for excision of the error (line 2). Based on early work (reviewed in (Kornberg and Baker, 1992)) and on more recent studies, we now realize that there are several ways to influence this critical balance between polymerization and excision (Table 2). Proofreading can be inactivated by amino acid substitutions in the exonuclease active site, or exonuclease activity can be inhibited if the end product of excision, a dNMP, binds to the exonuclease active site. Proofreading can be reduced by amino acid substitutions in replicative polymerases that prevent movement of the frayed primer terminus to the exonuclease active site (so-called “switching mutants” (e.g., see (Jin et al., 2005) and references therein), or by amino acid substitutions in the polymerase active site that promote mismatch extension (e.g., see (Nick McElhinny et al., 2008) and references therein). Proofreading can be suppressed by increasing the concentration of the next correct nucleotide to be incorporated after a misinsertion (dCTP for the examples in Fig. 3A), thereby promoting mismatch extension at the expense of excision (Ninio, 1975). Lastly, in some circumstances, mismatches escape proofreading by tricking the replicative polymerases. A well-known example involves 8-oxo-guanine, a common lesion resulting from oxidative stress. Replication of template 8-oxo-G can generate 8-oxoG•dA mismatches whose geometry is similar to that of a correct base pair, such that the mismatch largely escapes proofreading (e.g., by replicative T7 DNA polymerase (e.g., see (Brieba et al., 2004)). Another example with high biological relevance involves proofreading of insertion-deletion mismatches during replication of repetitive sequences (Fig. 3A, line 3). Proofreading does correct misaligned intermediates containing extra bases in one strand or the other near the primer terminus, as illustrated by the higher indel error rates of exonuclease-deficient pols δ, ε and γ when compared to their proofreading proficient counterparts (Fortune et al., 2005; Longley et al., 2001; Shcherbakova et al., 2003b). However, the efficiency of proofreading of indels decreases as the length of a repetitive sequence increases (e.g., Fig. 3B). This is because in a long repetitive sequence, the mismatch generated by strand slippage (i.e., the unpaired base) is likely to be located upstream of the polymerase active site, such that it does not strongly reduce the rate of polymerization (Fig. 3A, line 3). Such diminished proofreading, in conjunction with a higher rate of strand slippage by polymerases (e.g., left panel in Fig. 3B and see (Garcia-Diaz and Kunkel, 2006)), contributes to the observation that long repetitive sequences are at risk for a high rate of replication slippage errors, as evidenced by the well known “microsatellite instability” phenotype of eukaryotic cells defective in DNA mismatch (see below). Based on the logic in Figure 3 and the parameters in Table 2, it is now very clear that, just as for nucleotide selectivity, the contribution of proofreading to replication fidelity can vary over a wide range (Fig. 2), from almost none (8-oxoG•dA mismatches) to several hundred fold (e.g., for bacteriophage T7 replication, e.g., see (Donlin et al., 1991)).
Figure 3. Exonucleolytic proofreading.
(A) Depiction of the principles that determine the efficiency of proofreading. A proofreading-proficient polymerase (blue) harbors its polymerase and exonuclease activities in separate domains (e.g., see RB69 pol in Fig. 1A), depicted as large and small ovals, respectively. The partitioning between these two activities determines the efficiency of proofreading. Also shown is the possibility that errors made by an exonuclease-deficient polymerase (yellow) may be proofread by a separate exonuclease, either that of a proofreading-proficient polymerase (as shown) of present in another protein. See text for a further description and (Nick McElhinny et al., 2006) and references therein] for additional discussion and information. (B) The left panel depicts the single base deletion error rates of proofreading-proficient (open bars) and proofreading-deficient (closed bars) T7 DNA polymerase when copying tracks of 3–8 consecutive template Ts. The right panel depicts the ratio of the error rates of the two polymerases, to illustrate the decreasing efficiency of proofreading as a function of increasing repetitive sequence track length. Reproduced from (Kroutil and Kunkel, 1998), with permission.
Table 2.
Variables that can modulate the efficiency of correcting replication errors.
| Reduced proofreading by: |
| Mutational inactivation of 3′ exonuclease |
| Inhibiting 3′ exonuclease activity - dNMPs |
| Suppressing proofreading by: |
| Reducing switching from polymerase to exonuclease active site |
| Promoting MM extension by: |
| Polymerase active site mutations |
| High concentration of next correct dNTPs |
| Mismatch mimicry of correct base pairing |
| Internalizing a mismatch in a repetitive sequence |
| Reduced mismatch repair by: |
| Mutational inactivation of MMR proteins |
| Cadmium inhibition of MMR in S. cerevisiae |
| Promoter hypermethylation to silence expression of human Mlh1 |
| Saturation of MMR |
| Rapid replication in proofreading-defective E. coli (MutD) |
| DNA damage |
| Imbalanced expression of MMR proteins |
| human Msh3 |
| S. cerevisiae Mlh1 |
“Extrinsic” proofreading may also contribute to genome stability
Interestingly, among many mammalian DNA polymerases, only those responsible for the bulk of chain elongation during replication (pols δ, ε, and γ) contain intrinsic 3′ exonucleolytic proofreading activity. Nonetheless, the exonuclease-deficient polymerases have very important roles in maintaining genome stability (Table 1). Are errors made by exonuclease-deficient polymerases subject to “extrinsic” proofreading by a separate exonuclease? The idea (Fig. 3A, line 4) is that, after making a mismatch, the polymerase would dissociate, allowing the exonuclease activity of another protein to excise the mismatch. Indeed, the major E. coli replicative polymerase, DNA polymerase III, harbors its polymerase and exonuclease activities in two different subunits (the α and ε subunits, respectively), and these two proteins work in concert to achieve high replication fidelity. Proofreading by a separate protein may also occur in eukaryotes. For example, yeast DNA polymerase α lacks its own proofreading activity yet synthesizes perhaps 10% of each Okazaki fragment on the lagging strand, i.e., about 5% of the human genome. Given a base substitution error rate of ~10−4 (Fig. 2), this amount of replication would generate 30,000 mismatches during each replication cycle. Can pol α errors be proofread by a separate exonuclease? This possibility was recently examined in a genetic study of yeast pol α with a Leu868Met substitution at the polymerase active site (Pavlov et al., 2006). L868M pol α copies DNA in vitro with normal activity and processivity but with reduced fidelity. In vivo, the pol1-L868M allele confers a mutator phenotype. This mutator phenotype is strongly increased upon inactivation of the 3′ exonuclease of pol δ but not that of pol ε. Among several possible (non-exclusive) explanations, the results support the hypothesis that the 3′ exonuclease of pol δ proofreads errors generated by pol α during initiation of Okazaki fragments. Given the existence of many other specialized, naturally proofreading-deficient DNA polymerases with even lower fidelity than pol α, intrinsic proofreading could be relevant to other DNA transactions that control genome stability (reviewed in (Nick McElhinny et al., 2006)), such as base excision repair and possibly translesion synthesis by Pol η (see below).
Replication asymmetry and fidelity
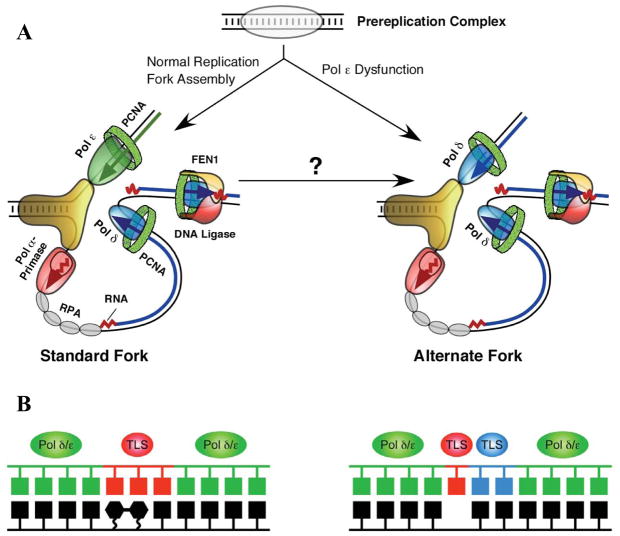
The two strands of duplex DNA are oriented anti-parallel to each other, and DNA polymerases copy DNA in only the 5′ to 3′ direction. Thus, replication of duplex DNA is intrinsically asymmetric. This asymmetry is illustrated by the simple model of a eukaryotic replication fork shown in Figure 4 (panel A, left). Recent evidence in budding yeast suggests that the leading strand is primarily replicated by pol ε (Pursell et al., 2007), whereas Okazaki fragments on the lagging strand are initiated by pol α-primase and then primarily completed by pol δ (Nick McElhinny et al., 2008). These enzymes differ from each other in primary sequence, in subunit composition, in interactions with other proteins, and in several biochemical properties, including processivity, proofreading capacity, fidelity and error specificity. It is therefore possible that the fidelity of leading and lagging strand replication may differ, perhaps even more so under nonstandard replication conditions arising under stress (Fig. 4A, right), either environmental (DNA lesions) or genetic (mutations in key genes). Evidence for differences in leading and lagging strand replication fidelity have been reported in E. coli (Fijalkowska et al., 1998), where both strands are replicated by the same polymerase acting as a multi-subunit dimer, DNA polymerase III holoenzyme.
Figure 4. Replication fork and translesion synthesis models.
(A) Current model of the eukaryotic DNA replication fork (left), with pol ε replicating the leading strand template and pols α and δ replicating the lagging strand template. On the right is an “alternative fork” that might result from stress. See text and (Kunkel and Burgers, 2008) for further discussion. Reproduced from (Kunkel and Burgers, 2008), with permission. (B) “One TLS polymerase” (left) and “Two TLS polymerase” (right) models for translesion DNA synthesis. See text for further discussion.
Fidelity of DNA repair polymerases
Efficient and accurate replication requires clean substrates, such that many organisms devote great attention and energy to repairing DNA lesions that can result from endogenous metabolic process and from exposure to physical and chemical agents in the external environmental. Many different repair processes exist and can be distinguished by lesion specificity, by the enzymes involved and by when they operate (reviewed in Friedberg]). For many of these repair pathways, e.g., BER, NER, NHEJ, MMR and ICL repair, excision of a lesion is followed by gap-filling DNA synthesis and ligation (Friedberg et al., 2006). The gap filling is conducted by polymerases that are highly accurate (e.g., pol δ for filling long gaps during NER and MMR) or moderately accurate (pol β for filling short gaps during BER). However, certain gap-filling transactions in cells may involve inaccurate DNA polymerases, e.g., pol κ in NER, pol ζ/pol θ in ICL repair, pol μ in NHEJ), such that DNA synthesis errors occurring during repair may contribute to mutagenesis. Perhaps the best examples are for pol ζ and pol η, both of which are implicated in somatic hypermutation of immunoglobulin genes, a process that involves processing uracil in DNA generated by cytosine deamination catalyzed by activation-induced cytosine deaminase (Diaz and Lawrence, 2005).
Fidelity of translesion synthesis polymerases
Replication forks can stall upon encountering lesions that distort helix geometry. Among several possible solutions that allow complete genome replication, one is translesion synthesis (TLS) by DNA polymerases. Two general models have been put forth for TLS, one involving a single TLS polymerase for lesion bypass (Fig. 4B, left), and another involving two TLS polymerases, one for insertion opposite a lesion and another for extending aberrant primer termini (Fig. 4B, right). Several specialized TLS polymerases have been discovered in the past decade, and these are evolutionarily conserved (Ohmori et al., 2001; Prakash and Prakash, 2002; Yang and Woodgate, 2007). In mammals they include family B pol ζ, family Y pols η, κ, and ι, and possibly family A pols θ and ν. These are the least accurate of polymerases (Fig. 2). They all lack proofreading activity and they also have lower nucleotide selectivity than the major replicative polymerases, as indicated by their higher error rates for base substitutions and indels (Fig. 2). The extreme case is for pol ι, a conserved family Y member that rarely generates certain mismatches (e.g., A•dC) but can preferentially misincorporate dG as compared to dA opposite template T (Fig. 2, e.g., see (Bebenek et al., 2001)). This rather amazing violation of Watson-Crick base pairing dogma leads one to wonder what the true physiological substrates and functions of pol ι might be in vivo, whether in TLS (Dumstorf et al., 2006) or in yet to be discovered DNA transactions. Structural and biochemical studies suggest that the low fidelity of family Y enzymes is partly due to relaxed geometric selectivity in the nascent base pair binding pocket, which is more open and solvent accessible than those of more accurate DNA polymerases. An example is shown in Figure 1C, which depicts the active site of a bacterial Y family polymerase, Sso Dpo4. Indeed, much of the seminal work on family Y polymerases has been performed using bacterial enzymes, which include two DNA damage-induced E. coli DNA polymerases, IV and V (Fuchs et al., 2004). Another TLS polymerase is the B-family member pol ζ. When copying undamaged DNA, Pol ζ has somewhat higher fidelity than the Y-family polymerases, but lower fidelity than the other B-family members (Fig. 2). The ability of Pol ζ to generate both base substitutions and indels at relatively high rates is consistent with its known participation in a large majority of spontaneous mutations, as well as in mutagenesis induced by a variety of DNA damaging agents (Lawrence, 2002). Pol ζ’s high base substitution error rate clearly demonstrates that it has relatively low nucleotide selectivity, consistent with a possible direct role in mutagenic misinsertion of dNTPs in vivo. Pol ζ also efficiently extends terminal mismatches when copying undamaged DNA, as well as efficiently extending damaged termini, the latter being consistent with a role for Pol ζ in the extension step of TLS in the 2-polymerase model (Fig. 4B). A similar role has also been proposed for Pol κ, which like Pol ζ, is promiscuous for mismatch extension (reviewed in (Prakash and Prakash, 2002). During DNA synthesis in vitro, Pol ζ also generates “complex” mutations that contain multiple substitutions and indels within a short tract of DNA (Sakamoto et al., 2007; Stone et al., 2009). Consistent with this property, Pol ζ also generates complex errors in vivo, which could be significant from an evolutionary perspective. The biological relevance of TLS is perhaps best illustrated by the role of Pol V in the mutagenic SOS response in E. coli, and the fact that loss of Pol η function in humans and in mice results in sensitivity to sunlight, predisposition to skin cancer and altered specificity of somatic hypermytation of immunoglobulin genes. The topics and the TLS ability and fidelity of various polymerases when encountering a wide range of structurally diverse lesions have been described in great detail elsewhere (e.g., see (Diaz and Lawrence, 2005; Friedberg et al., 2005; Fuchs et al., 2004 ; Prakash and Prakash, 2002; Yang and Woodgate, 2007).
DNA Mismatch repair
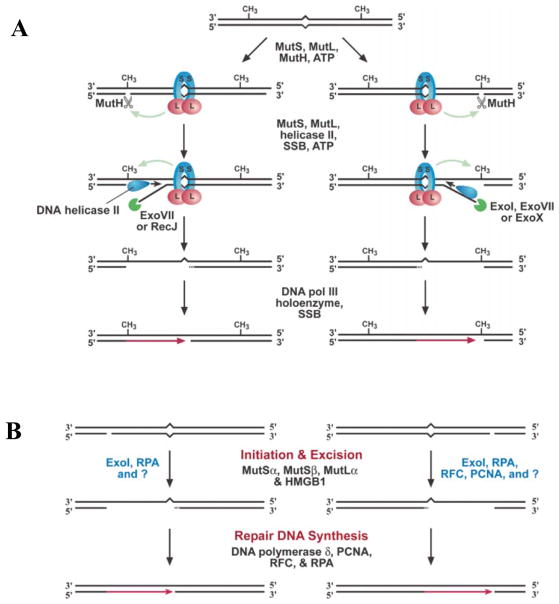
Replication errors are corrected by DNA mismatch repair (MMR, reviewed in (Hsieh and Yamane, 2008; Iyer et al., 2006; Kunkel and Erie, 2005). The reactions and proteins catalyzing MMR are evolutionarily conserved from E. coli (Fig. 5A) through humans (Fig. 5B). MMR requires initial recognition of mismatches by bacterial MutS protein or its eukaryotic homologs (Msh2-Msh6 or Msh2-Msh3). This is followed by binding of a second protein, MutL or its eukaryotic homologs, either Mlh1-Pms1 (Pms2 in humans), Mlh1-Mlh2 or Mlh1-Mlh3. These MutS and MutL proteins bind and hydrolyze ATP, and in so doing, these complexes undergo conformational changes that help coordinate the multiple protein partnerships and reactions needed to find the strand-discrimination signal, incise the nascent strand, excise the replication error, correctly synthesize new DNA and then ligate the nascent strand. In addition to their functions in repairing replication errors, some MMR proteins also participate in other DNA transactions. These include critical environmental stress-response pathways such as repair of double-strand DNA breaks and DNA damage surveillance to signal apoptosis. As a consequence, loss of MMR is associated with elevated mutation rates and altered survival in response to DNA damage. These in turn can give rise to microbial populations with increased fitness to survive adverse environmental conditions, and somatic cells with increased resistance to chemotherapeutic agents and increased probability of tumorigenesis. MMR proteins also prevent recombination between DNA sequences with imperfect homology, thereby influencing speciation. Some MMR proteins participate in meiotic recombination, such that loss of MMR results in infertility. MMR also modulates somatic hypermutation of immunoglobulin genes, such that loss of MMR protein functions alters the specificity of SHM.
Figure 5. DNA mismatch repair.
Models for DNA mismatch repair in E. coli (A) and eukaryotes (B). See text and for description. Reproduced from (Iyer et al., 2006), with permission.
Modulating MMR efficiency
Given these many functions and biological effects, intensive studies of MMR have revealed several ways to modulate MMR activity (Table 2). These include partial or complete inactivation by mutations in various MMR genes (reviewed in (Hsieh and Yamane, 2008; Iyer et al., 2006; Kunkel and Erie, 2005), cadmium inhibition of MMR in budding yeast (Jin et al., 2003), silencing Mlh1 expression by promoter hypermethylation (e.g., see (Herman et al., 1998), saturating MMR repair capacity under conditions of stress (Schaaper and Radman, 1989), and imbalanced expression of certain MMR proteins that can reduce MMR efficiency (see (Drummond et al., 1997; Shcherbakova et al., 2001) and references therein). Indeed, just as for nucleotide selectivity and proofreading, the contribution of MMR to replication fidelity can vary over a wide range (Fig. 2). On average, complete loss of bacterial MutS-dependent MMR or eukaryotic Msh2-dependent MMR elevates point mutation rates about 100- to 1000-fold. On the edges of the MMR efficiency continuum are some mismatches that are poorly corrected, e.g., ~5-fold for 8-oxo-G•A mismatches (Pavlov et al., 2003), and others that are repaired incredibly efficiently, e.g., exceeding 10,000-fold for single base deletion mismatches in long homo-nucleotide runs. The latter illustrate that MMR is the major guardian against the instability of repetitive sequence elements, which as explained above, are prone to slippage and poorly proofread.
Conclusions
In the half century since the DNA double helix was described, we have come to more fully, but still incompletely, appreciate the elegant complexity of the DNA transactions required to replicate genomes with the fidelity needed to maintain genetic identity in the face of environmental insults, coupled with the advantage of some promiscuity for survival and evolution. This flexibility stems from the wide variability in the contributions of the three processes that determine DNA synthesis fidelity (Fig. 2), and the fact that proofreading and MMR, the two main replication-error correction mechanisms that ensure genome stability, are not essential for cellular survival. Thus, neither proofreading nor MMR contributes to replication of RNA viruses, and these viruses have very high mutation rates that fit their lifestyle (Drake and Holland, 1999). Some DNA viruses use proofreading to achieve lower mutation rates, but do not take advantage of their host’s MMR machinery (e.g., bacteriophage T4 (Santos and Drake, 1994). As a consequence, they have mutations rates per base pair that are higher than organisms that do use MMR. Interestingly, given the differences in genome size, the mutation rate per genome is relatively constant among DNA based organisms, at 0.003 (Drakes rule (Drake, 1991, 1999). Also of interest is the fact that the genomes of certain bacteria do not encode obvious homologs of the major MMR genes. This leads one to wonder whether they forego MMR altogether, or correct replication errors in a manner not yet discovered.
Acknowledgments
I thank Alan Clark and Mercedes Arana for thoughtful comments on the manuscript. The research conducted in the author’s laboratory is supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Projects Z01 ES065070 and Z01 ES065089).
References
- Beard WA, Wilson SH. Structural insights into the origins of DNA polymerase fidelity. Structure. 2003 doi: 10.1016/s0969-2126(03)00051-0. [DOI] [PubMed] [Google Scholar]
- Bebenek K, Kunkel TA. Streisinger revisited: DNA synthesis errors mediated by substrate misalignments. Cold Spring Harb Symp Quant Biol. 2000;65:81–91. doi: 10.1101/sqb.2000.65.81. [DOI] [PubMed] [Google Scholar]
- Bebenek K, Kunkel TA, editors. Functions of DNA Polymerases in DNA Repair and Replication. 2004. [DOI] [PubMed] [Google Scholar]
- Bebenek K, Tissier A, Frank EG, McDonald JP, Prasad R, Wilson SH, Woodgate R, Kunkel TA. 5′-Deoxyribose phosphate lyase activity of human DNA polymerase iota in vitro. Science (New York, NY. 2001;291:2156–2159. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- Bessman MJ, Kornberg A, Lehman IR, Simms ES. Enzymic synthesis of deoxyribonucleic acid. Biochim Biophys Acta. 1956;21:197–198. doi: 10.1016/0006-3002(56)90127-5. [DOI] [PubMed] [Google Scholar]
- Brieba LG, Eichman BF, Kokoska RJ, Doubliâe S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23(17):3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DJ, Cimprich KA. DNA damage tolerance: when it’s OK to make mistakes. Nat Chem Biol. 2009;5:82–90. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M, Lawrence C. An update on the role of translesion synthesis DNA polymerases in Ig hypermutation. Trends Immunol. 2005;26:215–220. doi: 10.1016/j.it.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Donlin MJ, Patel SS, Johnson KA. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry. 1991;30:538–546. doi: 10.1021/bi00216a031. [DOI] [PubMed] [Google Scholar]
- Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci U S A. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW. The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann N Y Acad Sci. 1999;870:100–107. doi: 10.1111/j.1749-6632.1999.tb08870.x. [DOI] [PubMed] [Google Scholar]
- Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc Natl Acad Sci U S A. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond JT, Genschel J, Wolf E, Modrich P. DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSalpha/hMutSbeta ratio and reduces the efficiency of base-base mismatch repair. Proc Natl Acad Sci U S A. 1997;94:10144–10149. doi: 10.1073/pnas.94.19.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstorf CA, Clark AB, Lin Q, Kissling GE, Yuan T, Kucherlapati R, McGregor WG, Kunkel TA. Participation of mouse DNA polymerase iota in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc Natl Acad Sci U S A. 2006;103:18083–18088. doi: 10.1073/pnas.0605247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht AR, Knill-Jones JW, Tsui WC. Kinetic basis of spontaneous mutation. Misinsertion frequencies, proofreading specificities and cost of proofreading by DNA polymerases of Escherichia coli. J Mol Biol. 1982;156:37–51. doi: 10.1016/0022-2836(82)90457-0. [DOI] [PubMed] [Google Scholar]
- Fijalkowska IJ, Jonczyk P, Tkaczyk MM, Bialoskorska M, Schaaper RM. Unequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1998;95:10020–10025. doi: 10.1073/pnas.95.17.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. Washington, D.C: ASM Press; 2006. [Google Scholar]
- Fuchs RP, Fujii S, Wagner J. Properties and functions of Escherichia coli: Pol IV and Pol V. Adv Protein Chem. 2004;69:229–264. doi: 10.1016/S0065-3233(04)69008-5. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Kunkel TA. Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem Sci. 2006;31:206–214. doi: 10.1016/j.tibs.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Goldsby RE, Lawrence NA, Hays LE, Olmsted EA, Chen X, Singh M, Preston BD. Defective DNA polymerase-delta proofreading causes cancer susceptibility in mice. Nat Med. 2001;7:638–639. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- Jansen JG, Fousteri MI, de Wind N. Send in the clamps: control of DNA translesion synthesis in eukaryotes. Mol Cell. 2007;28:522–529. doi: 10.1016/j.molcel.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Jin YH, Carg P, Stith CMW, Al-Refai H, Sterling J, Murray LJW, Kunkel TA, Resnick MA, Burgers PM, Gordenin DA. The multiple bilogical roles of the 3′-.5′ exonuclease of yeast DNA polymerase delta require switching between the polymerase and exonuclease domains. Molecular and Cellular Biology. 2005 doi: 10.1128/MCB.25.1.461-471.2005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YH, Clark AB, Slebos RJ, Al-Refai H, Taylor JA, Kunkel TA, Resnick MA, Gordenin DA. Cadmium is a mutagen that acts by inhibiting mismatch repair. Nat Genet. 2003;34:326–329. doi: 10.1038/ng1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Delaney JC, Essigmann JM, Kool ET. Probing the active site tightness of DNA polymerase in subangstrom increments. Proc Natl Acad Sci U S A. 2005;102:15803–15808. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool ET. Active site tightness and substrate fit in DNA replication. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Baker T. DNA Replication. 2. New York: W.H. Freeman and Company; 1992. [Google Scholar]
- Kroutil LC, Kunkel TA. DNA Replication Errors Involving Strand Misalignments. In: Wells RD, Warren ST, editors. Genetic Instabilities and Hereditary Neurological Diseases. San Diego: Academic Press; 1998. pp. 699–716. [Google Scholar]
- Kunkel TA, Bebenek K. DNA Replication Fidelity. Annu Rev Biochem. 2000:69. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Pavlov YI, Bebenek K. Functions of human DNA polymerases eta, kappa and iota suggested by their properties, including fidelity with undamaged DNA templates. DNA Repair (Amst) 2003;2:135–149. doi: 10.1016/s1568-7864(02)00224-0. [DOI] [PubMed] [Google Scholar]
- Lawrence CW. Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair. 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Kunkel TA. Fidelity of DNA synthesis. Annual Review Biochemistry. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Monnat RJ., Jr DNA polymerases and human disease. Nat Rev Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. Journal of Biological Chemistry. 2001;276:38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AF, Garcia-Diaz M, Bebenek K, Davis BJ, Zhong X, Ramsden DA, Kunkel TA, Pedersen LC. Structural insight into the substrate specificity of DNA Polymerase mu. Nature structural & molecular biology. 2007;14:45–53. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- Morrison A, Sugino A. The 3′-->5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Pavlov YI, Kunkel TA. Evidence for extrinsic exonucleolytic proofreading. Cell cycle (Georgetown, Tex. 2006;5:958–962. doi: 10.4161/cc.5.9.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57:587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Frahm C, Nick McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Mian IM, Kunkel TA. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr Biol. 2003;13:744–748. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- Petruska J, Goodman MF. Enthalpy entropy compensation in DNA melting thermodynamics. Journal of Biological Chemistry. 1995;270:746–750. doi: 10.1074/jbc.270.2.746. [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes and Development. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science (New York, NY. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto AN, Stone JE, Kissling GE, McCulloch SD, Pavlov YI, Kunkel TA. Mutator alleles of yeast DNA polymerase zeta. DNA Repair (Amst) 2007;6:1829–1838. doi: 10.1016/j.dnarep.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos ME, Drake JW. Rates of spontaneous mutation in bacteriophage T4 are independent of host fidelity determinants. Genetics. 1994;138:553–564. doi: 10.1093/genetics/138.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper RM, Radman M. The extreme mutator effect of Escherichia coli mutD5 results from saturation of mismatch repair by excessive DNA replication errors. EMBO J. 1989;8:3511–3516. doi: 10.1002/j.1460-2075.1989.tb08516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova PV, Bebenek K, Kunkel TA. Functions of eukaryotic DNA polymerases. Sci Aging Knowledge Environ. 2003a;2003:RE3. doi: 10.1126/sageke.2003.8.re3. [DOI] [PubMed] [Google Scholar]
- Shcherbakova PV, Hall MC, Lewis MS, Bennett SE, Martin KJ, Bushel PR, Afshari CA, Kunkel TA. Inactivation of DNA mismatch repair by increased expression of yeast MLH1. Mol Cell Biol. 2001;21:940–951. doi: 10.1128/MCB.21.3.940-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J Biol Chem. 2003b;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- Steitz TA. DNA- and RNA-dependent DNA polymerases. Curr Opin Struct Biol. 1993;3:31–38. [Google Scholar]
- Stone JE, Kissling GE, Lujan SA, Rogozin IB, Stith CM, Burgers PM, Kunkel TA. Low-fidelity DNA synthesis by the L979F mutator derivative of Saccharomyces cerevisiae DNA polymerase {zeta} Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Wing RA, Bailey S, Steitz TA. Insights into the replisome from the structure of a ternary complex of the DNA polymerase III alpha-subunit. J Mol Biol. 2008;382:859–869. doi: 10.1016/j.jmb.2008.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci U S A. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]