Summary
Mitochondria are critical for supplying energy to the cell, but during catabolism this organelle also produces reactive oxygen species that can cause oxidative damage. Accordingly, quality control of mitochondria is important to maintain cellular homeostasis. It has been assumed that autophagy is the pathway for mitochondrial recycling, and that the selective degradation of mitochondria via autophagy (mitophagy) is the primary mechanism for mitochondrial quality control, although there is little experimental evidence to support this idea. Recent studies in yeast identified several mitophagy-related genes and have uncovered components involved in the molecular mechanism and regulation of mitophagy. Similarly, studies of Parkinson disease and reticulocyte maturation reveal that Parkin and Nix, respectively, are required for mitophagy in mammalian cells, and these analyses have revealed important physiological roles for mitophagy. Here, we review the current knowledge on mitophagy, in particular on the molecular mechanism and regulation of mitophagy in yeast. We also discuss some of the differences between yeast and mammalian mitophagy.
Introduction
Mitochondrial oxidative phosphorylation supplies energy that is essential for many cellular activities. On the other hand, mitochondria are the major source of cellular reactive oxygen species (ROS) that cause oxidative damage to mitochondrial DNA, protein and lipid. These types of mitochondrial damage have the potential to cause the further production of additional ROS through a defective electron transport chain. Accordingly, damaged, aged or excess mitochondria are a risk factor for the cell, and proper elimination of such organelles is important to maintain optimal cellular homeostasis. Indeed, the accumulation of damaged mitochondria induce apoptosis by releasing cytochrome c and correlate with aging or the development of certain neurodegenerative diseases (Wallace, 2005). Recent evidence form yeast to mammal suggests that autophagy is the primary mechanism to eliminate dysfunctional, aged or excess mitochondria. In fact, the autophagy-dependent degradation of mitochondria (mitophagy) is integrally related with cellular physiology and disease. For example, during erythroid cell maturation, mitochondria are eliminated by mitophagy when a Nix-related loss of mitochondrial membrane potential is induced, although the mechanism involved in sequestering mitochondria in response to Nix has not been determined and it may be independent of mitochondrial depolarization (Schweers et al., 2007; Sandoval et al., 2008; Zhang et al., 2009). A loss-of-function mutation of the PARK2 and PARK6 gene encoding Parkin and PINK1, respectively, causes Parkinson disease; Parkin translocates from the cytosol to impaired mitochondria in a PINK1-dependent manner and promotes their autophagic degradation (Narendra et al., 2008; Vivesbauza et al., 2009). Here we discuss the molecular mechanism of mitophagy based mainly on the evidence from yeast studies.
Mitophagy, a type of selective autophagy
Macroautophagy is a protein degradation process that involves the formation of a double-membrane compartment, termed the autophagosome, which engulfs cytoplasmic components non-selectively and fuses with the lysosome/vacuole for degradation (Matsuura et al., 1997). This process plays a role in various aspects of cell physiology such as survival during starvation, intracellular clearance of dysfunctional or superfluous proteins and organelles, proper development and aging (Mizushima et al., 2008). In contrast to macroautophagy, the cytoplasm to vacuole targeting (Cvt) pathway, pexophagy (selective peroxisome degradation via autophagy) and mitophagy are categorized as selective types of autophagy (Farre et al., 2009; Tolkovsky, 2009). Studies in Sac-charomyces cerevisiae and other fungi allowed the isolation of 33 AuTophaGy-related (ATG) genes. Fifteen of these genes are essential for both macroautophagy and selective autophagy and are categorized as part of the core autophagic machinery (Xie and Klionsky, 2007). Other genes have a role in certain types of autophagy. For example Atg19, a receptor protein for the Cvt pathway, binds the Ape1 complex composed of prApe1 and Ams1, to form the Cvt complex. Then Atg11, an adaptor protein for selective autophagy, interacts with Atg19 and recruits the Cvt complex to the phagophore assembly site (PAS), where the phagophores, the initial sequestering membrane structure, are generated (Shintani et al., 2002). Similarly, during pexophagy in Pichia pastoris, Atg30 localizes to peroxisomes, where it subsequently binds Ag11, allowing recruitment of peroxisomes to the PAS (Farre et al., 2008). As shown below, recent studies identified mitophagy-specific ATG genes and revealed that mitophagy also occurs by a pathway that is similar to the Cvt pathway or pexophagy. This finding finally confirmed that mitophagy is a type of selective autophagy.
Mitophagy and ATG genes
Because mitophagy is a type of autophagy, the core autophagic machinery is used in common with other types of autophagy. The requirement of several ATG genes for mitophagy has been reported from several groups (Kiššová et al., 2004; 2007; Tal et al., 2007; Zhang et al., 2007; Kanki and Klionsky, 2008) and very recently, mitophagy defects were analysed in all of the atg knockout strains in S. cerevisiae (Kanki et al., 2009a; Okamoto et al., 2009a). Table 1 summarizes the requirement for the ATG genes for mitophagy, the Cvt pathway and non-selective autophagy (Kiššová et al., 2004; 2007; Tal et al., 2007; Xie and Klionsky, 2007; Zhang et al., 2007; Kanki et al., 2009a; Nakatogawa et al., 2009; Okamoto et al., 2009a). ATG genes that play a fundamental role in autophagy such as that encoding the protein kinase Atg1 and its binding partner Atg13, the genes for the ubiquitin-like protein modification machinery (ATG3, 4, 5, 7, 8, 10, 12 and 16), those that encode components that are involved in supplying lipids to the phagophore (ATG2, ATG9 and ATG18) and those that encode part of phosphatidylinositol 3-kinase complex I that is required for vesicle nucleation (ATG6 and ATG14) are essential for all types of autophagy, suggesting that the core autophagic machinery is used in common between mitophagy and other types of autophagy. In contrast, ATG11, ATG20 and ATG24, which are required for both the Cvt pathway and pexophagy but not for macroautophagy (Nice et al., 2002), are required for mitophagy, suggesting that selective autophagy uses certain components in common that are in part different from those used in macroautophagy.
Table 1.
ATG gene requirement for non-selective (macroautophagy) and selective autophagy (Cvt pathway and mitophagy).
| Macroautophagy | Cvt pathway | Mitophagy | |
|---|---|---|---|
| ATG1 | ++ | ++ | ++ |
| ATG2 | ++ | ++ | ++ |
| ATG3 | ++ | ++ | ++ |
| ATG4 | ++ | ++ | ++ |
| ATG5 | ++ | ++ | ++ |
| ATG6 | ++ | ++ | ++ |
| ATG7 | ++ | ++ | ++ |
| ATG8 | ++ | ++ | ++ |
| ATG9 | ++ | ++ | ++ |
| ATG10 | ++ | ++ | ++ |
| ATG11 | − | ++ | ++ |
| ATG12 | ++ | ++ | ++ |
| ATG13 | ++ | ++ | ++a |
| ATG14 | ++ | ++ | ++ |
| ATG15b | ++ | ++ | ++ |
| ATG16 | ++ | ++ | ++ |
| ATG17 | ++ | − | ++ |
| ATG18 | ++ | ++ | ++ |
| ATG19 | − | ++ | − |
| ATG20 | − | ++ | ++ |
| ATG21 | − | ++ | ++a |
| ATG22 | − | − | − |
| ATG23 | − | ++ | + |
| ATG24 | − | ++ | ++ |
| ATG26 | − | − | − |
| ATG27 | + | − | ++ |
| ATG29 | ++ | − | ++a |
| ATG31 | ++ | − | +a |
| ATG32 | − | − | ++ |
| ATG33 | − | − | + |
There is some discrepancy in the results from two independent reports (Kanki et al., 2009a; Okamoto et al., 2009a).
This gene encodes a lipase; the cargo can be delivered into the vacuole, but cannot be degraded.
Phenotypes of indicated gene knockout strain: ++, severe defect; +, partial defect; −, no defect.
Recently, it is reported that there is a mechanism for Atg5- and Atg7-independent macroautophagy in mammalian cells; during reticulocyte maturation, mitochondria can be eliminated through canonical macroautophagy, or in an Atg5- and Atg7-independent manner (Nishida et al., 2009; Zhang et al., 2009). At present, there are no data that demonstrate that mitophagy can occur independent of Atg5 or Atg7 in yeast.
ATG32 and ATG33
From the genomic screen for yeast mutants defective in mitophagy, ATG32/YIL146C and ATG33/YLR356W were identified as mitophagy-related genes (Kanki et al., 2009a,b; Okamoto et al., 2009a). Both ATG32 and ATG33 are mitophagy-specific genes that are not required for macroautophagy and other types of selective autophagy. The Atg32 protein is 59 kDa and is located in the mitochondrial outer membrane with its N- and C-terminal domains oriented towards the cytoplasm and the inter-membrane space, respectively. Similar to the Cvt pathway or pexophagy, mitophagy uses Atg11 as an adaptor protein for recognizing cargo. Atg32 appears to function as a mitochondrial receptor and binds Atg11 during mitophagy to recruit mitochondria to the PAS (Kanki et al., 2009b; Okamoto et al., 2009a). Interestingly, Atg32 has a WXXI/L/V motif that is present in yeast Atg19, and in the mammalian protein p62/SQSTM1, and which functions as an Atg8 or LC3 binding sequence, respectively (Okamoto et al., 2009a). Atg32 binds Atg8 through this motif, and this binding is required for efficient sequestration of mitochondria by the phagophore (Okamoto et al., 2009b).
The Atg33 protein is 20 kDa and also localizes in the mitochondrial outer membrane. Although the deletion of ATG33 blocks mitophagy to half the level of the wild type when induced by starvation, it blocks mitophagy almost completely when induced at stationary phase (Kanki et al., 2009a). This finding leads to the hypothesis that Atg33 is required to detect or present aged mitochondria for mitophagy when cells have reached the stationary phase.
Other genes affecting mitophagy in yeast
Two groups simultaneously utilized large-scale genetic screens for mitophagy-defective mutants in yeast and identified either 23 or 32 genes, including ATG32 and/or ATG33, which are completely or partially required for mitophagy (Kanki et al., 2009a; Okamoto et al., 2009a). Although half of the genes identified by these screens are involved with membrane trafficking pathways that impinge on autophagy-related pathways, the other half includes several genes encoding mitochondrial or cytosolic proteins. Some of these genes may be involved in the regulation of mitophagy, although further analyses are required. Interestingly, one screen identified DNM1 that encodes a mitochondrial dynamin-related GTPase required for mitochondrial fission (Kanki et al., 2009a). This finding is in agreement with previous reports that the fragmentation of mitochondria is a prerequisite for mitophagy in mammalian cells (Twig et al., 2008), and the dnm1Δ strain inhibits the mitophagy induced by mdm38 conditional knockout in yeast (Nowikovsky et al., 2007).
Induction and regulation of mitophagy in yeast
Because mitophagy contributes both to eliminating damaged mitochondria and reducing the amount of mitochondria to adjust its volume in accordance with cellular energy requirements, induction and regulation of mitophagy in yeast is not a simple pathway, nor is it totally understood. There are several lines of evidence from yeast that suggest damaged mitochondria are eliminated by mitophagy. For example, interference with F1Fo-ATPase biogenesis in a temperature sensitive fmc1 mutant (Priault et al., 2005), or osmotic swelling of mitochondria caused by depletion of the mitochondrial K+/H+ exchanger Mdm38 (Nowikovsky et al., 2007) induce mitophagy. On the other hand, mitochondrial depolarization caused by an uncoupler such as carbonyl cyanide m-chlorophenylhydrazone (CCCP) does not induce mitophagy in wild-type yeast (Kiššová et al., 2004; Kanki et al., 2009a), although mitophagy can be induced by the same stimulus in mammalian cells (Narendra et al., 2008; Sandoval et al., 2008). This finding suggests that induction of mitophagy in yeast requires not only mitochondrial depolarization but also an additional unidentified factor(s) or signalling.
Mitophagy can be induced by nitrogen starvation or treating with the TOR kinase inhibitor rapamycin after pre-culturing yeast in a non-fermentable medium that induces the proliferation of mitochondria (e.g. where lactate or glycerol are the sole carbon source) (Tal et al., 2007; Kanki and Klionsky, 2008; Kanki et al., 2009c). Although both non-specific macroautophagy and mitophagy can be induced by nitrogen starvation or rapamycin, these events may be regulated independently based on the following two lines of evidence: first, mitophagy is blocked under even strong macroautophagy-inducing nitrogen starvation conditions, if the carbon source makes mitochondria essential for metabolism (Kanki and Klionsky, 2008). Second, N-acetylcysteine (NAC), a compound that increases the cellular reduced glutathione (GSH) pool, prevents mitophagy induced by nitrogen starvation or rapamycin, presumably because the cellular redox imbalance affects mitophagy induction, but has no effect on non-specific macroautophagy (Deffieu et al., 2009; Kiššová and Camougrand, 2009). Mitophagy is also induced at stationary phase when yeast cells are cultured in a medium with a non-fermentable carbon source (Tal et al., 2007; Kanki and Klionsky, 2008; Okamoto et al., 2009a). In this case, it is thought that mitochondria are eliminated by mitophagy to adapt the cells to the stationary phase condition where the cellular energy requirement is reduced (Kanki and Klionsky, 2008).
Pathway for delivering mitochondria into the vacuolar lumen
In yeast, the PAS is usually formed next to the vacuole surface, and most of the Atg proteins accumulate at the PAS during selective and non-selective autophagy (Suzuki et al., 2007). In the Cvt pathway and mitophagy, both cargos, the Cvt complex and mitochondria, are delivered to the PAS by Atg11 before Cvt vesicle and mitophagosome formation, respectively. Although it is believed that non-selective autophagy and the Cvt pathway use the same site for biogenesis of the PAS, the Cvt pathway and mitophagy apparently use a different site (Kanki et al., 2009b); this fits with the concept that cargo are selectively sequestered during specific types of autophagy, and different cargo molecules or bulk cytoplasm are excluded from the resulting vesicles. The remainder of the mitophagy pathway is still controversial. Some researchers find microautophagy-like mitochondria uptake directly at the vacuole limiting membrane, based on electron microscopy (Kiššová et al., 2007; Nowikovsky et al., 2007). In contrast, others observe macroautophagy-like mitophagosomes in the cytosol (Kanki et al., 2009a,b; Okamoto et al., 2009a).
Summary of mitophagy
A summary of mitophagy in yeast is shown in Fig. 1. Mitochondrial damage caused by dysfunction of Mdm38 or Fmc1 reduces the mitochondrial membrane potential, which along with additional factors induces mitophagy. Nitrogen starvation or the TOR kinase inhibitor rapamycin also induce mitophagy, whereas the cellular pool of reduced glutathione, which affects the mitochondrial redox status, negatively regulates mitophagy induction by nitrogen starvation or rapamycin. Culturing cells to stationary phase in non-fermentable medium can also induce mitophagy. Atg33 is presumably required to detect or present aged mitochondria for degradation at stationary phase. Once mitophagy is induced, the mitochondrial protein Atg32 binds Atg11. Atg11 recruits mitochondria to the PAS, where mitophagy-specific uptake may occur. At the PAS, Atg32 binds Atg8. Atg20 and Atg24, which colocalize at the PAS (Nice et al., 2002), are required for mitophagy via an unidentified mechanism. The last step, delivering mitochondria into the vacuolar lumen, is unclear and may occur via microautophagy, macroautophagy or both.
Fig. 1.
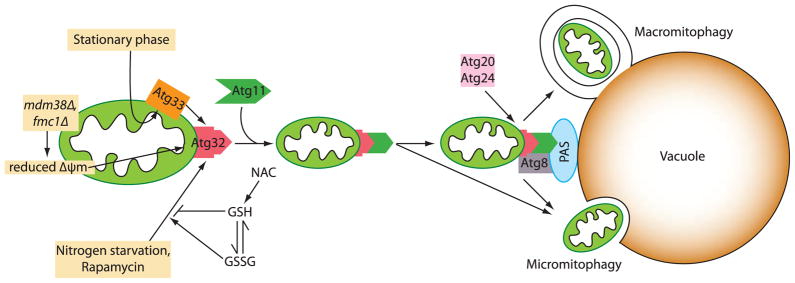
A schematic model of mitophagy in yeast. Mitophagy is induced by a reduction of the mitochondrial membrane potential, nitrogen starvation, rapamycin or entry into stationary phase. The reduced glutathione pool (GSH) inhibits mitophagy. Once mitophagy is induced, the mitochondrial receptor Atg32 binds the adaptor protein Atg11. Atg11 recruits mitochondria to the PAS where Atg32 can bind Atg8 to promote the generation of the phagophore, which encloses the mitochondria. Mitochondria may be delivered into the vacuole via a macroautophagy-like or microautophagy-like mechanism.
Future directions
Recent progress in mitophagy studies such as identification of Atg32 in yeast or characterization of Parkin and Nix in mammalian cells has uncovered some of the molecular mechanism of mitophagy. There are, however, many questions still to be addressed including the following: (i) Disappointingly, mammalian homologues or counterparts of the Atg proteins required for mitophagy (Atg11, 20, 24, 32 and 33) have not yet been identified. We think it is likely that there are analogues of these proteins; for example, Atg11 is required for all types of selective autophagy so far characterized in yeast. Accordingly, the identity of the mitochondrial receptor and adaptor for mitophagy in mammalian cells remains an open question. Similarly, it is not even known whether mammalian cells follow a similar pathway with yeast or utilize a completely different mechanism for autophagic mitochondria degradation. (ii) It is apparent that one of the physiological roles of mitophagy in mammalian cells is maintaining the quality of mitochondria, because mitochondria with an altered membrane potential are eliminated by mitophagy. Although mitochondrial damage caused by conditional mutants of MDM38 or FMC1 induce mitophagy, chemical disruption of the mitochondrial membrane potential does not appear to induce mitophagy to the same extent in yeast. In addition, deletion of the mitophagy-specific gene ATG32 does not affect cell growth on non-fermentable medium or increase cellular ROS production (Kanki et al., 2009b; Okamoto et al., 2009a). These findings make it unclear how mitophagy contributes to the quality control of mitochondria in yeast. Thus, further studies on the physiological role of mitophagy in yeast are needed. (iii) The initial step for mitochondria delivery to the vacuole is the interaction of Atg32 and Atg11. However, how this interaction is regulated and what triggers it are still unclear. The signalling pathway upstream of this interaction that connects nitrogen starvation or stationary phase stimuli to the mitophagy machinery remains to be identified.
Acknowledgments
This work was supported by Grant-in-Aid for Young Scientists Start-up (21890177) to T.K. and National Institutes of Health Grant GM53396 to D.J.K.
References
- Deffieu M, Bhatia-Kiššová I, Salin B, Galinier A, Manon S, Camougrand N. Glutathione participates in the regulation of mitophagy in yeast. J Biol Chem. 2009;284:14828–14837. doi: 10.1074/jbc.M109.005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Krick R, Subramani S, Thumm M. Turnover of organelles by autophagy in yeast. Curr Opin Cell Biol. 2009;21:522–530. doi: 10.1016/j.ceb.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009a;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009b;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Kang D, Klionsky DJ. Monitoring mitophagy in yeast: the Om45-GFP processing assay. Autophagy. 2009c;5:1186–1189. doi: 10.4161/auto.5.8.9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiššová I, Deffieu M, Manon S, Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- Kiššová I, Salin B, Schaeffer J, Bhatia S, Manon S, Camougrand N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autoph-agy. 2007;3:329–336. doi: 10.4161/auto.4034. [DOI] [PubMed] [Google Scholar]
- Kiššová IB, Camougrand N. Glutathione participates in the regulation of mitophagy in yeast. Autophagy. 2009;5:872–873. doi: 10.4161/auto.9065. [DOI] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem. 2002;277:30198–30207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Nowikovsky K, Reipert S, Devenish RJ, Schweyen RJ. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 2007;14:1647–1656. doi: 10.1038/sj.cdd.4402167. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009a;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. A landmark protein essential for mitophagy: Atg32 recruits the autophagic machinery to mitochondria. Autophagy. 2009b;5:1203–1205. doi: 10.4161/auto.5.8.9830. [DOI] [PubMed] [Google Scholar]
- Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 2005;12:1613–1621. doi: 10.1038/sj.cdd.4401697. [DOI] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem. 2007;282:5617–5624. doi: 10.1074/jbc.M605940200. [DOI] [PubMed] [Google Scholar]
- Tolkovsky A. Mitophagy. Biochim Biophys Acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Zhang J, Randall MS, Loyd MR, Dorsey FC, Kundu M, Cleveland JL, Ney PA. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114:157–164. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qi H, Taylor R, Xu W, Liu LF, Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007;3:337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]


