SUMMARY
Genome-wide transcriptional profiling was used to characterize the molecular underpinnings of neocortical organization in rhesus macaque, including cortical areal specialization and laminar cell type diversity. Microarray analysis of individual cortical layers across sensorimotor and association cortices identified robust and specific molecular signatures for individual cortical layers and areas, prominently involving genes associated with specialized neuronal function. Overall, transcriptome-based relationships were related to spatial proximity, being strongest between neighboring cortical areas and between proximal layers. Primary visual cortex (V1) displayed the most distinctive gene expression compared to other cortical regions in rhesus and human, both in the specialized layer 4 as well as other layers. Laminar patterns were more similar between macaque and human compared to mouse, as was the unique V1 profile that was not observed in mouse. These data provide a unique resource detailing neocortical transcription patterns in a non-human primate with great similarity in gene expression to human.
INTRODUCTION
The mammalian neocortex is characterized by its stereotyped laminar cytoarchitecture and regional variations in cellular architecture that differentiate cortical areas. As emphasized by Brodmann over a century ago through the creation of cytoarchitectonic cortical maps (Brodmann, 1909), cortical organization is conserved across species, particularly between humans and non-human primates (reviewed in (Zilles and Amunts, 2010)). Gene expression is increasingly used as an empirical means of differentiating and delineating cortical areas, for example through identification of area-specific gene markers (Takahata et al., 2009), or boundary mapping based on differences in neurotransmitter receptor expression (Zilles et al., 2004). Whole genome transcriptional profiling has particular potential to elucidate cortical areal specification and specialization through identification of differentially regulated genes and molecular pathways that underlie cytoarchitectural and functional areal identity (Johnson et al., 2009).
The major factor that differentiates different cortical areas is their distinct laminar organization, reflecting the composition of specific cell types within each layer. Work in rodents has shown that the specific cell types that make up different cortical layers have robust and selective molecular signatures. Many gene markers have been identified through mining genome-wide cellular resolution gene expression data resources in the Allen Mouse Brain Atlas (Lein et al., 2007)(http://www.brain-map.org) and by using targeted approaches (Molyneaux et al., 2007). In addition, transcriptional profiling using DNA microarrays or RNA sequencing has been successful in identifying molecular signatures for discrete cortical layers in mice (Belgard et al., 2011; Hoerder-Suabedissen et al., 2009; Wang et al., 2009) using punches or laser microdissection, as well as in specific excitatory and inhibitory cortical cell types using selective genetic or tracer-based cell labeling and live isolation methods (Arlotta et al., 2005; Doyle et al., 2008; Sugino et al., 2006).
In contrast, other studies aiming to identify cortical area-enriched gene expression in humans and non-human primates were performed using macrodissected whole cortex, which yielded few genes that robustly differentiate between cortical areas (Khaitovich et al., 2004; Yamamori and Rockland, 2006). One likely reason for this is methodological variability associated with regional dissections, as precise dissections have yielded significantly more regional differences in the Vervet neocortex (Jasinska et al., 2009) and in developing and adult human brain (Johnson et al., 2009). Additionally, since gene markers differentiating cortical areas have been readily identified in mouse via cellular resolution in situ hybridization databases (Lein et al., 2007), the paucity of areal gene markers identified in primate transcriptional profiling studies might be due to dilution effects resulting from the high degree of cellular heterogeneity in whole cortical samples. Therefore, a more precise approach targeting more homogeneous cortical cell populations may reveal more robust areal signatures as well.
Rhesus macaque provides a tractable non-human primate model system to analyze the transcriptional organization of the primate neocortex. Macaque is genetically and physiologically similar to humans, with a sequence identity of approximately 93% (Gibbs et al., 2007). Many elements of cortical cytoarchitecture are similar in macaque and human, including specialized primary visual cortex and dorsal and ventral visual streams. In this study, we aimed to understand organizational principles of the primate neocortex using transcriptional profiling analysis of individually isolated cortical layers from a variety of well-defined cortical regions in the adult rhesus macaque, and to compare rhesus gene expression patterns in homologous cortical areas and cell types in human and mouse. The entire microarray data set is also available through the NIH Blueprint NHP atlas website (http://blueprintnhpatlas.org).
RESULTS
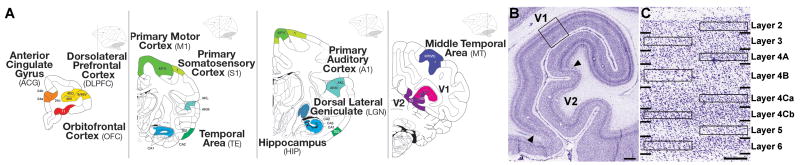
To analyze transcriptional profiles associated with major laminar and areal axes of cortical organization, laser microdissection (LMD) was used to selectively isolate individual cortical layers in ten discrete areas of the neocortex from two male and two female adult rhesus monkeys. As shown schematically in Figure 1A, these areas spanned primary sensorimotor cortices (S1, M1, A1, and V1), higher order visual areas (V2, MT and TE), and frontal cortical areas (DLPFC, OFC and ACG). In each cortical region, samples were isolated from layers definable on the basis of lightly stained Nissl sections used for the sample preparation, taking care to avoid layer boundaries. In most areas, 5 layers were isolated (L2, L3, L4, L5 and L6), although in M1, OFC and ACG no discernible L4 could be isolated. 8 layers were sampled in V1 (Figure 1B, C) to include the functionally specialized and cytoarchitecturally distinct sublayers of L4 (4A, 4B, 4Ca and 4Cb). For a non-neocortical comparator data set, samples were also isolated from subfields of the hippocampus (CA1, CA2, CA3 and dentate gyrus) and from the magno-, parvo- and koniocellular layers of the dorsal lateral geniculate nucleus (LGN). Collectively, the selected regions allowed for interrogation of differences in gene expression between cortical areas and layers located distal or proximal to each other, and from regions that comprise specific functional types or streams. Representative pre- and post-cut images from each structure are shown in Figure S1, and stereotaxic locations of sampled cortical regions in Table S1. RNA was isolated from LMD samples, and 5ng total RNA per sample was amplified to generate sufficient labeled probe for use on Affymetrix rhesus macaque microarrays.
Figure 1. Experimental Paradigm for Profiling Macaque Cortical Regions and Layers.
(A) Reference atlas plates from rostral (upper left) to caudal (lower right) illustrating approximate locations of ten cortical regions targeted for microarray profiling, as well as the hippocampus and thalamic dorsal lateral geniculate nucleus.
(B) A Nissl-stained section of visual cortex centered on the calcarine fissure, corresponding to the lower right panel in (A).
(C) High magnification image of V1 (boxed area in (B)) showing detailed laminar architecture (numbers) and approximate delineations used for conservative isolation of individual cortical layers using laser microdissection (boxes). V1, primary visual cortex; V2, secondary visual cortex. Scale bars: 1mm in B, 250μm in C. Panels in (A) modified with permission from Paxinos, Huang and Toga (Paxinos et al., 2000). Detailed anatomical sampling locations shown in Figure S1, and representative sample isolations in Table S1.
Multiple analytical methods were used independently to identify the most robust patterns of gene expression. Principle component analysis (PCA) can often illustrate the major organizational features of microarray data sets (Colantuoni et al., 2011), and we initially applied it to the whole sample set comprising 225 cortical, hippocampal and thalamic samples across all 52,865 probes. A significant proportion of the variance was accounted for by the first three components (12.5%, 8.7% and 6.8% respectively; Figure S2). As shown in Figure 2A, samples from major structures (cortex, hippocampus, and thalamus) cluster together, have highly distinct molecular signatures and appear well-segregated. Considering the cortical samples alone, the first three components accounted for a similar proportion of variance (13.6%, 8.5% and 6.6% respectively), and plotting samples by areal or laminar class revealed striking organization along two orthogonal axes reflecting the areal (Figure 2B) and laminar (Figure 2C) dimensions of the neocortex. Remarkably, the spatial relationships between neocortical samples are recapitulated by the transcriptional relationships between samples. Samples align in a rostral to caudal orientation by cortical area along the first principle component horizontally in Figure 2B, and appear tightly clustered in their native laminar order along the second principle component vertically in Figure 2C.
Figure 2. Major Organizational Features of the Rhesus Neocortical Transcriptome.
(A-C) PCA of cortical, hippocampal and thalamic samples (A) or cortical samples alone colored by cortical region (B) or layer (C). The first three components in (A) accounted for 12.5%, 8.7% and 6.8% of variance respectively. The first three components in B and C accounted for 13.6%, 8.5% and 6.6% respectively.
(D) 3-way ANOVA of the cortical samples showing number of differentially expressed genes versus statistical significance for cortical regions, layers and animals. The high degree of overlap between genes showing differential expression across these three variables is shown in the Venn diagram in E (ANOVA p< 10-12 for each variable).
(F) Cluster dendrograms representing probe assignment to modules in WGCNA.
(G) Module eigengene plots for a male-enriched module (lightyellow, top panel) and an individual animal-related module (grey60, lower panel). Samples in (G) are organized by cortical region from caudal (V1) to rostral (ACG), and by layer within each region from superficial L2 to deep L6). Male samples in upper panel have grey background. Samples in lower panel are identified by animal (#1-5), and all samples from animal #2 have light green background. PCA and ANOVA details are provided in Figure S2 and Table S2.
To identify differentially expressed genes, three-way ANOVA of the cortical data set identified large numbers of probes that vary between cortical regions (6,170 at p<10-12), layers (4,923) and individual animals (2,347; Figure 2D, E and Table S2). Importantly, there was a high degree of overlap between the sets of genes varying by cortical region and layer, suggesting that a substantial proportion of the genes differentiating cortical areas vary within specific cortical layers. Gene set analysis of both areal and laminar genes showed enrichment for genes associated with axonal guidance signaling and ephrin receptor signaling, synaptic long term potentiation (LTP) and neuronal activities (Table S2). Gene expression patterns associated with gender and individual animals were also identified by ANOVA (Figure S2), and individual-associated differences were enriched with genes related to metabolism, mitochondria and antigen presentation (Table S2). Gender-specific gene expression was observed both on sex and autosomal chromosomes (Fig S2), and there was significant overlap (p<10-9) between the individual-related genes identified here and gender-related genes identified in human brain (Kang et al., 2011).
We next applied WGCNA to identify sets, or modules, of highly co-expressed genes by searching for genes with similar patterns of variation across samples as defined by high topological overlap (Zhang and Horvath, 2005). Applied to the entire set of neocortical samples, WGCNA revealed a series of gene modules (named here as colors) related to different features of the data set (Figure 2F,G, also Figure 3B,D and Figure 5B,C). Gene assignment to modules and gene ontology analysis for the whole cortex network are shown in Table S3. The majority of these modules correlated with laminar and regional patterns as described below. Several modules were related to gender and individual differences, as previously observed in humans (Oldham et al., 2008). In Figure 2G the lightyellow module was strongly enriched in male versus female samples (upper panel), while the grey60 module was selectively lowest in samples originating from one particular animal. The top (hub) genes in the lightyellow module were on the Y chromosome, including the putative RNA helicaseDDX3Y and the 40S ribosomal protein RPS4Y1.
Figure 3. Robust Transcriptional Signatures of Cortical Laminar Structure.
(A) 1D clustering of genes showing differential laminar expression (ANOVA p< 10-12) across all cortical samples, with selected enriched gene sets for each cluster (BF corrected p<.01).
(B) Module versus layer relationships based on whole cortex WGCNA, with individual modules showing strong correlations to individual cortical layers (red indicates high correlations). Red arrows under module names indicate modules shown in panel C.
(C) Identification of genes selectively expressed in specific cortical layers. Displayed are genes with correlation coefficient >0.7 (L2) or >0.6 (L3-6) to artificial templates (red bars) in each layer across all cortical areas (see Methods).
(D) Layer-enriched network modules. Plotted are the top 30 (black, salmon) or top 20 (greenyellow, royalblue, red) hub genes from 5 modules showing different patterns of laminar enrichment. Individual gene profiles in C and D were normalized by the mean expression value for that gene for display on same scale. ANOVA (E) and WGCNA (F) of V1 samples only.
(E) 2D clustering of genes showing differential laminar expression among V1 samples (ANOVA, p<10-3), with selected gene set enrichment results (BF corrected p<0.1).
(F) Module versus layer relationships based on V1 WGCNA. Dendrograms in B, E, F show strongest relationships between proximal layers, and a distinct signature associated with the specialized L4 sublayers of V1. Module assignment and gene set enrichment for WGCNA, ANOVA and template analyses are provided in Tables S3-8.
Figure 5. Molecular Signatures of Cortical Regions.
(A) 2D cluster of genes differentially expressed between cortical regions (ANOVA, p<10-12), with selected enriched gene sets for specific clusters (BF corrected p<.01).
(B) Module versus region relationships based on whole cortex WGCNA. Individual modules show strong correlations to subsets of cortical regions, and proximal regions show the strongest similarity.
(C) Module eigengene plots for tan (upper left, caudal to rostral high gradient), purple (upper right, rostral to caudal high gradient), blue (lower left, V2, MT, DLPFC, OFC high), and pink (TE, A1, ACG high) modules. Further ANOVA is provided in Table S9 and Figure S3.
Cortical laminar variation
The most striking features were the robust molecular signatures associated with different cortical layers. As shown in Figure 3, a wide variety of transcriptional patterns were associated with individual cortical layers or subsets of layers. For example, ANOVA of laminar expression in all cortical regions and clustering of these genes identified large gene sets enriched in specific subsets of (generally proximal) layers (Figure 3A). Notably, the majority of these laminar patterns are consistent across different cortical areas, reflecting conserved laminar and cellular architecture across the cortex. Gene set analysis suggests these layer-associated clusters are associated with neuronal function, including neuronal activity, LTP/LTD, calcium, glutamate and GABA signaling (Figure 3A and Table S4). Consistent with functional studies of superficial layer synaptic plasticity, genes and pathways involved in LTP and calcium signaling were most represented in L2 and L3. Pathways related to cholesterol metabolism were enriched in deeper layers, likely reflecting the greater proportion of oligodendrocytes closer to the underlying white matter. Similarly, many of the gene modules identified through WGCNA of all cortical samples were correlated with specific cortical layers (Figure 3B). By ANOVA-based clustering and WGCNA, proximal layers showed the strongest correlations, with superficial L2 and L3 highly correlated with one another, and the deeper L4-6 highly correlated as well (dendrograms in Figure 3B, E,F).
Individual layers showed highly specific gene expression signatures. Layer-enriched expression patterns were identified by searching for genes with high correlation to layer-specific artificial template patterns (Lein et al., 2004)(Table S5). Figure 3C shows cohorts of genes with remarkably layer-specific expression that was relatively constant across all cortical areas. These observations demonstrate the specificity of the laminar dissections with minimal interlaminar contamination, and also the constancy of laminar gene expression across the neocortex.
WGCNA gene modules derived from the whole cortex network also showed highly layer-enriched expression, demonstrating the robustness of our findings. For example, the black module contains genes enriched in superficial L2 (hub genes plotted in Figure 3D, top row). While some layer-specific genes could be identified by targeted analyses, the dominant patterns were more complex, with most network modules being associated with combinations of layers, typically proximal to one another. For example, individual modules were enriched in L2-4 (salmon), L3-5 (greenyellow), L4-5 (royalblue) and in a gradient increasing from L2 to L6 (red). This tendency for coexpression between adjacent layers is also apparent in the heatmap representation of gene clusters in Figure 3A and 3E. Gene ontology (GO) analysis of these modules provides some insight into their functional relevance (Table S3). The greenyellow module was enriched for genes associated with axons and neuron projections, potentially related to long-range pyramidal projection neurons in L3 and L5. The red module showed enrichment in genes associated with myelination, consistent with the presence of oligodendrocytes in deep layers, and this module was highly correlated with oligodendrocyte-associated gene networks in other studies (data not shown, (Oldham et al., 2008)).
Interestingly, the expanded L4 of V1 displayed a distinct signature from the rest of L4 (see top of middle box in Fig 3A). To explore this further, we performed ANOVA and WGCNA selectively on samples from V1 (Figure 3E, F and Tables S6 and S7). A comparison between V1 ANOVA-derived laminar differential expression and membership in whole cortex WGCNA modules is in Table S8. Similar to the whole cortex analysis, robust clusters and network modules were associated with individual cortical layers. As shown in the unsupervised hierarchical 2D clustering of ANOVA results in Figure 3E, individual samples from each layer cluster together, and neighboring cortical layers are most similar to one another. Interestingly, L4A clusters with more superficial layers, while L4B, L4Ca and L4Cb display a distinct transcriptional pattern, most easily seen by the dendrograms based on ANOVA and network analysis in Figures 3E and 3F.
To investigate whether layer specificity of gene expression may relate to selective patterns of connectivity, we examined the relationship between thalamocortical inputs and their targets in V1. L4Ca and L4Cb receive input selectively from magnocellular (M) and parvocellular (P) divisions of the LGN, respectively. Hypothesizing that there may be substantial shared gene expression patterns selective for specific pairs of connected neurons, we searched for genes that were differentially expressed between the thalamic inputs and between the cortical targets. 1,002 probes were differentially expressed between L4Ca and L4Cb (t-test, p<.01), and 825 probes between M and P. Surprisingly, these gene sets did not significantly overlap (13/1827, p=.08). Although the possibility certainly exists that specific ligand-receptor pairs are associated with this selective connectivity, it would appear that the specificity of these connections is not associated with specific large-scale correlated gene expression patterns.
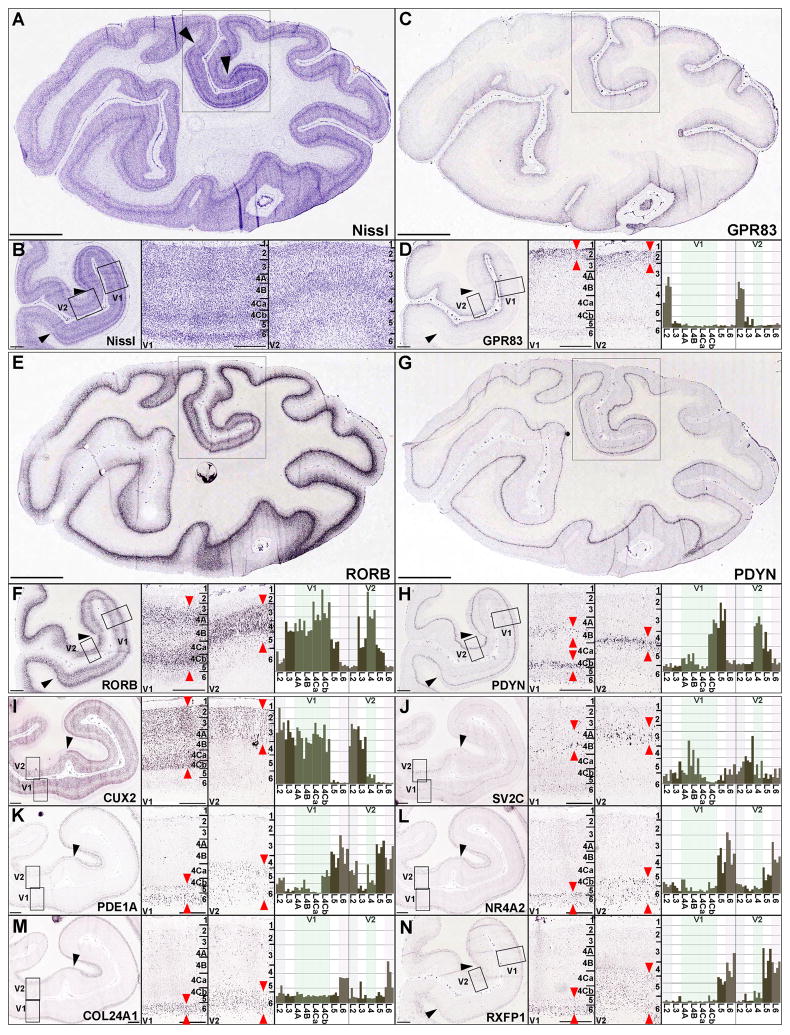
To validate the specificity of the microarray findings and test hypotheses about laminar enrichment based on ANOVA and WGCNA, we examined a set of genes displaying layer-enriched patterns using in situ hybridization (ISH) in areas V1 and V2 (Figure 4). Overall the laminar specificity of gene expression and variations between cortical areas predicted by microarrays were confirmed by cellular-level analysis, and illustrate the high information content of layer-specific expression profiling and gene specificity of the microarray probesets. For example, GPR83 is selectively expressed in L2 of all cortical areas, both by microarray and ISH analysis (Figure 4C, D). Laminar specificity was confirmed for RORB (L3-5; Figure 4E, F), PDYN (L4-5; Figure 4G, H), CUX2 (L2-4; Figure 4I), and SV2C (L3-4 enriched; Figure 4J). Specificity for deep cortical layers was prominent, as shown for PDE1A (L5-6; Figure 4K), NR4A2 (L5-6; Figure 4L), COL24A1 (L6; Figure 4M) and RXFP1 (L5-6; Figure 4N). Differences in laminar specificity were sometimes apparent between V1 and V2 (generally V1 versus all other areas); CUX2 was expressed in L2 through L4Cb in V1 but more limited to L2 and L3 in V2 (Figure 4I), and SV2C was highest in L4B in V1, but highest in L3 in area V2 (Figure 4J).
Figure 4. Layer-specific Gene Expression in Visual Cortical Areas.
(A and B) Coronal Nissl-stained section of caudal macaque neocortex containing areas V1 and V2 at low magnification (A) and higher magnifications (B). Left panel in (B) corresponds to boxed region in (A); right panels show high magnification views of areas V1 and V2 corresponding to boxes in left panel.
(C, E, G) Low magnification images of ISH labeling for GPR83 (C), RORB (E) and PDYN (G).
(D, F, G) Corresponding medium magnification image (left panel), high magnification views of V1 and V2 (middle panels), and histogram plots of gene expression for layer-specific samples in V1 and V2 (4 replicates per layer, right panels).
(F-N) ISH and microarray data for CUX2 (I), SV2C (J), PDE1A (K), NR4A2 (L), COL24A1 (M), and RXFP1 (N), displayed in the same format as upper panels. Black arrowheads delineate boundaries of V1, numerals in high magnification ISH and Nissl panels denote laminar architecture of V1 and V2, and red arrows denote regions of enriched expression. Scale bars: Low magnification images in A, C, E, G: 5mm; medium magnification images in B, D, F, H, I, J, K, L, M, N: 1mm, high magnification images: 500μm.
Cortical areal variation
Both ANOVA (Figure 5A) and WGCNA analysis (Figure 5B) identified gene clusters enriched in specific subsets of cortical regions. As illustrated in the dendrograms from both methods, the strongest relationships between cortical areas were based on areal proximity rather than functionally connectivity. For example, the caudal visual areas V1, V2 and MT showed highly correlated patterns of gene expression, while the functionally related but distal visual region TE had greater transcriptional similarity to its proximal neighbor A1 in temporal cortex. Strong relationships were observed for the adjacent primary motor and sensory cortices M1 and S1, and for the frontal DLPFC and OFC regions. Differentially expressed genes showed enrichment in specific subsets of (generally proximal) cortical areas (Figure 5A), generally related to neuronal development and function (axon guidance, neuronal activities, LTP/LTD; Table S9). Areal expression also had a strong laminar signature, easily visualized by grouping these ANOVA-derived genes by cortical layer (Figure S3).
Parallel relationships between cortical areas were observed by WGCNA demonstrating the robustness of these observations (Figure 5B), with individual gene modules showing enrichment in specific cortical regions (Figure 5C). Module eigengenes revealed additional patterning, including rostrocaudal gradients and laminar components to areal patterning. For example the tan module (Figure 5C, upper left) reflected a caudal low to rostral high patterning enriched in deep L5 and L6. Another gene module (purple, upper right) had an opposite gradient from high caudal to low rostral, in this case enriched in L3 and L4. Other modules were more area-specific: in V2, MT, DLPFC and OFC (blue) or lowest in V1, V2 and MT, with enrichment in L2 and L3 (pink).
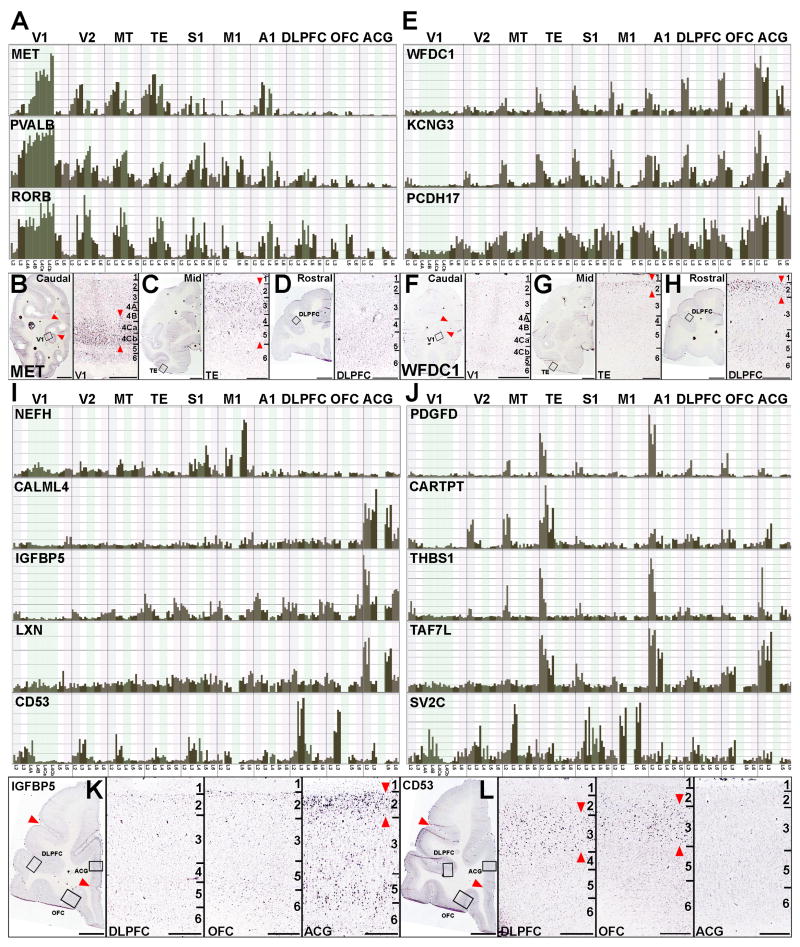
Individual genes showed a wide range of areal patterns reflecting the modules described above, as well as patterns related to individual cortical areas or combinations of areas. Example gene patterns derived from the clustering analyses above, as well as analysis of the genes showing maximal cross-area fold changes, are shown in Figures 6 and 8. A large cohort of genes displayed rostrocaudal gradients. For example, MET, PVALB and RORB were expressed most strongly in caudal V1 and decreased moving rostrally. Typically this gradient expression also had a laminar component. For example, MET, which has been associated with autism (Campbell et al., 2006) was expressed preferentially in L4 of V1, expanded to include L2-4 in temporal areas, and had no detectable expression in frontal cortices (Figure 6A-D), consistent with observations in fetal human brain (Mukamel et al., 2011). Conversely, WFDC1 (Figure 6E-H) was expressed most strongly in frontal cortical areas (e.g. DLPFC) and lowest in caudal V1, with expression primarily restricted to L2.
Figure 6. Diversity of Regional Gene Expression Patterns.
(A) Caudal high to rostral low gradients. Normalized expression histograms for MET, PVALB, and RORB across cortical areas and layers.
(B-D) ISH for MET in caudal (B), mid rostrocaudal (C) and rostral (D) sections at low (left panels) and high magnification (right panels, corresponding to boxed areas in left panels). E. Caudal low to rostral high gradients, with normalized expression histograms for WFDC1, KCNG3 and PCDH17.
(F-H) ISH for WFDC1 in caudal (F), mid rostrocaudal (G) and rostral (H) sections at low (left panels) and high magnification (right panels, corresponding to boxed areas in left panels). I. Genes displaying robust region-specific expression, including NEFH in M1, CALML4, IGFBP5, and LXN in ACG, and CD53 in DLPFC and OFC. J. Complex regional gene expression patterns for PDGFD, CARTPT, THBS1, TAF7L, and SV2C. K, L. ISH for frontal cortex enriched genes in (I), including selective IGFBP5 expression in ACG relative to DLPFC and OFC (K) and selective expression of CD53 in DLPFC and OFC relative to ACG (L). High magnification panels correspond to boxed regions in low magnification panels. Red arrowheads in low magnification images in B and F delineate boundaries of area V1. Red arrowheads in high magnification ISH figures delineate predominant laminar patterns. Scale bars: low magnification in B-D, F-H, K, L: 5mm; high magnification: 500μm.
Figure 8. V1-selective Macaque Patterns of Gene Expression and Cross-species Comparisons.
(A-F) Microarray (lower panels) and ISH (upper panels) analysis of genes enriched (A-C) or lowest (D-F) in macaque V1.
(A-C) Enriched expression in V1 for ASAM, VAV3 and ESRRG, primarily in L4. Low magnification ISH centered on calcarine sulcus (left), and high magnification images of V1 and V2 corresponding to boxed areas. Layers showing higher expression in V1 are delineated by red arrows in ISH panels. Lower panels show sample-level histogram plots of normalized expression intensities across cortical areas (V1 samples delineated by red boxes).
(D-F) Genes selectively lowest in V1, including MEPE (D), RBP4 (E) and CTGF (F). Areas of high expression are denoted by red arrows in V2 by ISH.
(G-H) Comparative analysis of V1-selective ISH patterns in macaque (upper panels), human (lower left) and mouse (lower right). Upper panels: ISH (upper) and microarray (lower) analysis of gene expression at low (left) and high magnification (V1, V2, right panels) for SYT6 (G), NPY2R (H) and HTR2C (I). Note higher expression (red arrows) in V1 for SYT6 and NPY2R, and lower expression in V1 for HTR2C. Lower panels in G-I show corresponding ISH data for adult human V1 and V2 and adult mouse V1 and S1. Red arrows delineate patterns with similar laminar and areal expression to macaque, while blue arrows denote differential expression between rodents and primates. Large black arrowheads in A, E, G, H, I delineate boundaries of area V1. Scale bars: Low magnification: 5mm; high magnification: 500μm (macaque, human), 250μm (mouse).
Many genes showed area-selective expression (Figure 6I-L). NEFH, an intermediate filament heavy chain subunit generally expressed in long range projection neurons, was selectively enriched in L5 of M1 where the longest, spinally-projecting neurons, Betz cells, are located. Other genes were selectively enriched in specific regions of frontal cortex, including ACG (CALML4, IGFBP5, and LXN) or DLPFC and OFC (CD53). ISH showed selective enrichment of IGFBP5 in ACG compared to DLPFC and OFC (Figure 6K). Conversely, ISH analysis of CD53 (Figure 6L) showed enrichment in dorsolateral, ventrolateral and ventromedial cortex relative to dorsomedial cortex and ACG.
Comparative analysis of laminar and areal expression patterns
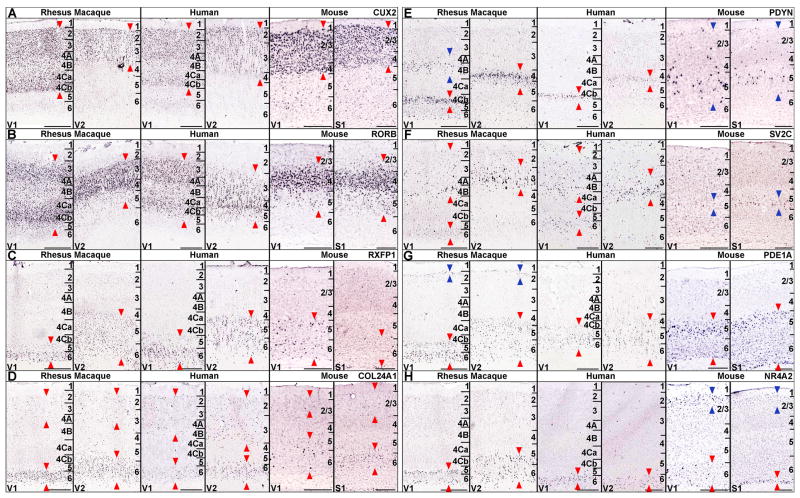
The laminar and areal patterning observed in macaque was then compared to homologous structures in human and mouse. This phylogenetic comparison is shown in Figure 7, made possible by the availability of mouse and human ISH data in the Allen Mouse Brain Atlas (http://mouse.brain-map.org) and Allen Human Brain Atlas (http://human.brain-map.org) generated using the same ISH technology platform as the macaque data. Some macaque data was also derived from the NIH Blueprint Non-Human Primate Atlas (www.blueprintnhpatlas.org). Most genes in Figure 4 above showed laminar expression patterns that were highly conserved between macaque and human, with lesser conservation in mouse. Laminar patterns were generally conserved between primates and mice in visual cortex for CUX2, RORB, RXFP1 and COL24A1 (left panels in Figure 7), although RXFP1 in mouse showed some regional differences between visual and somatosensory cortices not apparent in the macaque (Figure 7C). Other genes with highly laminar patterns showed major differences between species, indicated by blue arrowheads in Figure 7E-G. PDYN (Figure 7E) was enriched in excitatory neurons in L4 and L5 in rhesus and human, but in neurons with a broader laminar distribution that co-label with GAD1 indicating expression in GABAergic interneurons in mice (data not shown). The synaptic vesicle protein SV2C was predominantly enriched in superficial L3 pyramidal neurons in most cortical areas in primates (e.g. V2), while it is fairly selective for deep L5 pyramidal neurons in mice. NR4A2 was expressed selectively in deep L5 and L6 in macaque and human, and in both L6 and L2/3 in mice (Figure 7H).
Figure 7. Cross-species Comparison of Laminar Gene Expression Patterns Between Adult Macaque, Human and Mouse Neocortex.
ISH data for CUX2 (A), RORB (B), RXFP1 (D), COL24A1 (D), PDYN (E), SV2C (F), PDE1A (G), and NR4A2 (H). Left panels show cellular expression patterns in macaque V1 and V2, middle panels show expression in human V1 and V2, and right panels show expression in P56 mouse V1 and S1.
(A-D) Genes displaying predominantly similar laminar expression patterns across all three species.
(E-H) Genes displaying qualitative differences in laminar patterns between macaque, human and mouse. Red arrows delineate macaque expression and conserved expression in human and mouse. Blue arrows delineate expression showing qualitative differences between species. Scale bars: 500μm (macaque, human), 250μm (mouse). Human ISH data from the Allen Human Brain Atlas, and mouse ISH data from the Allen Mouse Brain Atlas.
While laminar distributions between V1 and V2 were highly conserved between macaque and human, differences were also noted. PDYN was expressed in L4B in macaque V1 in addition to the dominant L4Cb/5 expression (Figure 7E), while in human only L 4Cb/5 expression was observed. Similarly, faint L2 expression in addition to L5/6 enrichment was observed for PDE1A in macaque but not human (Figure 7G). In both cases, only the fainter expression in macaque was not observed in human, leaving the possibility that these differences relate less to true biological differences than to detection sensitivity on postmortem human tissues as compared to rapidly frozen rhesus macaque specimens.
The most robust patterns of areal specificity, both in terms of numbers of genes and their relative fold differences, were related to the highly specialized area V1 (Figure 8). Both selective enrichment in V1 and selective lack of expression in V1 were observed, with a sharp boundary corresponding to the cytoarchitectural boundary observed by Nissl staining. This areal patterning was typically restricted to particular cortical layers as well. Some of this selective expression related to the expanded input L4 in V1. For example, ASAM, VAV3 and ESRRG were enriched primarily in L4 of V1 (Figure 8A-C). However, selective enrichment or decreased expression was seen in all cortical layers, including L2 and L3 (MEPE and RBP4; Figs. 8D, E), L5 (HTR2C; Figure 8I), and L6 (CTGF, SYT6 and NPY2R; Figures 8F-H).
The V1-selective patterning appeared to be highly conserved between macaque and human, while significant differences were observed between primates and mice (Figure 8G-I). For example, the enrichment of SYT6 (Figure 8G) and NPY2R (Figure 8H) in L6 of V1 relative to V2 was conserved between macaque and human, as was absence in L5 of V1 for HTR2C (Figure 8I). NPY2R expression showed a completely different pattern in mice, restricted to sparse (presumably GABAergic) neurons scattered across the cortex. Conversely, for both SYT6 and HTR2C, laminar restriction to L6 and L5 respectively was conserved in mice, but with no selective enrichment or lack of expression in V1. Thus these V1-specific gene expression differences correlate with primate-specific cytoarchitectural and functional specialization, rather than with the functional sensory modality subserved by visual cortex.
DISCUSSION
The basic laminar structure of the neocortex is highly conserved across mammalian species, reflecting a general preservation of the constituent cell types and local circuitry (Brodmann, 1909). However, the specifics of laminar structure of the neocortex vary across both cortical region and species, with primates showing both a general expansion of superficial cortical layers and a massive expansion of cortical area with particular functional and cytoarchitectural specializations that is most dramatic in humans (Krubitzer, 2009). Understanding molecular differences between cortical layers and cell types across cortical regions, and the degree to which gene regulation is similar in homologous structures in humans and model organisms, may help explain features of cortical structure and function and the gene networks that underlie them. The current study aimed to examine organizational principles of the transcriptome of non-human primate neocortex by first determining the whole-genome mRNA profiles of adult rhesus macaque across cortical areas and individual layers within each area. We then performed a phyletic comparison of these laminar and areal gene expression patterns to those of human and mouse cortices. Our analyses show that these data provide a rich platform for species-specific phenotype discovery.
Individual Cortical Layers Display Highly Discrete Molecular Signatures
Individual layers of macaque cortex have highly distinct patterns of gene expression sufficient to unequivocally identify and cluster samples together based on the layer of origin across independent biological replicates, similar to recent observations in mouse cortex (Belgard et al., 2011) and validating the sampling strategy of profiling individual layers. These differential laminar patterns are both robust and widespread. For example, 5,537 probes showed differential expression among cortical layers in V1 where the greatest number of sublayers were assayed (ANOVA p<.01), with 3,353 genes showing at least 2-fold difference between two layers and 4,980 genes showing >1.5 fold differences. These data appeared to be highly reliable, as these laminar differences were consistent across animals and were readily confirmed by ISH. Very small numbers of individual genes show layer-specific expression; rather, most laminar genes appeared to be expressed in more complex patterns most often involving enrichment in multiple proximal layers. Interestingly, gene set annotation of different laminar gene clusters tended to give similar results for each cluster, namely enrichment in gene categories associated with neuronal development (including axon guidance), activity and plasticity. This suggests that the specific properties of particular neuronal subtypes are the product of unique combinations of members of the same gene families underlying those properties (e.g. ion channels, neurotransmitter receptors, axon guidance molecules, etc).
Network-based analytical techniques can be very effective at extracting biologically relevant information embedded in large, high-dimensional data sets. We applied WGCNA to identify groups of co-regulated genes, or “modules,” with high topological overlap across the data set (Langfelder et al., 2008; Zhang and Horvath, 2005). This approach has been successful at identifying biologically interpretable gene clusters in brain microarray data, including the identification of gene modules related to neural cell types (Oldham et al., 2008) and subcellular compartments (Winden et al., 2009). Applied either to the entire cortex data set or selectively to laminar samples from V1, many gene modules were identified whose expression was enriched in particular cortical layers. Individual modules were either enriched in specific layers (e.g. black module enriched in L2), or more commonly across multiple contiguous layers. Similar to previous WGCNA of human neocortex (Oldham et al., 2008), a robust oligodendrocyte module was identified with a laminar pattern similar to their cellular distribution from low numbers in superficial layers to higher in deep layers approaching the white matter.
Cortical Specializations Have Distinct Molecular Signatures
Cytoarchitectural variation in cortical laminar architecture and cellular makeup have been the basis for parcellation of cortical areas for over a hundred years (Brodmann, 1909), yet identification of genes with clear areal specificity has proven to be remarkably difficult (Yamamori and Rockland, 2006). V1 in primates is easily distinguished by the relative expansion and specialization of the input L4 compared to other areas, and most genes described with areal specificity thus far differentiate primary visual cortex from other areas. For example, OCC1 (FSTL1) was identified as a V1-enriched transcript, which is additionally regulated by light-driven activity through direct retino-thalamo-cortical activation (Takahata et al., 2009). Importantly, the majority of studies to date have used samples containing the entire neocortex from a particular brain region (Abrahams et al., 2007; Johnson et al., 2009; Khaitovich et al., 2004; Takahata et al., 2009; Watakabe et al., 2009). This type of design, while permitting analysis of broad cell classes (Oldham et al., 2008), likely under-represents differential areal gene expression through a dilution effect due to the high degree of cellular heterogeneity in the cerebral cortex. We took advantage of laser microdissection from tissue sections to selectively isolate specific cortical areas and their component layers on the basis of cellular cytoarchitecture, thereby providing a great improvement in precise regional anatomical specificity over gross dissections. These dissections were consistent across animals, although it should be noted that we balanced achieving the finest areal specificity with our ability to clearly differentiate areas based on Nissl cytoarchitecture on fresh frozen tissue sections alone. Consequently, while consistent and well-separated from one another, in some instances these areas contain further subdivisions that may be molecularly distinct from one another as well (e.g. anterior cingulate cortex). We found a large number of differentially expressed genes between cortical areas, with a high degree of overlap between genes with laminar enrichment and areal enrichment. Furthermore, all of the genes we analyzed for areal enrichment by ISH, selected for maximal fold change between areas, were highly enriched in specific cortical layers as well. Together this suggests that much of what differentiates cortical areas is differential expression in specific layers (i.e. in specific excitatory neuron populations).
Specializations in cortical cellular and functional architecture were reflected by differential gene expression. For example, L5 of primary motor cortex, containing the large projection neurons (corticospinal Betz cells), showed highest expression of the neurofilament heavy chain (NEFH) which is expressed in large caliber and long range projection neurons (Elder et al., 1998; Jacobs et al., 1996). Consistent with previous analysis of whole cortex (Oldham et al., 2008) , we find V1 to have the most distinct areal molecular profile, with differential gene expression patterns that changed sharply at the Nissl-defined boundaries between V1 and V2. As anticipated, this difference was due in part to the expanded sublayers of L4, which were highly distinctive both at the transcriptome-wide level and at the level of individual genes as shown by ISH. For example, several genes with novel selective expression in V1 L4 were identified, including adipocyte-specific adhesion molecule (ASAM), a type I transmembrane immunoglobulin protein that may participate in cell-cell adhesion (Raschperger et al., 2004), the guanine nucleotide exchange factor VAV3 which has been implicated in Purkinje cell dendritogenesis (Quevedo et al., 2010), and the orphan estrogen-related receptor gamma ESRRG. Surprisingly, many of the most robust V1-selective genes were outside of L4, most notably in L6 where the synaptic vesicle fusion-related gene SYT6 and the neuropeptide Y receptor NPY2R were highly enriched. Finally, V1 appears to be demarcated equally by selective enrichment and selectively decreased gene expression, as for the matrix extracellular phosphoglycoprotein MEPE in L2 and the serotonin receptor HTR2C in L5. From a molecular perspective then, the cytoarchitectural and functional specialization of primate V1 appears to be mediated by complex differences in gene expression across many different excitatory neuronal subtypes.
Molecular Similarities Between Cortical Areas and Layers Reflect Spatial Proximity
An unanticipated finding from this study is that molecular similarities are strongest between spatial neighbors, both between cortical areas and between cortical layers. There are a number of potential explanations for this finding. One possibility, particularly for cortical layers, is that these similarities reflect a “spill-over” of cell types between layers, since layer boundaries are not sharp, cellular segregation by layer may not be complete, and our isolations were not cell type-specific. However, this seems unlikely for several reasons. First, we were careful to avoid laminar borders (see Figure S1). Furthermore, we were able to identify genes with nearly binary layer-specific expression, while most genes with laminar specificity appeared to be expressed across multiple contiguous layers at similar expression levels. These observations would appear to be inconsistent with spill-over of a small proportion of cells of a particular type across layers, although it is certainly possible that gradients of glial or inhibitory cell subtypes account for some proportion of adjacent layer similarity.
An alternate explanation for proximity relationships is that they reflect developmental origin, or lineage, an interpretation that is supported by our results. The development of laminar cortical structure involves the sequential generation of excitatory neurons in an “inside-out” fashion (Bystron et al., 2008; Rakic, 1974). Cell division of precursor pools occurs in the ventricular and subventricular zones, and later-generated cells destined for more superficial cortical layers migrate over earlier-generated deep layer neurons. Ultimately, this process results in the segregation of different cortical cell types into discrete layers, with corticocortical pyramidal projection neurons dominating superficial L2 and L3, corticofugal projection neurons dominating deep L5 and L6, and local circuit stellate neurons in L4. Despite morphological and projectional similarities between deep and superficial neurons, relationships between layers based on gene expression clearly reflected physical proximity. This organizational principle was highly robust, and was seen using a variety of analytical methods including PCA (based on all 52,865 probe sets on the arrays), ANOVA and unsupervised hierarchical clustering (based on 3,000-5,000 probe sets with significant differential expression), and WGCNA-derived gene networks (based on >18,000 probe sets). Since physical proximity between cortical layers also reflects temporal proximity in terms of the developmental genesis of neurons from the neocortical germinal zones, this suggests that the global mRNA signatures for cortical layers bear a developmental imprint resulting from the sequential generation from increasingly differentiated cortical progenitor cells. Similar conclusions have been made by others comparing transcriptional profiles of different brain regions in rodents (Zapala et al., 2005).
Our selection of cortical areas allowed a discrimination between molecular similarities based on proximity, functional type (sensory, motor, association), or functional stream (e.g. dorsal (MT) versus ventral (TE) visual streams). Similar to findings for layers, cortical areas cluster by proximity more so than by functional type or functional stream. The caudally located visual areas V1, V2 and dorsal stream area MT cluster together, while ventral stream area TE is most similar to the proximally located primary auditory cortex (A1). The adjacent S1 and M1 areas are highly similar despite different cytoarchitecture and function. Furthermore, WGCNA identified modules of covarying genes with rostrocaudal gradients. These patterns are highly reminiscent of molecular gradients of transcription factors in the early developing neocortex that are important for proper areal patterning (Bishop et al., 2002; O’Leary and Sahara, 2008). Therefore, although individual cortical areas have molecular signatures that relate to their distinct cellular makeup or functional properties, broad molecular coherence between cortical areas more closely reflect spatial, nearest neighbor relationships.
Molecular similarities between nearby cortical areas may be important from the perspective of selection pressure for wiring economy in cortico-cortical connectivity (Bullmore and Sporns, 2009; Raj and Chen, 2011). Cortical networks show features of small-world networks (Bassett and Bullmore, 2006), with dense local clustering of connections and relatively few long-range connections that serve to minimize wiring and energy costs while maintaining dynamical complexity (Sporns and Zwi, 2004). An intriguing idea is that cortical areas with strong molecular similarities preferentially wire together during development. In support of this idea, the top enriched GO categories for genes that vary by cortical area were axon guidance and ephrin receptor signaling, while gene clusters showing enrichment in proximal cortical areas were enriched for axon guidance molecules as well.
Comparative Analysis of Laminar and Areal Molecular Signatures in Primates and Mice
It has been argued extensively that species differences may be largely a product of differences in gene regulation as opposed to gene sequence or structure (King and Wilson, 1975). Consistent with this idea, a number of genes with specific cellular distributions were seen to vary across species, suggesting alterations in cis-regulation at the level of specific cortical cell types. While it is possible that differences in species-specific probe sequences may contribute to differences observed across species in some cases, several overall patterns were observed across the genes examined. In general, rhesus patterns closely matched human expression patterns, both in their laminar (cellular) distributions and their areal specificity for V1 versus V2. Several differences were noted, including a lack of PDYN labeling in human compared to macaque in L4A, the same layer where other molecular differences have been noted in humans compared to other primates (Preuss and Coleman, 2002). However, these differences involved low expressing cells that may not be detected in human postmortem tissues with much longer postmortem intervals than experimental model system-derived tissues. On the other hand, substantially greater differences were observed for specific cortical laminar gene expression patterns between primates and mice, ranging from partially matching laminar patterns to completely different cell populations labeled. For example, SV2C is expressed preferentially in L3 pyramidal neurons in primates, and in L5 pyramidal neurons in mice. Prodynorphin (PDYN), which produces dynorphin and other kappa opioid receptor peptide agonists, is expressed in L4Cb and L5 in primate V1, but only in scattered GABAergic interneurons in mice. This difference suggests alterations in cis-regulation, potentially supported by the finding that the promoter region of PDYN has been shown to vary across primates and human populations through positive natural selection (Rockman et al., 2005) . A similar shift from L6 neurons to sparse, putative GABAergic neurons in V1 is seen for the neuropeptide Y receptor NPY2R. These types of species differences are particularly important, as cell type class-shifting in gene usage, particularly for genes such as neurotransmitter receptors, could have profound effects on cortical function.
Finally, the differential molecular signatures identified in V1 in rhesus macaque and human were not observed in mouse visual cortex. For example, SYT6 is highly enriched in L6 in V1 relative to other cortical areas, and shows a sharp transition at the V1/V2 boundary in macaque and human. In mouse Syt6 is similarly enriched in L6, but with no discernable difference in expression between primary visual cortex and other cortical regions. Similarly, the serotonin receptor HTR2C, implicated in schizophrenia, bipolar disorder and major depression (Iwamoto et al., 2009) is expressed selectively in L5 in macaque, human and mouse. However, in mouse this pattern is in all regions, while in primates HTR2C is selectively decreased in V1. These observations suggest that cortical specialization may occur through variations on a basic cellular and molecular cortical architectural template.
The strong similarities in molecular architecture indicate that rhesus macaque is a highly predictive non-human primate model system for human neocortical structure and corresponding gene expression, at least for homologous functional areas of the neocortex. These data provide an information-rich resource, and it will be important in the future to extend these studies to human neocortex to understand the molecular underpinnings of human-specific neocortical specialization.
EXPERIMENTAL PROCEDURES
Animals
Adult (mean age ± SEM = 8.5 ± 1.0 years) male and female Rhesus monkeys (Macaca mulatta) were used for the study. All animals were housed at the New Iberia Primate Research Center (New Iberia, LA). Animals had negative histories for Cercopithecine Herpes virus I, measles, pox viruses, rabies and tuberculosis.All animal handling procedures were approved by Institutional Animal Care and Use Committees at Merck and Co. and the New Iberia Primate Research Center.
Brain Tissue Collection
Animals were euthanized with an overdose of sodium pentobarbital and phenyltoin sodium, immediately after which the brain was removed,placed into cold (4°C) phosphate buffered saline (pH 7.4), and then placed ventral side up in a Rhesus brain matrix (EMS, Hatfield PA). Coronal slabs (6 mm thick) were made by placing razor blades (EMS) into slots on the matrix and gently depressing the blades through the tissue. Each slab was marked for orientation, placed on a metal disk that was embedded in dry ice until frozen, and stored at -80°C in bar-coded bags. The mean (± SEM) time between euthanasia and freezing was 49 ± 2.1 min.
Laser Microdissection and RNA Isolation
Slabs from frozen male and female (n=2-3 animals/gender) brains were serially cryosectioned at 14 μm onto PEN slides for LMD (Leica Microsystems, Inc., Bannockburn, IL) and a 1:10 Nissl series was generated for neuroanatomical reference. After drying for 30 minutes at room temperature, PEN slides were frozen at -80°C. Slides were later rapidly fixed in ice cold 70% ethanol, lightly stained with cresyl violet to allow cytoarchitectural visualization, dehydrated, and frozen at -80°C. LMD was performed on a Leica LMD6000 (Leica Microsystems, Inc.), using the cresyl violet stain to identify target brain regions. Quadruplicate biological replicates were collected from ten cortical regions (4-8 individual layers), four hippocampal subfields, and three layers of the LGN (Table S1). Images of pre- and post-laser microdissection are shown in Figure S1.
Microdissected tissue was collected directly into RLT buffer from the RNeasy Micro kit (Qiagen Inc., Valencia, CA) supplemented with ß-mercaptoethanol. Samples were volume-adjusted with RLT Buffer to 75μl, vortexed, centrifuged, and frozen at -80°C. RNA was isolated for each brain region following the manufacturer’s directions. RNA samples were eluted in 14μl and 1μl was run on the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA) using the Pico 6000 assay kit. Samples were quantitated using the Bioanalyzer concentration output. The average RNA Integrity Number (RIN) of all 225 passed experimental samples was 6.7.
mRNA Profiling
Sample amplification, labeling, and microarray processing were performed by the Rosetta Inpharmatics Gene Expression Laboratory (Seattle, WA). Samples passing RNA QC were amplified and profiled as described (Winrow et al., 2009) with a few modifications. Briefly, samples were amplified and labeled using a custom 2 cycle version, using 2 kits of the GeneChip® HT One-Cycle cDNA Synthesis Kit from Affymetrix. Five nanograms of total RNA was added to the initial reaction mix together with 250 ng of pBR322 (Invitrogen). As little as 2ng was used in some cases where tissue was extremely limited. Hybridization was performed in three batches to GeneChip® Rhesus Macaque Genome Arrays from Affymetrix containing 52,803 probesets/sequences. To control for batch effects, common RNA pool control samples were amplified and hybridized in each batch (3 replicates per 96 well batch). Profile quality was assessed using standard Affymetrix quality control metrics as well as by PCA. A total of 8 outliers were identified, and these samples were recollected and hybridized successfully.
A total of 258 samples passed sample QC, including 225 experimental samples and 33 control samples. The experimental samples include two male and two female profiles for each region (except three missing samples; Table S1). The data discussed in this publication were deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE31613.
Differential Gene Expression Data Normalization and Analysis
Each batch was normalized within itself using RMA (Irizarry et al., 2003), and batch effects were removed by subtracting the difference for each probe between controls of one batch from the controls of each other batch. Following this correction, no correlation with batch was observed among all samples within the four primary principal components which explain approximately 40% of the cumulative variance (Figure S2A, B and data not shown).
ANOVA, principal component and agglomerative clustering was performed using Matlab2007a. Gene set annotation analysis was performed by comparing input sets to GeneGo (genego.com), Ingenuity (ingenuity.com), KEGG (genome.jp/keg) and PANTHER (pantherdb.org) pathway sets. Bonferroni (BF) corrected hypergeometric p-values of less than 0.1 were considered as significant overlap between sets.
Genes correlated to templates were identified using Microsoft Excel, based on correlation function scores across all cortical samples between an artificial template set with values of 100 for one cortical layer of interest versus 0 for all other layers.
Weighted Gene Co-expression Network Analysis (WGCNA)
WGCNA (Langfelder et al., 2008; Zhang and Horvath, 2005) was used to identify clusters of coregulated genes across the entire neocortical sample set or just within the laminar samples in area V1. Outlier samples were removed based on inter-array correlations (IAC) < 2 standard deviations from the mean IAC, and cross-batch normalization was performed using the R package “ComBat” (http://statistics.byu.edu/johnson/ComBat/). 182 samples were included in the whole cortex analysis, and 30 samples in the V1 analysis, using probes present in at least half of the samples (18,080 for whole cortex, 15,234 for V1). A signed weighted network (Zhang and Horvath, 2005) was constructed for each data set. Using a dynamic tree-cutting algorithm (Langfelder et al., 2008), we identified 20 modules in the entire neocortical data set, and 36 modules in V1 only data set. The Module Eigengene (ME), defined as the first principle component of a given module, was used to represent the characteristic anatomical expression pattern of individual modules (Oldham et al., 2008).
In Situ Hybridization
Non-isotopic colorimetric in situ hybridization (ISH) was performed as described previously (Lein et al., 2007). Briefly, following cryosectioning of fresh frozen samples at 20μm, tissue sections were fixed, acetylated, and subsequently dehydrated. Digoxigenin-based riboprobe labeling coupled with TSA amplification and alkaline-phosphatase-based colorimetric detection was used to label target mRNAs in expressing cells.
Riboprobes were designed to overlap probe designs for homologous genes in mouse and human used in the Allen Mouse Brain Atlas (http://mouse.brain-map.org) and Allen Human Brain Atlas (http://human.brain-map.org/), and cross-species comparisons were made to data publicly available in those databases. A subset of rhesus macaque ISH data shown was generated in 4-year old adult male specimens as part of the NIH Blueprint NHP atlas (http://www.blueprintnhpatlas.org/). Additional ISH data were generated on tissue sections collected from the frontal pole, medial/temporal areas and caudal/visual areas in two adult specimens from this study.
Supplementary Material
Acknowledgments
This work was sponsored by Merck Research Labs, the Allen Institute for Brain Science and NIH Grant 5R37 MH060233-11 (DHG, RL). The authors wish to thank the Allen Institute founders, Paul G. Allen and Jody Patton, for their vision, encouragement, and support. We thank Mike Citron for specimen identification, Ken Lodge for providing pilot tissue and sample collection methods, and Dr. Jane Fontenot, Dana Hasselschwert and Marcus Louis for assistance with tissue collection. Thanks to Crissa Wolkey for sample processing and Rachel Dalley and Sheila Shapouri for LMD images. We wish to acknowledge Paul Wohnoutka, Amanda Ebbert and Lon Luong for supporting data production, Chinh Dang for supporting database needs, Kelly Overly for contracting assistance, David Haynor for discussions on project design and Christof Koch for critical reading of the manuscript. Finally, thanks to Affymetrix for preferred pricing on rhesus microarrays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahams BS, Tentler D, Perederiy JV, Oldham MC, Coppola G, Geschwind DH. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc Natl Acad Sci U S A. 2007;104:17849–17854. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Bullmore E. Small-world brain networks. Neuroscientist. 2006;12:512–523. doi: 10.1177/1073858406293182. [DOI] [PubMed] [Google Scholar]
- Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, Garcia-Moreno F, Molnar Z, Margulies EH, Ponting CP. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KM, Rubenstein JL, O’Leary DD. Distinct actions of Emx1, Emx2, and Pax6 in regulating the specification of areas in the developing neocortex. J Neurosci. 2002;22:7627–7638. doi: 10.1523/JNEUROSCI.22-17-07627.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodmann K. Vergelichende Lokalisationslehre der Grosshirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaus. Leipzig: Barth; 1909. [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, Elia M, Schneider C, Melmed R, Sacco R, et al. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci U S A. 2006;103:16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder GA, Friedrich VL, Jr, Kang C, Bosco P, Gourov A, Tu PH, Zhang B, Lee VM, Lazzarini RA. Requirement of heavy neurofilament subunit in the development of axons with large calibers. J Cell Biol. 1998;143:195–205. doi: 10.1083/jcb.143.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Hoerder-Suabedissen A, Wang WZ, Lee S, Davies KE, Goffinet AM, Rakic S, Parnavelas J, Reim K, Nicolic M, Paulsen O, Molnar Z. Novel markers reveal subpopulations of subplate neurons in the murine cerebral cortex. Cereb Cortex. 2009;19:1738–1750. doi: 10.1093/cercor/bhn195. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Kato T. Serotonin receptor 2C and mental disorders: genetic, expression and RNA editing studies. RNA Biol. 2009;6:248–253. doi: 10.4161/rna.6.3.8370. [DOI] [PubMed] [Google Scholar]
- Jacobs AJ, Swain GP, Selzer ME. Developmental increases in expression of neurofilament mRNA selectively in projection neurons of the lamprey CNS. J Comp Neurol. 1996;364:383–401. doi: 10.1002/(SICI)1096-9861(19960115)364:3<383::AID-CNE1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Jasinska AJ, Service S, Choi OW, DeYoung J, Grujic O, Kong SY, Jorgensen MJ, Bailey J, Breidenthal S, Fairbanks LA, et al. Identification of brain transcriptional variation reproduced in peripheral blood: an approach for mapping brain expression traits. Hum Mol Genet. 2009;18:4415–4427. doi: 10.1093/hmg/ddp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Krubitzer L. In search of a unifying theory of complex brain evolution. Ann N Y Acad Sci. 2009;1156:44–67. doi: 10.1111/j.1749-6632.2009.04421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lein ES, Zhao X, Gage FH. Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J Neurosci. 2004;24:3879–3889. doi: 10.1523/JNEUROSCI.4710-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Mukamel Z, Konopka G, Wexler E, Osborn GE, Dong H, Bergman MY, Levitt P, Geschwind DH. Regulation of MET by FOXP2, genes implicated in higher cognitive dysfunction and autism risk. J Neurosci. 2011;31:11437–11442. doi: 10.1523/JNEUROSCI.0181-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary DD, Sahara S. Genetic regulation of arealization of the neocortex. Curr Opin Neurobiol. 2008;18:90–100. doi: 10.1016/j.conb.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Huang XF, Toga AW. The rhesus monkey brain in stereotaxic coordinates. San Diego, CA: Academic Press; 2000. [Google Scholar]
- Preuss TM, Coleman GQ. Human-specific organization of primary visual cortex: alternating compartments of dense Cat-301 and calbindin immunoreactivity in layer 4A. Cereb Cortex. 2002;12:671–691. doi: 10.1093/cercor/12.7.671. [DOI] [PubMed] [Google Scholar]
- Quevedo C, Sauzeau V, Menacho-Marquez M, Castro-Castro A, Bustelo XR. Vav3-deficient mice exhibit a transient delay in cerebellar development. Mol Biol Cell. 2010;21:1125–1139. doi: 10.1091/mbc.E09-04-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Chen YH. The wiring economy principle: connectivity determines anatomy in the human brain. PLoS One. 2011;6:e14832. doi: 10.1371/journal.pone.0014832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- Raschperger E, Engstrom U, Pettersson RF, Fuxe J. CLMP, a novel member of the CTX family and a new component of epithelial tight junctions. J Biol Chem. 2004;279:796–804. doi: 10.1074/jbc.M308249200. [DOI] [PubMed] [Google Scholar]
- Rockman MV, Hahn MW, Soranzo N, Zimprich F, Goldstein DB, Wray GA. Ancient and recent positive selection transformed opioid cis-regulation in humans. PLoS Biol. 2005;3:e387. doi: 10.1371/journal.pbio.0030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Zwi JD. The small world of the cerebral cortex. Neuroinformatics. 2004;2:145–162. doi: 10.1385/NI:2:2:145. [DOI] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Takahata T, Komatsu Y, Watakabe A, Hashikawa T, Tochitani S, Yamamori T. Differential expression patterns of occ1-related genes in adult monkey visual cortex. Cereb Cortex. 2009;19:1937–1951. doi: 10.1093/cercor/bhn220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WZ, Oeschger FM, Lee S, Molnar Z. High quality RNA from multiple brain regions simultaneously acquired by laser capture microdissection. BMC Mol Biol. 2009;10:69. doi: 10.1186/1471-2199-10-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Komatsu Y, Sadakane O, Shimegi S, Takahata T, Higo N, Tochitani S, Hashikawa T, Naito T, Osaki H, et al. Enriched expression of serotonin 1B and 2A receptor genes in macaque visual cortex and their bidirectional modulatory effects on neuronal responses. Cereb Cortex. 2009;19:1915–1928. doi: 10.1093/cercor/bhn219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winden KD, Oldham MC, Mirnics K, Ebert PJ, Swan CH, Levitt P, Rubenstein JL, Horvath S, Geschwind DH. The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol. 2009;5:291. doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winrow CJ, Tanis KQ, Rigby AM, Taylor RR, Serikawa K, McWhorter M, Tokiwa GY, Marton MJ, Stone DJ, Koblan KS, Renger JJ. Refined anatomical isolation of functional sleep circuits exhibits distinctive regional and circadian gene transcriptional profiles. Brain Res. 2009;1271:1–17. doi: 10.1016/j.brainres.2009.02.083. [DOI] [PubMed] [Google Scholar]
- Yamamori T, Rockland KS. Neocortical areas, layers, connections, and gene expression. Neurosci Res. 2006;55:11–27. doi: 10.1016/j.neures.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Zapala MA, Hovatta I, Ellison JA, Wodicka L, Del Rio JA, Tennant R, Tynan W, Broide RS, Helton R, Stoveken BS, et al. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci U S A. 2005;102:10357–10362. doi: 10.1073/pnas.0503357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- Zilles K, Amunts K. Centenary of Brodmann’s map--conception and fate. Nat Rev Neurosci. 2010;11:139–145. doi: 10.1038/nrn2776. [DOI] [PubMed] [Google Scholar]
- Zilles K, Palomero-Gallagher N, Schleicher A. Transmitter receptors and functional anatomy of the cerebral cortex. J Anat. 2004;205:417–432. doi: 10.1111/j.0021-8782.2004.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.