Abstract
The human mind and body respond to stress1, a state of perceived threat to homeostasis, by activating the sympathetic nervous system and secreting the catecholamines adrenaline and noradrenaline in the ‘fight-or-flight’ response. The stress response is generally transient because its accompanying effects (for example, immunosuppression, growth inhibition and enhanced catabolism) can be harmful in the long term2. When chronic, the stress response can be associated with disease symptoms such as peptic ulcers or cardiovascular disorders3, and epidemiological studies strongly indicate that chronic stress leads to DNA damage4,5. This stress-induced DNA damage may promote ageing6, tumorigenesis4,7, neuropsychiatric conditions8,9 and miscarriages10. However, the mechanisms by which these DNA-damage events occur in response to stress are unknown. The stress hormone adrenaline stimulates β2-adrenoreceptors that are expressed throughout the body, including in germline cells and zygotic embryos11. Activated β2-adrenoreceptors promote Gs-protein-dependent activation of protein kinase A (PKA), followed by the recruitment of β-arrestins, which desensitize G-protein signalling and function as signal transducers in their own right12. Here we elucidate a molecular mechanism by which β-adrenergic catecholamines, acting through both Gs-PKA and β-arrestin-mediated signalling pathways, trigger DNA damage and suppress p53 levels respectively, thus synergistically leading to the accumulation of DNA damage. In mice and in human cell lines, β-arrestin-1 (ARRB1), activated via β2-adrenoreceptors, facilitates AKT-mediated activation of MDM2 and also promotes MDM2 binding to, and degradation of, p53, by acting as a molecular scaffold. Catecholamine-induced DNA damage is abrogated in Arrb1-knockout (Arrb1−/−) mice, which show pre served p53 levels in both the thymus, an organ that responds prominently to acute or chronic stress1, and in the testes, in which paternal stress may affect the offspring’s genome. Our results highlight the emerging role of ARRB1 as an E3-ligase adaptor in the nucleus, and reveal how DNA damage may accumulate in response to chronic stress.
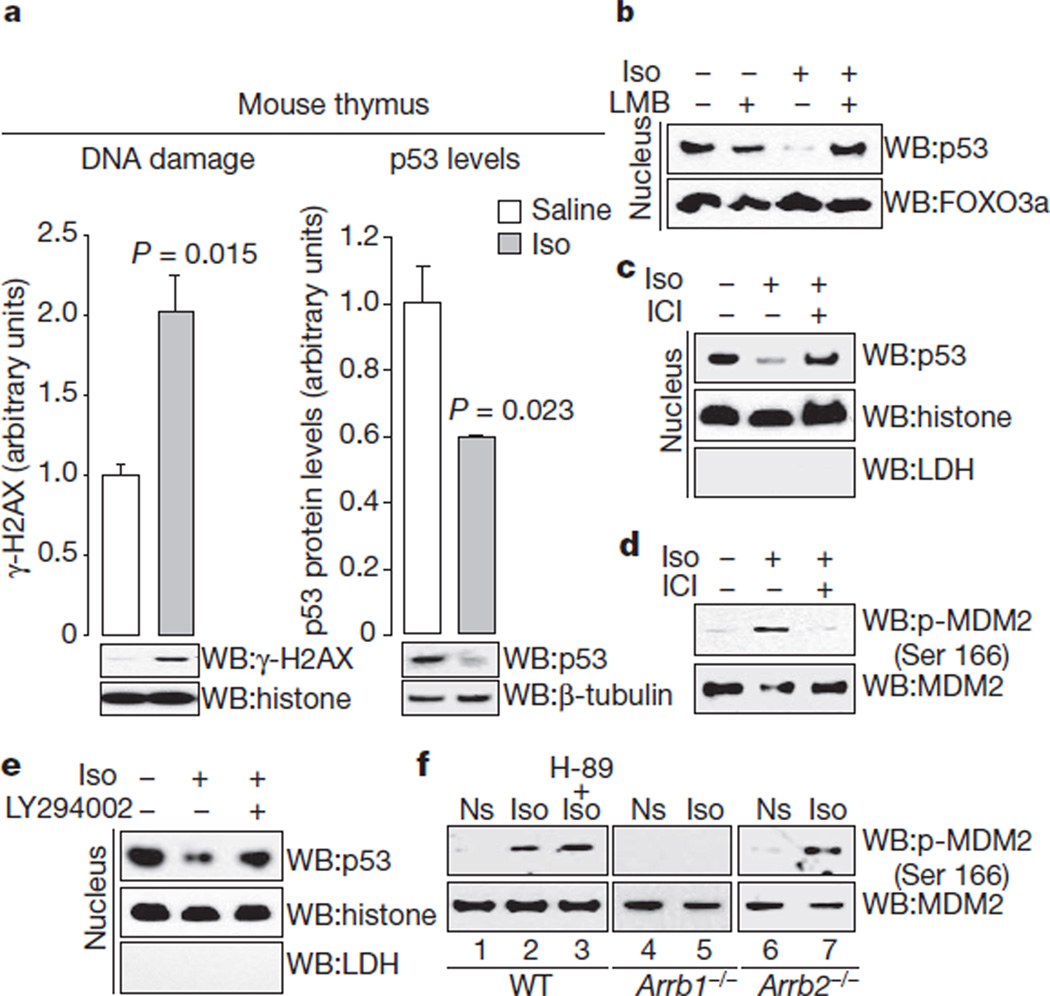
As a model of chronic stress and prolonged stimulation of β2-adrenoreceptors7,13, wild-type mice were infused for four weeks with either saline or the β2-adrenoreceptor-agonist isoproterenol, a syn thetic analogue of adrenaline. First, we tested whether this regimen affects DNA damage by examining phosphorylation of histone H2AX (γ-H2AX), one of the earliest indicators of DNA damage14. Isoproterenol infusion leads to DNA damage in the thymus (Fig. 1a, left panel). Accumulation of DNA damage indicates compromised genome maintenance. To investigate the potential mechanism, we examined p53 levels in the thymus and found that isoproterenol infusion leads to decreased levels of p53 (Fig. 1a, right panel). Consistent with the effects of isoproterenol in vivo, chronic stimulation of β2-adrenoreceptors with β-adrenergic catecholamines (isoproterenol, adrenaline or noradrenaline) leads to accumulation of DNA damage and a decrease in p53 levels in cultured U2OS cells (Supplementary Fig. 1a–c), which endogenously express wild-type p53 and only the β2-subtype of β-adrenoreceptors (Supplementary Fig. 2a–c). Moreover, the p53 in these cells, as well as in all other cell lines used in these studies (fibroblasts and HEK-293 cells), was demonstrated to be functional by a variety of techniques (Supplementary Fig. 3a–k), and all cell lines endogenously expressed only the β2-subtype of β-adrenoreceptors (Supplementary Fig. 2a–c).
Figure 1. Chronic catecholamine stimulation leads to p53 degradation and accumulation of DNA damage via ARRB1/AKT-mediated activation of MDM2.
a, Isoproterenol infusion leads to accumulation of DNA damage and decreased p53 levels. Mice (n 3–5 for each condition) were infused with saline or isoproterenol (30 mg kg−1 d−1) for 4 weeks. All bars represent mean ± s.e.m. Histone, histone H2B; Iso, isoproterenol; WB, western blot. b, Isoproterenol-induced p53 reduction is dependent on nuclear export. This effect is specific to p53, in that another nuclear-cytosol shuttling molecule, FOXO3a, is not affected. LMB, leptomycin B. c, Preincubation with the β2-adrenoreceptor-selective antagonist ICI 118,551 (ICI) blocks isoproterenol-induced nuclear export of p53. Lactate dehydrogenase (LDH) is a cytosolic marker and histone is a nuclear marker. d, Isoproterenol stimulation leads to MDM2 phosphorylation at Ser 166, and is blocked by preincubation with ICI 118,551. e, Inhibition of the PI3K/AKT cascade abolishes isoproterenol-stimulated decreases in p53 levels in U2OS cells. LY294002 is a PI3K inhibitor. f, Isoproterenol stimulation leads to Gs-independent, ARRB1-dependent MDM2 phosphorylation at Ser 166. Ns, not stimulated.
The isoproterenol-induced reduction in p53 levels results from p53 degradation, and is abolished by proteasome inhibition (Supplementary Fig. 1d). Because nuclear export of p53 has been shown to be involved in its degradation15, we examined p53 localization. Subcellular fractionation shows that isoproterenol stimulation leads to a decrease in nuclear p53 and an increase in cytosolic p53 (Supplementary Fig. 1e, lower panels), thus, isoproterenol stimulation leads to p53 nuclear export. Immunocytochemical examination also shows increased levels of cytosolic p53 after isoproterenol stimulation (Supplementary Fig. 1e, upper panels). Isoproterenol concentrations as low as 1 nM lead to p53 nuclear export, resulting in a decrease in total p53 levels (Supplementary Fig. 1f). The importance of nuclear export in modulating p53 levels was investigated by treating cells with leptomycin B, an inhibitor of nuclear export. Leptomycin B pretreatment reverses isoproterenol-induced nuclear export of p53 (Fig. 1b).
To examine whether isoproterenol-induced effects were specifically mediated by β2-adrenoreceptors, U2OS cells were stimulated with isoproterenol in the presence or absence of the subtype-selective β2-adrenoreceptor antagonist ICI 118,551. Preincubation with ICI 118,551 abrogates the isoproterenol-induced decrease in p53 levels (Fig. 1c). During in vivo experiments, isoproterenol infusion leads to accumulation of DNA damage in the cerebellum, where β2-adrenoreceptors are the major subtype of β-adrenoreceptor16 (Supplementary Fig. 1g). Furthermore, targeted disruption of the Adrb2 gene in mice markedly reduces accumulation of DNA damage upon isoproterenol infusion (Supplementary Fig. 1h). Taken together, these data indicate that stimulation of the β2-adrenoreceptor results in the nuclear export and degradation of p53 in a specific manner.
The E3 ligase MDM2 has been shown to have an important role in the regulation of p53 nuclear export and degradation15. Consistent with this, leptomycin B abrogates the ability of MDM2 to degrade p53 (ref. 15). Before MDM2-mediated ubiquitination of p53, the phosphoinositide 3-kinase (PI3K)/AKT cascade phosphorylates MDM2, activating its E3 ligase function17. To examine whether stimulation of β2-adrenoreceptors leads to MDM2 phosphorylation via the PI3K/AKT cascade, wild-type mouse embryonic fibroblasts (MEFs) were stimulated with isoproterenol in the presence or absence of ICI 118,551. Isoproterenol stimulation leads to MDM2 phosphorylation at Ser 166, an AKT phosphorylation site, and the effect is antagonized by ICI 118,551 (Fig. 1d and Supplementary Fig. 1i). To confirm that MDM2 is phosphorylated by the PI3K/AKT cascade upon isoproterenol stimulation, U2OS cells were stimulated with isoproterenol in the presence of either the PI3K inhibitor wortmannin or the AKT inhibitor AKTi. MDM2 phosphorylation is abolished by either wortmannin or AKTi (Supplementary Fig. 1j). Furthermore, a PI3K inhibitor also abolishes catecholamine-induced lowering of p53 levels in the nucleus (Fig. 1e and Supplementary Fig. 1k). The importance of MDM2 phosphorylation at Ser 166 was demonstrated by the overexpression of a phosphomimetic mutant at Ser 166 (MDM2-S166D)17, which facilitates the degradation of p53 when compared to wild-type MDM2 (Supplementary Fig. 1l). These data implicate the PI3K/AKT cascade downstream of the β2-adrenoreceptor as a mediator of p53 stability through the phosphorylation of MDM2.
Upon activation of β2-adrenoreceptors, the PI3K/AKT cascade can be stimulated by both the Gs-PKA18 and β-arrestin-mediated signalling pathways19,20. To elucidate which pathway was involved, we examined the effect of β2-adrenoreceptor stimulation in wild-type MEFs in the presence of H-89, a PKA inhibitor, or in Arrb1−/− or Arrb2-knockout (Arrb2−/−) MEFs. In wild-type MEFs, H-89 does not inhibit isoproterenol-stimulated MDM2 phosphorylation (Fig. 1f, lane 3). In contrast, the isoproterenol effect is abrogated in Arrb1−/− (Fig. 1f, lane 5), but not in Arrb2−/−, MEFs (Fig. 1f, lane 7). Furthermore, rescuing Arrb1 expression in Arrb1−/− MEFs restores the effects of isoproterenol stimulation (Supplementary Fig. 1m), and the effects of isoproterenol on MDM2 phosphorylation are abrogated in Arrb1−/− mice (Supplementary Fig. 1n). These data elucidate a Gs-independent, ARRB1-dependent signalling pathway that regulates the activation state of MDM2 through the PI3K/AKT cascade.
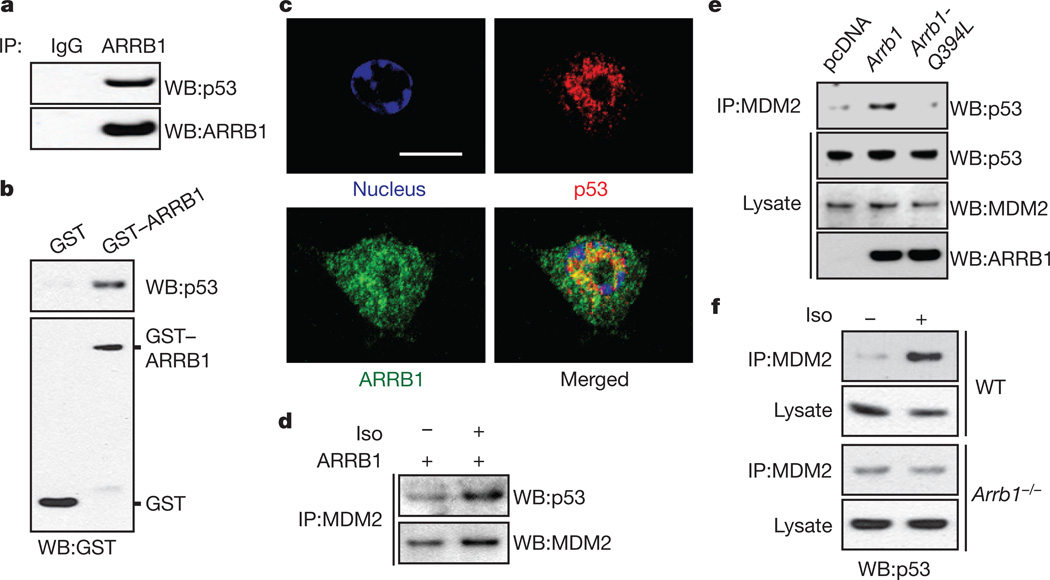
β-Arrestins can serve as adaptors for E3 ligases and their substrates21. Because we found that MDM2 activation by the PI3K/AKT cascade is an ARRB1-dependent event, we examined the binding between β-arrestins and p53, a known MDM2 substrate. In HEK-293 cells stably overexpressing ARRB1 or ARRB2, p53 binds preferentially to ARRB1, an isoform localized to both the cytosol and nucleus22, but not to ARRB2, which predominantly localizes to the cytosol22–25 (Supplementary Fig. 4a). Binding between these two molecules at endogenous levels is also observed in HEK-293 cells and brain homogenates (Fig. 2a and Supplementary Fig. 4b). The binding seems to be direct, because purified ARRB1 tagged with glutathione-S-transferase (GST-ARRB1) binds to p53 in vitro (Fig. 2b). To examine the effect of β2-adrenoreceptor stimulation on this complex, we treated untransfected HEK-293 cells, which endogenously express only the β2 subtype of β-adrenoreceptors (Supplementary Fig. 2b, c), with isoproterenol. Stimulation of β2-adrenoreceptors does not affect the binding of ARRB1 to p53 (Supplementary Fig. 4c). Subsequently, we mapped the binding sites in ARRB1 and p53 by performing sequential deletions, followed by immunoprecipitation from HEK-293 cells (Supplementary Fig. 4d, e). We identified the amino terminus of ARRB1 (amino acids 1–186) as critical for binding to p53. In p53, a domain comprising amino acids 101–186 is required for binding to ARRB1. Consistent with these results, a synthetic ARRB1-binding peptide (ARRB-BP), which binds to the N terminus of ARRB1 and induces a conformational change26, disrupts the interaction between ARRB1 and p53 (Supplementary Fig. 4f).
Figure 2. ARRB1 functions as an E3 ligase adaptor for MDM2 and p53 upon catecholamine stimulation.
a, Endogenous binding of p53 and ARRB1. Cell lysates from HEK-293 cells were used for immunoprecipitation (IP) with an anti-ARRB1 (K-16) antibody or normal IgG, and analysed by immunoblotting with anti-p53 (DO-1) antibody. b, In vitro binding of p53 and ARRB1. Purified p53 was incubated with either GST or GST-ARRB1 and precipitated with glutathione beads. Precipitates were analysed by immunoblotting with an anti-p53 (DO-1) antibody. c, Confocal analysis of co-localization of p53 and ARRB1 in non-treated RAW 264.7 macrophages, which endogenously express high levels of both p53 and ARRB1. Scale bar, 10 µm. d, Isoproterenol stimulation facilitates the binding of MDM2 to p53 in ARRB1 over expressed cells. e, Nuclear ARRB1 facilitates MDM2 binding to p53. U2OS cells were transfected with either empty vector (pcDNA), Arrb1 or Arrb1-Q394L. f, ARRB1 facilitates isoproterenol-induced MDM2 binding to p53. WT, wild-type.
Co-immunoprecipitation experiments after subcellular fractionation show that more than 90% of the binding between ARRB1 and p53 occurs in the nucleus (Supplementary Fig. 4g). Additionally, confocal analysis reveals that endogenous ARRB1 and p53 co-localize in the nucleus (Fig. 2c) and a ternary complex between ARRB1, MDM2 and p53 was observed in p53-null NCI-H1299 cells transfected with wild-type p53 (Supplementary Fig. 4h). The potential effects of nuclear ARRB1 on MDM2 binding to p53 were investigated in U2OS cells transfected with either Arrb1 or Arrb1-Q394L, in which a single amino acid, Gln 394, has been mutated to Leu to create a nuclear export signal in ARRB122,23. Overexpression of ARRB1 facilitates the binding of MDM2 to p53, enhancing basal β2-adrenoreceptor-stimulated ARRB1 signalling (Fig. 2d); however, the effect is abolished with ARRB1-Q394L (Fig. 2e). This result indicates that ARRB1 facilitates an MDM2-p53 interaction in the nucleus. The role of endogenous ARRB1 as a facilitator of MDM2-p53 complex formation under isoproterenol-stimulated conditions is further demonstrated in Arrb1−/− MEFs (Fig. 2f), in which loss of ARRB1 prevents the increased interaction of MDM2 and p53 after isoproterenol stimulation, when compared to wild-type cells.
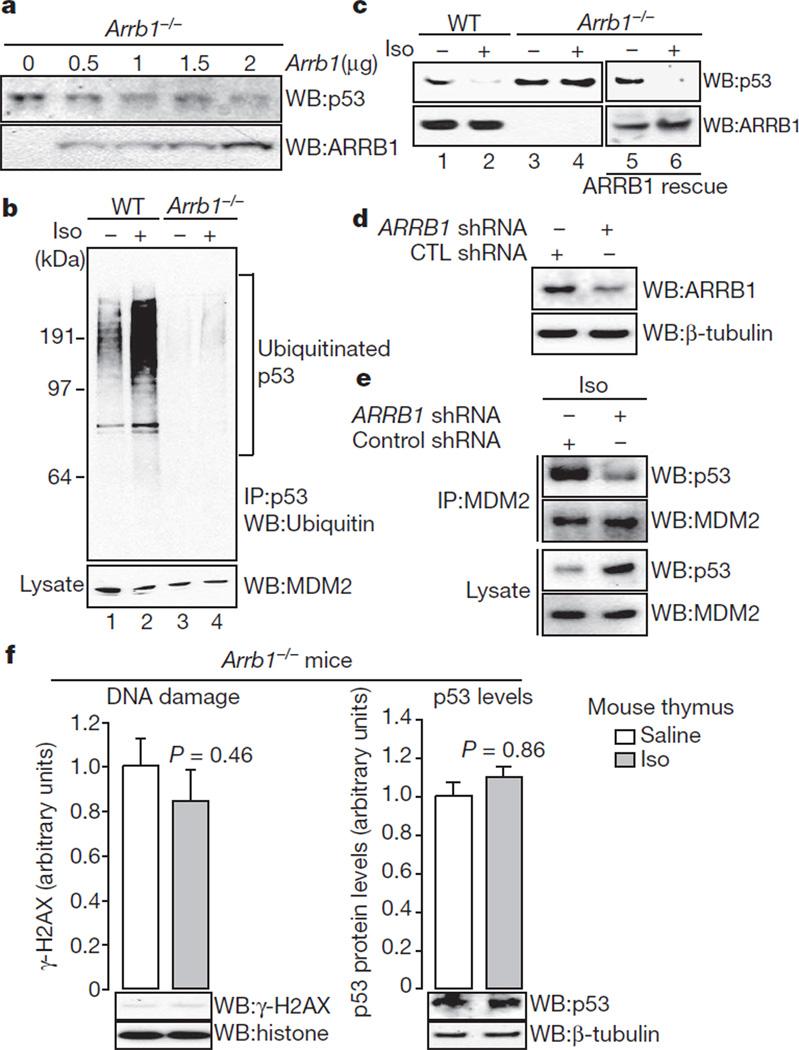
Next we examined whether ARRB1 expression affects p53 levels by comparing different clonal populations of wild-type and Arrb1−/− MEFs. Arrb1−/− MEFs show increased p53 levels (Supplementary Fig. 5a). Furthermore, rescuing ARRB1 expression in Arrb1−/− MEFs decreases p53 levels in a dose-dependent manner (Fig. 3a). Differences in p53 levels under basal conditions seem to be the result of decreased p53 ubiquitination in Arrb1−/− MEFs (Fig. 3b, lanes 1 and 3). Furthermore, consistent with β2-adrenoreceptor-induced degradation of p53, isoproterenol stimulation promotes p53 ubiquitination, but the effect is markedly decreased in Arrb1− MEFs(Fig.3b, lanes 2 and 4). To address further whether ARRB1 facilitates the ubiquitination of p53 by MDM2, we conducted in vitro ubiquitination assays (Supplementary Fig. 5b). Addition of ARRB1 facilitates MDM2-mediated ubiquitination of p53 and the effect is abolished with ARRB-BP. Together, these data indicate that upon catecholamine stimulation, ARRB1 promotes the interaction of MDM2 and p53 by acting as an E3 ligase adaptor that facilitates ubiquitination of p53.
Figure 3. ARRB1 facilitates catecholamine-induced p53 degradation by MDM2.
a, Rescuing ARRB1 expression in Arrb1−/− MEFs decreases p53 levels. Arrb1−/− MEFs were transiently transfected with Arrb1 and cell lysates were examined by immunoblotting. b, Isoproterenol stimulation leads to ubiquitination of p53 in a ARRB1-dependent manner. Wild-type and Arrb1−/− MEFs were stimulated with 10 µM isoproterenol for 24 h. Cell lysates were immunoprecipitated with an anti-p53 antibody (FL-393) and analysed by immunoblotting with an anti-ubiquitin (P4D1) antibody. c, Isoproterenol stimulation leads to ARRB1-dependent p53 degradation. d, Reduction in ARRB1 levels induced by small hairpin RNA (shRNA) in U2OS cells. e, Suppression of ARRB1 suppresses isoproterenol-induced binding of MDM2 to p53 and restores p53 levels in U2OS cells. U2OS cells were treated with either control shRNA or ARRB1 shRNA for 72 h, followed by 24 h stimulation with 10 µM isoproterenol. f, Levels of p53 in isoproterenol-infused Arrb1−/− mice remain constant and there is no accumulation of DNA damage. Arrb1−/− mice (n 3–4 for each condition) were treated as in Fig. 1a. All bars represent mean ± s.e.m.
Cytosolic ARRB1 mediates catecholamine-induced activation of AKT and MDM2, whereas nuclear ARRB1 serves as an adaptor for MDM2-dependent ubiquitination of p53. Consistent with these results, isoproterenol stimulation leads to a lowering of p53 levels (Fig. 3c, lane 2). By contrast, p53 levels remain constant in Arrb1−/− MEFs (Fig. 3c, lane 4). Furthermore, rescuing ARRB1 expression in Arrb1−/− MEFs by transient transfection restores isoproterenol-stimulated degradation of p53 (Fig. 3c, lane 6), and suppression of ARRB1 by RNA interference results in suppression of isoproterenol-stimulated MDM2-p53 complex formation, and a lowering of p53 levels in U2OS cells (Fig. 3d, e).
To differentiate the cytosolic (catecholamine-induced MDM2 phosphorylation) and nuclear (E3 ligase adaptor) functions of ARRB1 in this cascade, a phosphomimetic mutant of MDM2 (MDM2-S166D) was co-transfected with either Arrb1 or Arrb1-Q394L into Arrb1−/− MEFs. This allowed us to focus on the nuclear function of ARRB1. Restoring ARRB1 expression with the wild type, but not with the Q394L mutant, facilitates the degradation of p53 (Supplementary Fig. 5c). Consequently, it seems that although the cytoplasmic pool of ARRB1 is sufficient to activate MDM2 through the PI3K/AKT pathway, the nuclear pool of ARRB1 is required to act as an E3 ligase adaptor for MDM2 towards p53.
To examine this cascade in vivo, we examined the effects of catecholamine on p53 levels and accumulation of DNA damage in the thymus of Arrb1−/− mice. The mice were infused for four weeks with either saline or isoproterenol. In contrast to wild-type mice (Fig. 1a), p53 levels are maintained upon isoproterenol infusion in Arrb1−/− mice, and accumulation of DNA damage is abrogated (Fig. 3f).
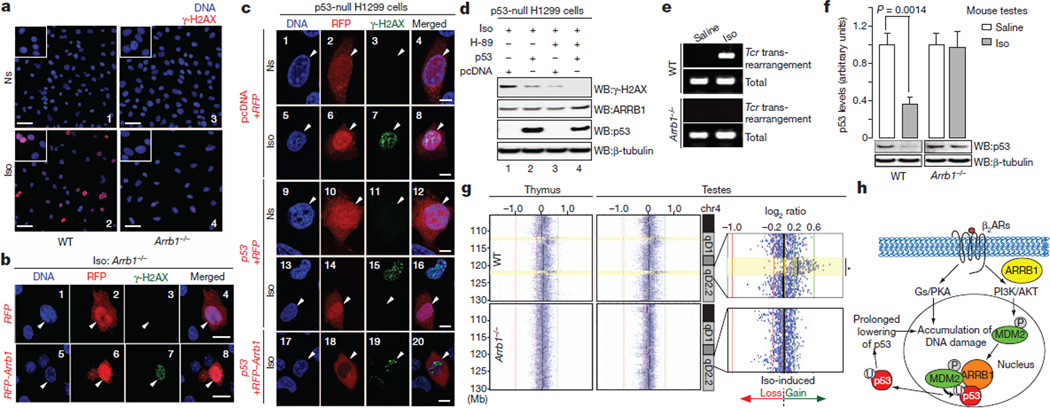
We have observed that isoproterenol infusion leads to lowering of p53 levels, and have elucidated a molecular mechanism whereby ARRB1 regulates MDM2-dependent degradation of p53 upon β2-adrenoreceptor stimulation. Next, we investigated further the effects of catecholamine-dependent p53 degradation on accumulation of DNA damage. To visualize the prevalence of DNA damage, we analysed the formation of γ-H2AX foci in both wild-type and Arrb1−/− MEFs. After chronic stimulation with isoproterenol, there is an increase in the formation of γ-H2AX foci (sevenfold) in wild-type MEFs, which is significantly reduced in Arrb1−/− MEFs (Fig. 4a, panels 2 and 4, and Supplementary Fig. 6a). Moreover, rescuing ARRB1 expression in Arrb1−/− MEFs restores the accumulation of isoproterenol-induced γ-H2AX foci (Fig. 4b, panels 5–8, and Supplementary Fig. 6a).
Figure 4. Chronic catecholamine stimulation leads to accumulation of DNA damage by an ARRB1- and p53-dependent mechanism.
a, Isoproterenol stimulation leads to formation of γ-H2AX foci in wild-type but not in Arrb1−/− MEFs. Wild-type and Arrb1−/− MEFs were chronically stimulated with 10 µM isoproterenol every 12 h for 3 days. Cells were immunostained and examined by confocal microscopy. Scale bar, 50 µm. b, Rescuing ARRB1 expression in Arrb1−/− MEFs restores isoproterenol-induced γ-H2AX foci. Two days after transfection, cells were stimulated and examined as described in a. RFP, red fluorescent protein. Scale bar, 10 µm. c, ARRB1-dependent regulation of p53 levels mediates the accumulation of isoproterenol-induced DNA damage. Two days after the transfection, cells were stimulated as in a. Transfected cells, indicated with arrowheads, were visualized with RFP. Scale bar, 10 µm. d, Isoproterenol-induced accumulation of DNA damage is synergistically suppressed by PKA inhibition and by rescuing p53 expression. e, Isoproterenol stimulated, ARRB1-dependent accumulation of DNA damage promotes rearrangements in chromosomes. Total indicates PCR amplification of a non-specific locus (see Methods). f, Isoproterenol infusion leads to decreased p53 levels in the testes from wild-type, but not Arrb1−/− mice. Wild-type and Arrb1−/− mice (n = 5 for each condition) were treated as in Fig. 1a. All bars represent mean ± s.e.m. g, Isoproterenol-infused mice develop chromosomal rearrangements in an ARRB1-dependent manner. Wild-type and Arrb1−/− mice were infused as in Fig. 1a. Genomic DNA from each organ was examined in an array-CGH. The data represent log2 ratio plots (isoproterenol/saline) of genomic content in chromosome 4 (chr4, 105–130 Mb), comparing isoproterenol-infused mice with saline-infused mice of a same genotype. *, significance threshold of 1.0 × 10−7 (rank segmentation algorithm). The direction of isoproterenol-induced chromosomal gain (green arrow) or loss (red arrow) is indicated. Yellow highlights represent sites of isoproterenol-induced rearrangements. h, Schematic diagram of β2-adrenoreceptor (β2AR)-dependent regulation of DNA damage in response to prolonged secretion of catecholamines during chronic stress.
To examine how exposure to stress hormones initiates DNA damage, p53-null NCI-H1299 cells were chronically stimulated with isoproterenol. This leads to accumulation of DNA damage (Fig. 4c, panels 1–8), indicating that DNA damage is triggered by p53-independent mechanisms after isoproterenol stimulation. One of the prominent cascades leading to DNA damage is the generation of reactive oxygen species through Gs-PKA signalling27. Accordingly, accumulation of isoproterenol-induced DNA damage is suppressed by inhibition of PKA (Fig. 4d, lanes 1 and 3). Consistent with the idea that ARRB1-mediated effects on DNA damage are due to altered p53 levels, rescuing p53 expression (Supplementary Fig. 3l) decreases isoproterenol-induced γ-H2AX foci (Fig. 4c, panels 9–16; Fig. 4d, lanes 1 and 2) and the p53 effect is antagonized by co-expression of ARRB1 (Fig. 4c, panels 17–20, and Supplementary Fig. 6b). These G-protein-mediated and ARRB1-mediated pathways may synergistically affect the accumulation of isoproterenol-induced DNA damage. Thus, combining PKA inhibition with rescue of p53 expression abrogates accumulation of DNA damage (Fig. 4d, lanes 1 and 4). Catecholamine-induced lowering of p53 levels may lead to increased survival of cells containing DNA damage, owing to an impaired DNA damage check-point and repair cascade28. This would then facilitate accumulation of DNA damage. Accordingly, U2OS cells were irradiated with ultra-violet light after isoproterenol stimulation. Chronic stimulation leads to increased FOS expression, an indicator of cell survival and proliferation (Supplementary Fig. 6c). Because DNA damage occurs under these conditions, FOS expression leads to proliferation of cells that contain DNA damage. Taken together, these data indicate that Gs-PKA-dependent signalling, which leads to the generation of reactive oxygen species27, and ARRB1-dependent p53 degradation, which results in impaired DNA checkpoint and repair mechanisms28, synergistically lead to accumulation of DNA damage and consequently may have effects on genomic integrity.
DNA damage may promote rearrangements in chromosomes. To quantify the occurrence of catecholamine-induced rearrangements, we analysed inter-chromosomal rearrangements between Tcrg (the T-cell-receptor-γ locus) and Tcrb (the T-cell-receptor-β locus) in thymocytes (see Methods and Supplementary Fig. 6d). Both wild-type and Arrb1−/− mice were infused for four weeks with either saline or isoproterenol, and genomic DNA was isolated from the thymus. Consistent with the accumulation of DNA damage (Figs 1a, 3f), isoproterenol infusion leads to an increase in these Tcr rearrangements in wild-type mice; however, the effects are no longer observed in Arrb1−/− mice (Fig. 4e and Supplementary Fig. 6e). This indicates that catecholamine-stress-hormone-dependent accumulation of DNA damage promotes rearrangements in chromosomes.
Because chronic isoproterenol stimulation affects DNA damage and chromosomal rearrangements, we examined whether this cascade also affects genome integrity in the testes, in which paternal stress may affect the offspring’s genome. Using the isoproterenol-infusion model of chronic stress7,13, we observed that isoproterenol stimulation leads to a lowering of p53 levels in the testes. The effects are abolished in Arrb1−/− mice (Fig. 4f). To examine these phenomena in a genome-wide context, we conducted an array-comparative genomic hybridization (array-CGH). In the same model of chronic stress7,13, genomic DNA was isolated from the testes and thymus, which allowed us to eliminate any changes due to meiotic recombination by considering only rearrangements that occurred in both organs (see study design in Supplementary Fig. 6f). These studies show that the only such rearrangement occurring upon isoproterenol-infusion results in a duplication of more than 1 megabase (Mb) in regions 4qD2.2 and 4qD1 in wild-type mice; however, these events are not observed in the testes from Arrb1−/− mice (Fig. 4g and Supplementary Fig. 6f). Quantitative PCR (qPCR) of the testicular genome from each mouse also confirms isoproterenol-induced duplication at 4qD2.2 in an ARRB1-dependent manner (Supplementary Fig. 6g). Taken together, these data support the hypothesis that β2-adrenoreceptor- and ARRB1-dependent signalling regulates catecholamine-induced degradation of p53, thus leading to the accumulation of DNA damage in both somatic and germline cells (Fig. 4h).
The stress response is conserved in mammals, and is probably required for survival. However, psychosocial stress in humans is not time-limited, because aspects of this type of stress response can be sustained over months or even years. This may lead to prolonged secretion of stress hormones and consequent adverse effects for the individual. Indeed, clinical studies have shown marked risk-reductions for prostate cancer, lung adenocarcinoma and Alzheimer’s disease associated with chronic β-blocker (β-adrenoreceptor-antagonist) therapy4,29,30. It also seems plausible that such hormonal influences on DNA damage may not be limited to the β2-adrenoreceptors.
METHODS
Reagents
Unless otherwise noted, chemicals were purchased from Sigma.
Antibodies
Antibodies used were as follows, indicated WB for western blotting, IP for immunoprecipitation and CM for confocal microscopy. PARP-1 (WB, 1:500 dilution, Alexis Biochemicals). FOXO3a (WB, 1:500), MDM2 phosphorylated at Ser 166 (WB, 1:1,000), FOS (WB, 1:500), PUMA (WB, 1:500), p21 (WB, 1:500), all from Cell Signaling. Mouse p53 (FL-393:IP, 2 µg; WB, 1:200; CM, 1:50), human p53 (DO-1: WB, 1:5,000), MDM2 (SMP14: IP, 1 µg), ARRB1 (K-16: IP, 1 µg), ubiquitin (P4D1: WB, 1:200), human β2-adrenoreceptor (H-20: WB, 1:3,000), mouse β2-adrenoreceptor (M-20: WB, 1:200), anti-goat IgG-HRP (WB, 1:10,000), all from Santa Cruz. ARRB1 (10: WB, 1:200; CM, 1:50; BD Biosciences). LDH (WB, 1:300; Calbiochem). Mdm2 (HDM2-323: WB, 1:200), β-tubulin I (SAP.4G5: WB, 1:10,000), both from Sigma. Histone H2B (WB, 1:5,000), histone H2B phosphorylated on Ser 14 (WB, 1:5,000), γ-H2AX (WB, 1:1,000; CM, 1:100), all from Millipore. 53BP1 (WB, 1:1,000; Novus). p21 (WB, 1:200; Rockland). Anti-mouse IgG-HRP (WB, 1:10,000), anti-rabbit IgG-HRP (WB, 1:10,000), both from GE Healthcare. Rabbit polyclonal ARRB1 antibody (A1CT: WB, 1:20,000) was generated as previously described31.
Primers
Tcrg a1: 5′-ACCATACACTGGTACCGGCA-3′, Tcrg b1: 5′-ACCCC TACCCATATTTTCTTAG-3′, Tcrb a2: 5′-TCTACTCCAAACTACTCCAG-3′, Tcrb b2: 5′-CCTCCAAGCGAGGAGATGTGAA-3′, non-specific locus (chr 7) forward: 5′-AGGCCTGGCTAGGCTTTTGGAATCTTTC-3′, non-specific locus (chr 7) reverse: 5′-TGCCAGTGCTGGTGCGTGTGCACGGCTGT-3′, qPCR chr4 qD2.2 forward: 5′-TGGTGCTGGCACAACTGGCA-3′, qPCR chr4 qD2.2 reverse: 5′-TGACGGTGTCTTTTGCCTTACAGAAGC-3′, qPCR control (chr1) forward: 5′-CCTCCCATCAACGTTCAGGAGCC-3′, qPCR control (chr1) reverse: 5′-ACTGCTTCTGCTCCAAACCCTGC-3′, p21 promoter forward: 5′-CCAGAGGATACCTTGCAAGGC-3′, p21 promoter reverse: 5′-TCTCTGT CTCCATTCATGCTCCTCC-3′.
Peptides
The synthesis of ARRB1-binding peptide (ARRB1-BP: V2Rpp) has been described elsewhere. The sequence of the peptide, with phosphorylation sites underlined, is: ARGRTPPSLGPQDESCTTASSSLAKDTSS (ref. 32).
Plasmids
The MDM2-S166D plasmid17 and p53 deletion constructs33 and were gifts from M. C. Hung and S. H. Snyder, respectively. Plasmids encoding shRNAs against p53 (psiRNA-mp53 and psiRNA-hp53), and the control psiRNA-LucGL3, were purchased from InvivoGen.
Experimental procedures
Each experiment was repeated at least three times with comparable results, unless indicated otherwise.
Immunoblotting
SDS polyacrylamide gel electrophoresis (SDS-PAGE) was performed on 1.0-mm-thick NuPAGE 4–12% Bis-Tris gels (Invitrogen) and separated proteins were transferred to nitrocellulose membranes by semi-dry transfer, using trans-blot transfer medium (Bio-Rad). Blots were blocked with blocking buffer (5% skimmed milk in PBS with 0.02% Tween-20) before incubation at 4 °C overnight with primary antibodies, diluted in blocking buffer as described above. Blots were washed three times for 5 min each in PBS with 0.02% Tween-20, and then incubated with secondary antibodies in blocking buffer. Blots were washed three times for 5 min each in PBS with 0.02% Tween-20, and developed by SuperSignal West Pico/Femto solution (Pierce). Each protein band of interest on the immunoblot was quantified by densitometry using the GeneTools program (SynGene).
Co-immunoprecipitation
Cells were lysed in a lysis buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 0.1% CHAPS buffer, 0.1 mg ml−1 BSA, 1 mM PMSF and 1 mM EDTA, with Halt protease and phosphatase inhibitor cocktail (Pierce)), and homogenized by passing through a 28-gauge needle 20 times. Crude lysates were cleared of insoluble debris by centrifugation at 14,000g. Extra lysis buffer was added to 100–500 µg of cell lysate to bring samples to a total volume of 1 ml. Immunoprecipitating antibody (1–2 µg) was added and incubated on a rotator at 4 °C overnight. On the following day, 25 µl (50% slurry) of the appropriate TrueBlot IP beads (eBioscience) was added and incubated on a rotator at 4 uC for 1 h. The beads were washed five times with the lysis buffer and quenched with 30 µl of SDS sample buffer (×2). For detection of p53 ubiquitination (Fig. 3b), 10 mM N-ethylmaleimide and 20 µM MG132 were added to the lysis buffer. Coimmunoprecipitation after cell fraction was conducted as previously described34. Briefly, cells were lysed in RIPA A buffer (0.3% Triton X-100,50 mM Tris (pH 7.4) and 1 mM EDTA), with rotation at 4 °C for 30 min. Cell lysates were centrifuged at 14,000g for 10 min and the supernatant was used as the cytosolic fraction. The nuclear fraction was extracted from the pellet with RIPA B buffer (1% Triton X-100, 1% SDS, 50mM Tris (pH7.4), 500 mM NaCl and 1 mM EDTA), affinity-precipitated with the indicated antibodies, and subjected to SDS-PAGE.
Subcellular fractionation
U2OS cells from a 10-cm plate were resuspended in 300 µl of buffer B (0.25 M sucrose, 10 mM Tris (pH7.4), 10 mM MgCl2, 10 mM KCl,1 mM DTT and protease inhibitor cocktail without EDTA) and homogenized with 150 strokes in a 1-ml dounce tissue grinder (Wheaton) using a tight pestle on ice. After centrifugation at 750g for 10 min, the supernatant was isolated to separate the cytosolic fraction. The pellet was washed twice with buffer B, and resuspended in 100–200 µl of buffer B. The suspension was analysed as the nuclear fraction. Cytosolic fractions and nuclei were also prepared by using Nuclei EZ prep nuclei isolation kit (Sigma), following the manufacturer’s protocol, and comparable results were obtained. To detect the effects of PI3K inhibition by LY294002 on isoproterenol-induced p53 nuclear export, U2OS cells were pre-incubated with 10 µM LY294002 for 30 min, and then stimulated with 10 µM isoproterenol for 1 h. To detect the effects of nuclear export on decreased nuclear p53, U2OS cells were pre-incubated with 10 nM leptomycin B for 30 min, and then stimulated with 10 µM isoproterenol for 1 h. To prepare a total-cell extract, cell pellets were lysed in a lysis buffer (50 mM Tris (pH7.4), 150 mM NaCl, 0.1% CHAPS, 0.1 mg ml−1 BSA, 1mM PMSF and 1mM EDTA, with Halt protease and phosphatase inhibitor cocktail (Pierce)) and homogenized by passing through a 28-gauge needle 20 times. Crude lysates were cleared of insoluble debris by centrifugation at 20,000g.
Cell culture conditions and treatments
Wild-type MEFs (passage number ~72), Arrb1−/− MEFs (passage number ~76) and Arrb2−/− MEFs (passage number ~49) were prepared according to the 3T3 protocol35,36. Established MEF cultures and RAW264.7 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) with 10% FBS and 2 mM L-glutamine at 37 °C with a 5% CO2 atmosphere in a humidified incubator. U2OS, HEK-293 and NCI-H1299 cells were maintained in modified Eagle medium (MEM) with 10% FBS and 2 mM L-glutamine, with the same conditions as above. U2OS and NCI-H1299 cells were transfected with FuGENE6 transfection reagent (Roche) following the manufacturer’s protocol. For RNA interference for ARRB 1, the vector system shRNA was used as previously described37,38. Briefly, U2OS cells in 10-cm plates were transfected with either 10 µg control shRNA plasmid (5′-ACGTGACACGTTCGGAGAATTGATATCCGTTC TCCGAACGTGTCACGTTT-3′) or 10 µg ARRB1 shRNA plasmid (5′-ATTCT CCGCGCAGAAGGCTTT GATATCCG AGCCTTCTGCGCGGAGAATTT-3′), and incubated for 72 h. HEK-293 cells and MEFs were transfected with lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol. Briefly, 2 µg of DNA was dissolved in 35 µl of serum- and antibiotic-free medium per well, in a 6-well plate. Lipofectamine 2000 (10 µl) was mixed with 25 µl of serum- and antibiotic-free medium, and incubated for 5 min. The prepared DNA and lipofectamine 2000 solutions were mixed and the mixture was incubated for 20 min at 18–23 °C. During the incubation, normal cell-culture medium was replaced with serum-and antibiotic-free medium. The transfection mixture was added to the cells in serum-free culture, and incubated overnight. On the following day, the medium was replaced with normal serum- and antibiotic-containing growth medium, and the cells were incubated for 48–72 h before testing.
Isoproterenol, epinephrine and norepinephrine were prepared fresh for each experiment by dissolving the bitartrate salts (Sigma) immediately before stimulation. To study chronic β-adrenergic effects, U2OS cells and MEFs were cultured until confluent, then stimulated with 10 µM isoproterenol for 24 h, unless otherwise indicated. To study γ-H2AX formation, cells were cultured until 40–50% confluent, then stimulated with 10 µM isoproterenol every 12 h for 3 days. H-89 (10 µM), leptomycin B (10nM), ICI 118,551 (10 µM), wortmannin (100 nM), 5-(2-benzothiazolyl)-3-ethyl-2-(2-(methylphenylamino)ethenyl)-1-phenyl-1H-benzimidazolium iodide (AKTi, 1 µM) or LY294002 (10 µM, Sigma) were added to the media 30 min before stimulation with isoproterenol. To study the effects of isoproterenol stimulation on cell proliferation after DNA damage, cells were ultraviolet-irradiated (50 J per m2) and incubated for 6 h, followed by stimulation with 10 µM isoproterenol every 12 h for 3 days. Cell lysates were examined by immunoblotting for FOS, an indicator of cell survival and proliferation39. To study phosphorylation of MDM2 at Ser 166 in MEFs, cells were serum-starved for 4 h, then stimulated with 10 µM isoproterenol for 1 h. To study this phosphorylation event in U2OS cells, cells were serum-starved for 36 h, then stimulated with 10 µM isoproterenol for 10 min.
In vitro ubiquitination assay
10 nM His-p53 (ProteinOne) was mixed with 200 ng E1 (BostonBiochem), 200 ng UbcH5b (BostonBiochem), 5 µg ubiquitin (BostonBiochem) and 25 nM MDM2 in 20 µl of reaction mixture (40 mM Tris (pH7.6), 2 mM ATP-Mg2+, 1mM dithiothreitol and 5 mM MgCl2). Purified recombinant ARRB1 (0, 50 or 500 nM) was added to the reaction mixture in the presence or absence of 300 nM ARRB1-BP. The sample was incubated for 60 min at 30 °C, resolved by SDS-PAGE and analysed by immunoblotting with anti-p53 antibody (DO-1).
Isoproterenol infusion
Wild-type (C57BL/6), Arrb1 knockout (Arrb1−/−)40 or β2-adrenoreceptor knockout (Adrb2−/−)41 mice were subcutaneously implanted with ALZET osmotic pumps to administer saline or isoproterenol (30 mg kg−1 d−1) continuously, dissolved in saline, for 28 days (mini-osmotic pump model 2004), following the manufacturer’s procedure. After administration, animals were killed and the indicated organs were dissected out. For protein preparation, dissected organ tissues were lysed and sonicated in RIPA buffer (50 mM Tris (pH 7.4), 500 mM NaCl, 1% SDS, 1% Triton X-100 and 1 mM EDTA, with Halt protease and phosphatase inhibitor cocktail). Genomic DNA was prepared from dissected organ tissues by DNeasy blood & tissue kit (Qiagen), following the manufacturer’s protocol. All animals used in these studies were adult male mice of 8–12 weeks of age. All mouse strains were backcrossed to the C57BL/6 background for ≥ 10 generations. Animals were handled according to approved protocols and animal welfare regulations of the Institutional Review Board at Duke University Medical Center.
Quantitative real-time PCR (qPCR)
qPCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems) and StepOne Real-time PCR system (Applied Biosystems) following the manufacturer’s protocol. To validate the array-CGH analysis, relative genomic content (copy number) was determined with the comparative CT(ΔΔCT) method42.
GST pulldown assay
Wild-type rat Arrb1 or human MDM2 were subcloned into the pGEX4T1 vector and prepared according to the manufacturer’s recommendations (Amersham Biosciences). The GST tag was cleaved with thrombin protease (Hematologic Technologies Inc.). p53 (1 nM) was co-incubated overnight with 10 nM of GST-ARRB1 or 10 nM of GST at 4 °C in 1 ml binding buffer (50 mM Tris (pH7.4), 150 mM NaCl, 0.1 mg ml−1 BSA and 10 µM D-myo-inositol 1,2,3,4,5,6-hexakisphosphate), and 20 µl of 50% glutathione-sepharose was then added to the mixture. The mixture was further incubated at 4 °C for 1 h with rotation. The beads were washed once with 1 ml binding buffer, separated by SDS-PAGE and analysed by immunoblotting.
Immunofluorescence experiments
Immunofluoresence using confocal microscopy was carried out as previously described43. For detection of γ-H2AX foci, we captured images of more than 20 fields per preparation, which were randomly chosen in a blind manner. Cells positive for γ-H2AX foci in each field were tallied and added together to determine the percentage. The total number of cells was counted with 4′,6-diamidino-2-phenylindole (DAPI) nuclear staining. For rescue experiments using the expression of RFP-ARRB1 (or RFP as a control) in MEFs, RFP-positive cells were counted for γ-H2AX foci. For p53 rescue experiments in NCI-H1299 cells, RFP-Arrb1 (or RFP) and p53 were co-transfected in a 1:3 ratio.
Detection of interchromosomal rearrangements between Tcrg and Tcrb
The trans-rearrangement between Tcrg and Tcrb loci were detected by nested PCR, using first the ‘a’ set of primers and then the ‘b’ set of primers, as previously described44 (Supplementary Fig. 6d). The number of rearrangements is expressed as the reciprocal of the highest dilution of DNA yielding an amplified product (for example, the number of trans-rearrangements per 1.5 × 105 cells (1 µg of DNA) is 1,000, yielding an amplifiable fragment at a 1:1,000 dilution (1 ng of DNA))44.
DNA preparation
DNA was prepared from the testes and thymus by using the DNeasy blood & tissue kit (Qiagen) following the manufacturer’s protocol. To enrich sperm from excised testis grafts, the testis was minced and the epithelial tissue, containing leydig and sertoli cells, was removed.
Array-comparative genomic hybridization (Array-CGH)
A tiling path CGH array for the genome analysis in mouse (UCSC Build mm9) was designed and constructed by NimbleGen Systems (NimbleGen). The resulting array contained 720,000 probes with a median probe spacing of 3,537 base pairs. Probes were synthesized using an isothermal format (melting temperature 76 °C), and varied in length from 50 to 75 base pairs. Genomic DNA from five mice for each experimental condition was pooled and examined. Genomic DNAs (1 µg) from test (isoproterenol-treated) and reference (saline-treated) mice were differentially labelled with 5′-Cy3 and 5′-Cy5 random nonamers (TriLink Biotechnologies), respectively, and hybridized to the oligoarray for 72 h using the MAUI hybridization station (BioMicro Systems Inc.). Image-capture of the hybridized arrays for fluorescent intensity extraction was performed using a Genepix 4100A scanner (Molecular Dynamics) and normalized using Nimblescan v2.5 microarray software (Nimblegen) before importing into Nexus Copy-Number (BioDiscovery) for analysis.
Array-CGH analysis
BioDiscovery’s rank segmentation algorithm, which is similar to circular binary segmentation45, was used to identify genomic rearrangements. The significance threshold was set as 1.0 × 10−7. The calling algorithm used cluster values and defined log2 thresholds of ±0.2. We applied a cutoff of ten oligomer clones showing the same trend in copy-number change to define chromosomal rearrangements. Black lines in the plot indicate a ‘cluster value’, which is the median log-ratio value of all the probes in that region. Isoproterenol-induced rearrangements in the testes were determined, and identical rearrangements were detected in the thymus (see study design in Supplementary Fig. 6f).
Radioligand binding experiments
For ICI 118,551 and CGP20712A affinity measurements, subtype-selective ligand affinities were determined from competition radioligand binding experiments, conducted according to previous works46,47. Briefly, 25 µg of cell membranes, prepared via differential centrifugation, were resuspended in assay buffer (50 mM Tris-HCl (pH7.4), 12.5 mM MgCl2, 2 mM EDTA and 1 mM ascorbic acid) containing 60 pM [125I]cyanopindolol (NEX189, 2,200 Ci mmol−1) and concentrations of ICI 118,551 or CGP 20712A ranging from 1 µM to 1 pM. Nonspecific binding was determined in the presence of 10 µM propranolol. After incubation at 25 °C for 90 min, membranes were collected and washed via vacuum filtration (Brandel) and the bound radioactivity was quantified using a Packard Cobra gamma counter (Perkin Elmer). Equilibrium inhibition constant (Ki) values were calculated from nonlinear regression analysis (Graphpad) using the method in ref. 48.
p53 reporter assay
U2OS cells were transfected with the p53-luc reporter plasmid (Stratagene) in the presence of serum, using FuGENE6 transfection reagent (Roche). Three hours after the transfection, media were changed to serum-free media containing 100 µM ascorbic acid and appropriate concentrations of iso-proterenol. Cells were incubated for 24 h and lysed in × 1 passive lysis buffer (PLB, Promega). The firefly luciferase reporter was analysed with addition of luciferase assay reagent II (Promega).
Chromatin immunoprecipitation assay (ChIP) and Re-ChIP
ChIP was performed as previously described49. In brief, both wild-type and Arrb1−/− MEFs were incubated with 50 µM etoposide for 20 h. After incubation, cells were treated with 2 mM disuccinimidyl glutarate (Pierce) to crosslink protein complexes, then treated with formaldehyde to link protein to DNA covalently. Cells were lysed and the nucleoprotein complexes were sonicated. DNA-protein complexes enriched by the initial immunoprecipitation with anti-ARRB1 (K-16) antibody were eluted from beads with elution buffer (1% SDS, 0.1 M NaHCO3), and further immunoprecipitated with anti-p53 (FL-393) antibody for Re-ChIP. The retrieved complexes were then analysed by PCR amplification of p53-binding elements in the p21 promoter.
Statistics
Unless otherwise noted, P values were calculated with Student’s t-test (two-tailed). Analysis of variance was performed with Prism (GraphPad).
Acknowledgements
R.J.L. is a Howard Hughes Medical Institute investigator. This work was supported byHL16037 and HL70631 (R.J.L.). We thank D.Addison and Q. Lennon for secretarial assistance; S. H. Snyderand M. A. Koldobskiy for providing p53 deletion constructs; B. K. Kobilkaand H. A. Rockman for providing Adrb2−/− mice; M. C. Hung for providing the MDM2-S166D plasmid; A. K. Shukla, A. Kahsai, J. Kim, J. Sun, S. M. DeWireand N. Odajima for discussion and comments.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions M.R.H. and R.J.L. designed experiments, directed the study and wrote the paper. M.R.H. performed most of the experiments, analysed the data and prepared the figures. R.J.L. supervised the study and provided financial support. J.J.K. performed some experiments, helped to analyse the data and helped to write the paper. E.J.W., S.R., K.X., S.K.S. and S.A. helped to analyse the data. E.J.W., S.R., R.T.S., B.W., C.M.L and S.A. performed some experiments. A.J.T. and S.G.G. performed the array-CGH and helped to analyse the data. W.G. and D.R.D. helped to characterize the functionality of p53 and to analyse the data.
The authors declare no competing financial interests.
References
- 1.Selye H. A syndrome produced by diverse nocuous agents. Nature. 1936;138:32. [Google Scholar]
- 2.Charmandari E, Tsigos C, Chrousos G. Endocrinology of the stress response. Annu. Rev. Physiol. 2005;67:259–284. doi: 10.1146/annurev.physiol.67.040403.120816. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein DS. Catecholamines and stress. Endocr. Regul. 2003;37:69–80. [PubMed] [Google Scholar]
- 4.Antoni MH, et al. The influence of bio-behavioural factors on tumour biology: pathways and mechanisms. Natl. Rev. 2006;6:240–248. doi: 10.1038/nrc1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flint MS, Baum A, Chambers WH, Jenkins FJ. Induction of DNA damage, alteration of DNA repair and transcriptional activation by stress hormones. Psychoneuroendocrinology. 2007;32:470–479. doi: 10.1016/j.psyneuen.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Lu T, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 7.Thaker PH, et al. Chronic stress promotes tumor growth and angiogenesis in a mouse model of ovarian carcinoma. Nature Med. 2006;12:939–944. doi: 10.1038/nm1447. [DOI] [PubMed] [Google Scholar]
- 8.Fratiglioni L, Paillard-Borg S, Winblad B. An active and socially integrated lifestyle in late life might protect against dementia. Lancet Neurol. 2004;3:343–353. doi: 10.1016/S1474-4422(04)00767-7. [DOI] [PubMed] [Google Scholar]
- 9.Kinney DK, Munir KM, Crowley DJ, Miller AM. Prenatal stress and risk for autism. Neurosci. Biobehav. Rev. 2008;32:1519–1532. doi: 10.1016/j.neubiorev.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nepomnaschy PA, et al. Cortisol levels and very early pregnancy loss in humans. Proc. Natl Acad. Sci. USA. 2006;103:3938–3942. doi: 10.1073/pnas.0511183103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Čikoš S, et al. Expression of beta adrenergic receptors in mouse oocytes and preimplantation embryos. Mol. Reprod. Dev. 2005;71:145–153. doi: 10.1002/mrd.20256. [DOI] [PubMed] [Google Scholar]
- 12.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. β-arrestins and cell signaling. Annu. Rev. Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 13.Ni Y, et al. Activation of β2-adrenergic receptor stimulates γ-secretase activity and accelerates amyloid plaque formation. Nature Med. 2006;12:1390–1396. doi: 10.1038/nm1485. [DOI] [PubMed] [Google Scholar]
- 14.Bonner WM, et al. GammaH2AX and cancer. NatureRev. Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freedman DA, Levine AJ. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rainbow TC, Parsons B, Wolfe BB. Quantitative autoradiography of beta 1- and beta 2-adrenergic receptors in rat brain. Proc. Natl Acad. Sci. USA. 1984;81:1585–1589. doi: 10.1073/pnas.81.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou BP, et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nature Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 18.De Gregorio G, et al. The p85 regulatory subunit of PI3K mediates TSH–cAMP– PKA growth and survival signals. Oncogene. 2007;26:2039–2047. doi: 10.1038/sj.onc.1210011. [DOI] [PubMed] [Google Scholar]
- 19.Goel R, Phillips-Mason PJ, Raben DM, Baldassare JJ. α-Thrombin induces rapid and sustained Akt phosphorylation by β-arrestin1-dependent and - independent mechanisms, and only the sustained Akt phosphorylation is essential for G1 phase progression. J. Biol. Chem. 2002;277:18640–18648. doi: 10.1074/jbc.M108995200. [DOI] [PubMed] [Google Scholar]
- 20.Buchanan FG, et al. Role of β-arrestin 1 in the metastatic progression of colorectal cancer. Proc. Natl Acad. Sci. USA. 2006;103:1492–1497. doi: 10.1073/pnas.0510562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovacs JJ, Hara MR, Davenport CL, Kim J, Lefkowitz RJ. Arrestin development: emerging roles for β-arrestins in developmental signaling pathways. Dev. Cell. 2009;17:443–458. doi: 10.1016/j.devcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott MG, et al. Differential nucleocytoplasmic shuttling of β-arrestins. Characterization of a leucine-rich nuclear export signal in β-arrestin2. J. Biol. Chem. 2002;277:37693–37701. doi: 10.1074/jbc.M207552200. [DOI] [PubMed] [Google Scholar]
- 23.Wang P, Wu Y, Ge X, Ma L, Pei G. Subcellular localization of β-arrestins is determined by their intact N domain and the nuclear export signal at the C terminus. J. Biol. Chem. 2003;278:11648–11653. doi: 10.1074/jbc.M208109200. [DOI] [PubMed] [Google Scholar]
- 24.Wang P, et al. β-arrestin 2 functions as a G-protein-coupled receptor-activated regulator of oncoprotein Mdm2. J. Biol. Chem. 2003;278:6363–6370. doi: 10.1074/jbc.M210350200. [DOI] [PubMed] [Google Scholar]
- 25.Boularan C, et al. β-arrestin 2 oligomerization controls the Mdm2-dependent inhibition of p53. Proc. Natl Acad. Sci. USA. 2007;104:18061–18066. doi: 10.1073/pnas.0705550104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nobles KN, Guan Z, Xiao K, Oas TG, Lefkowitz RJ. The active conformation of β-arrestin1: direct evidence for the phosphate sensor in the N-domain and conformational differences in the active states of β-arrestins1 and-2. J. Biol. Chem. 2007;282:21370–21381. doi: 10.1074/jbc.M611483200. [DOI] [PubMed] [Google Scholar]
- 27.Yan L, et al. Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell. 2007;130:247–258. doi: 10.1016/j.cell.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 28.Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nature Rev. Mol. Cell Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 29.Schuller HM. Mechanisms of smoking-related lung and pancreatic adenocarcinoma development. Nature Rev. Cancer. 2002;2:455–463. doi: 10.1038/nrc824. [DOI] [PubMed] [Google Scholar]
- 30.Khachaturian AS, et al. Antihypertensive medication use and incident Alzheimer disease: the Cache County Study. Arch. Neurol. 2006;63:686–692. doi: 10.1001/archneur.63.5.noc60013. [DOI] [PubMed] [Google Scholar]
- 31.Attramadal H, et al. β-arrestin2, a novel member of the arrestin/β-arrestin gene family. J. Biol. Chem. 1992;267:17882–17890. [PubMed] [Google Scholar]
- 32.Xiao K, Shenoy SK, Nobles K, Lefkowitz RJ. Activation-dependent conformational changes in β-arrestin 2. J. Biol. Chem. 2004;279:55744–55753. doi: 10.1074/jbc.M409785200. [DOI] [PubMed] [Google Scholar]
- 33.Koldobskiy MA, et al. p53-mediated apoptosis requires inositol hexakisphosphate kinase-2. Proc. Natl Acad. Sci. USA. 2010;107:20947–20951. doi: 10.1073/pnas.1015671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hara MR, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nature Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 35.Kohout TA, Lin FS, Perry SJ, Conner DA, Lefkowitz RJ. β-Arrestin 1 and 2 differentially regulate heptahelical receptor signaling and trafficking. Proc. Natl Acad. Sci. USA. 2001;98:1601–1606. doi: 10.1073/pnas.041608198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 38.Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl Acad. Sci. USA. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seshadri T, Campisi J. Repression of c-fos transcription and an altered genetic program in senescent human fibroblasts. Science. 1990;247:205–209. doi: 10.1126/science.2104680. [DOI] [PubMed] [Google Scholar]
- 40.Conner DA, et al. β-Arrestin1 knockout mice appear normal but demonstrate altered cardiac responses to β-adrenergic stimulation. Circ. Res. 1997;81:1021–1026. doi: 10.1161/01.res.81.6.1021. [DOI] [PubMed] [Google Scholar]
- 41.Chruscinski AJ, et al. Targeted disruption of the β2 adrenergic receptor gene. J. Biol. Chem. 1999;274:16694–16700. doi: 10.1074/jbc.274.24.16694. [DOI] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Kovacs JJ, et al. β-arrestin-mediated localization of smoothened to the primary cilium. Science. 2008;320:1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lista F, Bertness V, Guidos CJ, Danska JS, Kirsch IR. The absolute number of trans-rearrangements between the TCRG and TCRB loci is predictive of lymphoma risk: a severe combined immune deficiency (SCID) murine model. Cancer Res. 1997;57:4408–4413. [PubMed] [Google Scholar]
- 45.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 46.Hausdorff WP, Hnatowich M, O’Dowd BF, Caron MG, Lefkowitz RJ. A mutation of the β 2-adrenergic receptor impairs agonist activation of adenylyl cyclase without affecting high affinity agonist binding. Distinct molecular determinants of the receptor are involved in physical coupling to and functional activation of Gs. J. Biol. Chem. 1990;265:1388–1393. [PubMed] [Google Scholar]
- 47.Baker JG. The selectivity of β-adrenoceptor antagonists at the human β1, β2 and β3 adrenoceptors. Br. J. Pharmacol. 2005;144:317–322. doi: 10.1038/sj.bjp.0706048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng Y, Prusoff WH. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition(I50) of anenzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 49.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-κB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]