Abstract
Protein tyrosine kinases (PTKs) coordinate a broad spectrum of cellular responses to extracellular stimuli and cell–cell interactions during development, tissue homeostasis, and responses to environmental challenges. Thus, an understanding of the regulatory mechanisms that ensure physiological PTK function and potential aberrations of these regulatory processes during diseases such as cancer are of broad interest in biology and medicine. Aside from the expected role of phospho-tyrosine phosphatases, recent studies have revealed a critical role of covalent modification of activated PTKs with ubiquitin as a critical mechanism of their negative regulation. Members of the Cbl protein family (Cbl, Cbl-b and Cbl-c in mammals) have emerged as dominant “activated PTK-selective” ubiquitin ligases. Structural, biochemical and cell biological studies have established that Cbl protein-dependent ubiquitination targets activated PTKs for degradation either by facilitating their endocytic sorting into lysosomes or by promoting their proteasomal degradation. This mechanism also targets PTK signaling intermediates that become associated with Cbl proteins in a PTK activation-dependent manner. Cellular and animal studies have established that the relatively broadly expressed mammalian Cbl family members Cbl and Cbl-b play key physiological roles, including their critical functions to prevent the transition of normal immune responses into autoimmune disease and as tumor suppressors; the latter function has received validation from human studies linking mutations in Cbl to human leukemia. These newer insights together with embryonic lethality seen in mice with a combined deletion of Cbl and Cbl-b genes suggest an unappreciated role of the Cbl family proteins, and by implication the ubiquitin-dependent control of activated PTKs, in stem/progenitor cell maintenance. Future studies of existing and emerging animal models and their various cell lineages should help test the broader implications of the evolutionarily-conserved Cbl family protein-mediated, ubiquitin-dependent, negative regulation of activated PTKs in physiology and disease.
Keywords: Cbl, E3 ubiquitin ligase, Tyrosine kinase binding domain, Ubiquitination, Signal transduction, Animal model
1. Introduction
Phosphorylation of proteins on tyrosine residues is an evolutionarily conserved mechanism used by higher eukaryotes to alter the activity of cellular pathways in response to changes in extracellular environment. This fundamentally simple biochemical process, mediated by protein tyrosine kinases (PTKs), serves as an initial trigger in cellular activation processes whose downstream components include other major signal transduction enzyme cascades such as serine/threonine kinases, lipid kinases, phosphatases, phospholipases, small GTPases and other pathways. PTK-mediated signaling ensures orderly cell–environment and cell–cell interactions during development, tissue homeostasis, tissue repair and remodeling, as well as protection against pathogens. It is therefore not surprising that deregulation of PTK-initiated signal transduction is a major cause of human diseases such as cancer [1]. Given their critical importance in signal transduction cascades that regulate normal development and homeostasis in higher organisms, as well as the causal linkage of their aberrant activities to human diseases, understanding the basis of PTK regulation is therefore of fundamental interest in biology and medicine.
2. Protein tyrosine kinases as key mediators of trans-membrane signaling
2.1. Both receptor and non-receptor protein tyrosine kinases mediate trans-membrane signaling
58 of the 90 PTKs in the human genome are trans-membrane receptors (referred to as receptor tyrosine kinases or RTKs) and are therefore directly poised for signal transduction in response to extracellular cues. Classical members of this family include growth factor receptors such as platelet-derived growth factor receptor (PDGFR) and epidermal growth factor receptor (EGFR). However, RTKs are now known to play crucial roles in essentially all developmental and homeostatic signaling pathways triggered by interactions with soluble or cell-surface associated ligands [1]. Nearly all of the remaining PTKs, the so-called non-receptor PTKs, are also involved in membrane receptor-initiated signaling through their constitutive or inducible association with cell surface receptors. For example, members of the Src-family of PTKs associate with antigen receptors on immune cells and members of the JAK-family of PTKs with cytokine receptors; in each case, non-covalently associated PTKs are essential for cellular activation upon receptor engagement, as demonstrated by naturally-occurring or engineered mutations of genes encoding these PTKs [2,3]. Thus, PTKs serve as essential membrane-proximal receptor signal transducers. Importantly, cell surface receptors with intrinsic or associated PTK activity commonly serve as biochemical or functional partners of other cell surface receptors such as G-protein coupled receptors (GPCRs), integrins and others, and are also activated by altered metabolic states of cells, such as oxidative stress. These collaborative interactions of PTKs further expand their critical roles to essentially every physiological response in higher eukaryotes [1].
2.2. Tyrosine phosphorylation-dependent signaling
Similar to serine/threonine phosphorylation, tyrosine phosphorylation can directly regulate enzymatic activity as exemplified by the phosphorylation of activation loops of PTKs. Importantly, phosphotyrosine (pY)-containing peptide sequences generated on PTKs themselves or on their interacting partners serve as docking sites that are recognized by Src-Homology 2 (SH2) and phosphotyrosine-binding (PTB) domains on signaling proteins. These interactions promote the recruitment of key regulatory and/or enzymatic components of signaling pathways such as Ras-Erk, PI3-kinase and PLCγ to membrane-associated activated receptors resulting in the activation of these downstream pathways [1]. Given the primary role of phosphorylation-dependent regulation of signaling protein complex formation and subsequent activation of enzymatic cascades, dephosphorylation by phosphotyrosine phosphatases (PTPs) is thought to provide one mechanism to negatively regulate PTK-dependent cellular activation. Notably, however, only in a limited number of cases have PTPs been categorically established as negative regulators of PTK signaling and paradoxically a number of PTPs mediate positive roles in PTK-dependent signaling cascades likely through dephosphorylation of sites that serve in negative regulation of PTK signaling [4]. Consistent with this dual role, recent cancer genome sequencing analyses have revealed that PTPs may function in both negatively (as putative tumor suppressors) and positively (as oncogenes) regulating oncogenesis [5].
2.3. Endocytic traffic of protein tyrosine kinase-associated receptors as a determinant of duration and diversification of signaling
Similar to ligand-binding cell surface receptors in general, PTK-associated receptors invariably undergo ligand-induced internalization through the endocytic pathway (Fig. 1). The rapid relocation of activated receptors from the cell surface into endocytic compartments has emerged as a fundamental mechanism to control signaling downstream of PTK-associated cell surface receptors. Studies over the last quarter century have elegantly demonstrated that a major consequence of the internalization of activated receptors is their trafficking to lysosomes where they undergo degradation [6,7]. This provides a mechanism of signal termination that appears to be essential for regulating signaling outputs from activated PTK-associated receptors under physiological conditions. Indeed, endocytosis-impaired receptors, such as EGFR with a deletion of its C-terminal sequences, become hyperactive and transforming [8]. Similarly, higher signaling potency of ErbB2, compared to its family members such as EGFR, has been attributed to its being internalization (and lysosomal degradation)-impaired [9,10].
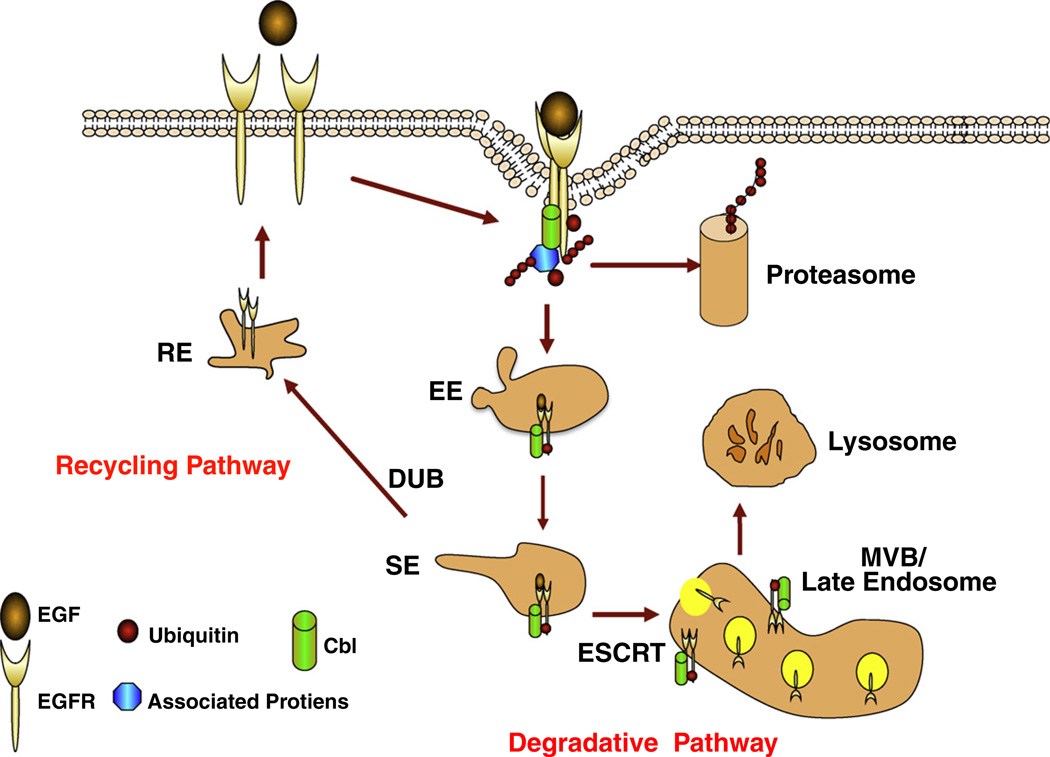
Fig. 1.
Alternate routes of endocytic traffic of activated tyrosine kinase coupled cell surface receptors and the role of Cbl-family protein dependent ubiquitination. The schematic shows the EGF-induced activation and endocytosis of EGF receptor (EGFR) as a prototype; for simplicity, only clathrin-dependent endocytic pathway is shown. EGF activation leads to receptor dimerization, auto-/trans-phosphorylation and recruitment of signaling proteins including Cbl proteins. The activated receptors accumulate in clathrin-coated pits followed by internalization into early endosomes (EE) and traffic to sorting endosomes (SE), late endosome/multi-vesicular body (MVB) and finally lysosomes (degradative pathway). Alternatively, internalized receptors can be sorted into recycling pathway and returned for further ligand binding and activation (Recycling Pathway). Cbl-dependent ubiquitination is thought to serve as an internalization signal and as a sorting signal at the level of MVBs. The latter role involves the recognition of the ubiquitin modification on the receptor by components of the ESCRT complexes to facilitate the sorting of ubiquitinated receptors into internal vesicles of the MVB, an event required for lysosomal degradation of activated receptors. Non-ubiquitinated receptors or those from which ubiquitin species have been removed by deubiquitinases (DUBs) are thought to enter the recycling pathway. Signaling proteins that interact with Cbl proteins and are ubiquitinated may also be targeted for an alternate mode of degradation in the proteasome.
Further studies have demonstrated that lysosomal degradation is not an invariant, or the only, consequence of the endocytosis of activated PTK-associated receptors. For example, while studies of EGFR using its prototypical ligand EGF have demonstrated lysosomal degradation as a major consequence, TGFα stimulation is associated with less lysosomal degradation of EGFR and a higher degree of its recycling back to the cell surface [11,12]. More recent analyses of all available EGF ligands have further established the distinct itineraries of internalized EGFR depending on the ligand [13]. In other studies, it has been established that concentrations of ligands can also dictate distinct itineraries of internalized receptors. For example, lower concentrations of EGF sufficient for induction of cell proliferation were associated with internalization of EGFR but not its degradation while higher concentrations of EGF promoted EGFR degradation [14]. These findings, together with studies of additional PTK-associated as well as non-PTK-associated receptors have contributed to the idea of endosomal signaling [15,16].
According to this newer concept, internalization of activated receptors by itself is not a signal attenuation mechanism. Instead, internalized receptors continue to signal within endosomal compartments until they are degraded in the lysosome or inactivated by other mechanisms and returned to the cell surface. The extent of receptor signaling within endosomes may indeed surpass that at the cell surface. In all cell models, the life of an activated RTK on the cell surface is very short (1–3 min) yet substantial degradation of internalized, activated receptor typically takes ten times longer: thus, activated RTKs spend nearly 90% of their lifespan within endosomes [6,7]. Studies in many cell systems have now shown that internalized RTKs continue to signal [15,17,18]. For example, signaling proteins such as Grb2 and Shc remain physically associated with activated EGFR and NGF receptors even after they traverse to endosomes [17–19]. Signaling from endosomes is indeed required for Trk receptor-mediated gene regulation in the neuronal cell body in response to stimulation at distant nerve endings [16].
Continued signaling ability of internalized PTK-associated receptors therefore provides a potential mechanism to regulate the duration of signal output depending on cellular needs. Thus, ligands (or concentrations of ligands) that promote lysosomal degradation can be anticipated to induce a relatively un-sustained signal output. In contrast, ligand conditions that do not promote lysosomal degradation can be anticipated to provide more sustained signaling outputs [6,7]. Studies of growth factor receptors in model cell systems have demonstrated that the transient vs. sustained activation of a single signaling axis can have dramatically different consequences for cell fate decisions. For instance, elegant studies using PC12 neuroblastoma cells have shown that transient Erk activation by EGF leads to a proliferative outcome while sustained Erk activation in response to NGF is associated with neuronal differentiation [20]. In a more recent example, maintenance of primary mammary epithelial organoid cultures in EGF versus amphiregulin determined the dominance of basal vs. luminal cell populations based on the extent of Erk activation [21].
In addition to regulating the duration and intensity of signaling outputs, evidence also supports the role of endosomal signaling by PTK-associated receptors in signal diversification. For example, inhibition of EGFR endocytosis with a dynamin-K44A mutant reduced the level of phosphorylation of EGFR, the p85 subunit of PI3-kinase and Erk1, but increased the phosphorylation of PLCγ1 and Shc [22]. Recent studies indicate that RTK internalization is required for full MAPK activation, apparently due to the localization of a critical MEK-MAPK tethering protein within endosomes [23,24]. Additional studies have uncovered endocytic pathways that deliver active receptor signaling complexes to ER and Golgi for Ras activation [25]. Indeed, the requirement of endocytosis to activate signaling pathways extends to other receptors, such as G-protein-coupled receptors and TGFβ receptor [26–28]. Notably, endocytosis of EGF-EGFR complexes in the presence of a washable tyrosine kinase inhibitor (TKI), followed by inhibitor washout, demonstrated that initiation of signaling directly from endosomes was sufficient to promote cell survival and cell-cycle entry [29,30]. The new paradigm of endosomal signaling by receptors activated by their ligands at the cell surface therefore provides a unique mechanism for spatio-temporal regulation to achieve optimal duration and diversification of signaling outputs.
Given the vastly distinct functional consequences of endosomal sorting of internalized PTK-associated receptors into alternative lysosomal vs. recycling fates, the regulatory mechanisms involved have been of great interest. Early studies demonstrated that EGF-dependent lysosomal targeting of EGFR was a saturable process, suggesting the likelihood that certain key regulatory proteins were rate-limiting [31]. Recent studies have established that this postulated mechanism for lysosomal sorting of activated PTK-associated cell surface receptors is mediated by members of the Cbl family of ubiquitin ligases. The next sections therefore focuses on a brief overview of the ubiquitin pathway followed by a discussion of how the Cbl family of proteins controls lysosomal sorting of activated PTK-associated receptors to provide a fundamental mechanism of negative regulation of signaling. We will then discuss the physiological implications of this mechanism and pathological consequences of its aberrations.
3. Ubiquitin machinery and the control of cellular signaling by protein tyrosine kinase-coupled cell surface receptors
3.1. Components of the ubiquitin pathway
The pathway of post-translational modification of proteins with ubiquitin involves a highly conserved enzymatic cascade [32–34]. A ubiquitin-activating or E1 enzyme initiates the cascade in which a series of enzymatic reactions lead to an ATP-dependent thioester bond formation with the C-terminus of a ubiquitin molecule. This activated E1-ubiquitin conjugate binds with high affinity to a ubiquitin-conjugating enzyme (UBC) and transfers active ubiquitin to E2, again linked via a thioester bond. The E2 enzymes stably associate with ubiquitin ligase enzymes (E3s) of which the two main classes are the catalytic variety of HECT-domain containing E3s and the non-catalytic category of RING finger domain (and related domains)-containing E3s. The E2 enzymes bound to HECT domain-containing E3s transfer the ubiquitin moiety to E3 (via a thioester bond), which in turn transfers ubiquitin to the ε-amine groups of lysine residues in target polypeptides through a stable iso-peptide linkage. The E3s of the RING finger domain category function as scaffolds, with the E3-bound E2 directly transferring ubiquitin to the target.
The E1–E2–E3 cascade helps establish the wide range of targets that can be subjected to ubiquitin modification relatively specifically [32–34]. Only two ubiquitin-directed functional E1s exist in mammals and it is likely that they function in distinct cellular processes, although their redundant roles remain to be fully explored. There are nearly three dozen E2s, a majority of them dedicated to the ubiquitin pathway. In contrast, several hundred ubiquitin pathway E3 proteins are encoded by the mammalian genome. The large number of E3s broadens the range of target proteins that are regulated by ubiquitin modification. E3s typically use distinct domains/motifs (and often several of them) to specifically interact with their substrates, with many of the interactions involving recognition of motifs generated by post-translational modifications (such as phosphorylation; this will be further illustrated with the interactions of Cbl proteins below). These specific interactions and their modulation under defined physiological conditions provide a mechanistic basis for ubiquitin modification of a limited group of substrates during specific cellular processes.
3.2. Types of ubiquitin modification and their consequences
In general, the major categories of ubiquitin modification of target proteins involve mono-ubiquitination and poly-ubiquitination with the latter involving chains linked through various lysine residues in ubiquitin (e.g., commonly observed K48- or K63-linked chains) or mixed chains [32–34]. While the precise regulatory mechanisms that result in a particular type of ubiquitin modification are not well understood, the type of ubiquitin modification dictates distinct functional consequences for the modified proteins. The common form of K48-linked poly-ubiquitin chains serves as a proteasomal targeting signal leading to degradation in the proteasome. In contrast, K63-linked poly-ubiquitin chains do not appear to function in proteasomal targeting. Both K63-linked chains as well as mono-ubiquitin modification, which is frequently seen in signal transduction, transcriptional and other regulatory pathways, dictate protein–protein interactions that do not lead to proteasomal degradation.
In the context of PTK-associated cell surface receptors, which is the main focus of this review, the receptors themselves or their associated adaptor proteins undergo K63-linked poly-ubiquitin and/or mono-ubiquitin modifications [35]. These modifications are recognized by a network of proteins with ubiquitin binding domains. These interactions provide a basis for recognition of ubiquitin-modified PTK-associated receptors by a series of multi-protein complexes referred to as the Endosomal Sorting Complex Required for Transport (ESCRT). The serial handover by ESCRT 0-III complexes is critical for sorting ubiquitin-modified receptors on outer limiting membrane of the multi-vesicular body (MVB) compartment of the endocytic pathway for delivery to inner vesicles of this organelle. This step is critical for further transport of receptors to the lysosome where they are degraded by lysosomal hydrolases (Fig. 1). While a greater structural and functional complexity is seen with the mammalian ESCRT machinery, the components and the architecture of ESCRT complexes is conserved from yeast to mammals. The ESCRT machinery has been thoroughly reviewed elsewhere [35–39], and will not be discussed in further detail here.
3.3. The role of the de-ubiquitination machinery
Over 100 proteases in the mammalian genome are members of several subfamilies of de-ubiquitinases (DUBs). These enzymes play multiple roles in regulating the ubiquitination process. Members of this group are essential for generating ubiquitin from its precursor covalent fusions with itself or other polypeptides as well as in regenerating ubiquitin from ubiquitin-modified proteins. Importantly, members of this enzyme family are involved in proteasomal degradation of ubiquitinated proteins as they play essential roles in processing and removing ubiquitin chains to facilitate the entry of ubiquitin-targeted polypeptides into proteasomal core, which harbors the catalytic proteases. Significantly, emerging evidence indicates key, and seemingly opposite, roles of DUBs in regulating endocytic traffic of PTK-associated receptors. Studies in yeast identified an essential role of the de-ubiquitinase Doa4/Ubp4 in vacuolar sorting of ubiquitinated receptors. Studies of the mammalian homolog of this DUB, Usp8/UBPY, in the context of RTKs have produced somewhat conflicting results. One set of studies indicates that Usp8/UBPY-mediated de-ubiquitination reduces the lysosomal degradation of RTKs such as EGFR [40]. Other studies have suggested an opposite role of facilitating RTK degradation [41]. More in depth analyses have begun to shed light on this complexity and apparently contradictory findings from different studies. It has now become clear that Usp8/UBPY also regulates the level of ubiquitination of other endocytic pathway regulators; such as the components of the receptor internalization and ESCRT machinery and alterations in Usp8/UBPY are associated with marked changes in the structure of endocytic compartments [42]. Similarly, endosomal deubiquitinase AMSH (Associated Molecule with the SH3-domain of STAM) has been shown to antagonize the ubiquitin-dependent lysosomal degradation of EGFR by trimming the K63-linked ubiquitin chains [43] while other studies have shown this DUB to be required for MVB-dependent EGFR degradation [43,44]. Furthermore, recent studies have shown that Usp8/UBPY and AMSH may regulate each other [45] as well as E3s such as the ErbB3-directedNrdp1 [46]; therefore, their direct versus indirect effects require further elucidation. Notably, Usp8/UBPY is transcriptionally up-regulated by PTK-coupled cytokine receptor activation and its inducible upregulation was shown to alter colony stimulating factor 3 receptor traffic [47]. Thus while initial studies support a role of the Usp8/UBPY and AMSH deubiquitinases in providing a layer of regulation in the ubiquitin-dependent lysosomal sorting and degradation of activated PTK-associated receptors, the generality of this concept needs to be established in the context of additional RTKs and the cellular and/or molecular contexts that lead to distinct outcomes of their DUB function in RTK traffic need to be delineated [48].
Further pointing to this complexity, ubiquitin specific protease 2a (USP2a), a DUB that can remove both K48- and K63-linked poly-ubiquitination chains from substrates, localizes at the early endosome and exhibits oncogenic transforming activity, de-ubiquitinates and stabilizes internalized EGFR and promotes its recycling back to the cell surface [49]. Recently, the ovarian tumor protease DUB family member Cezanne-I (also known as A20) was demonstrated to directly interact with EGFR, induce de-ubiquitination of the receptor and curtail its ligand-induced degradation; notably, Cezanne-1 was overexpressed in breast cancer patients and its mRNA overexpression predicted aggressive disease [50]. It is pertinent that Cezanne-1/A20 is an established regulator of NF-κB signaling and other pathways [51], more in depth analyses into its RTK regulatory function are warranted.
As the relative substrate selectivity and subcellular localization of DUBs during dynamic cellular processes remain relatively poorly characterized, further studies of their roles that reflect the regulation of the endocytic itinerary of PTK-associated cell surface receptors versus other functions are needed. The roles of DUBs demonstrated by these isolated initial examples also highlight the need for more unbiased studies of the larger DUB family. Such analyses should help uncover the potential roles of DUBs in the endocytic traffic of the wider group of PTK-associated receptors with attention to the tissue-specific and developmental patterns of expression of DUBs as these become more clearly defined, and may help elucidate novel mechanisms of PTK mediated oncogenesis that can be targeted for new therapeutic interventions.
3.4. Direct targeting of PTKs by the ubiquitin machinery
Following the well-established antagonistic relationship of phosphorylation and dephosphorylation as physiological switches, early studies of PTK regulation naturally focused on phosphotyrosine phosphatases (PTPs). As indicated above, PTPs provide one mechanism for negative regulation of PTK signaling although the vast majority of PTPs appear to have complex roles, including positive roles in PTK signaling. Early studies of the first PTK, Src, and subsequent studies of other cellular PTKs have also shown these cellular signaling switches to be potent proto-oncogenes whose oncogenic activity can be unleashed by overexpression, mutational or heterologous fusion-dependent activation, and other mechanisms [52]. Thus, it is not surprising that ubiquitination, an ancient pathway of regulating protein function, has evolved as an additional biochemical mechanism of PTK regulation.
Although ligand-induced ubiquitination of a RTK, PDGFR, was observed in early studies [53] the potential for PTK ubiquitination as a regulatory mechanism became clearly established in the early to mid-1990s through the convergence of a number of studies on the proto-oncogene Cbl, which is the focus of this review and is discussed in detail below.
The idea that direct interaction with Cbl or its family members (below), targets PTKs for ubiquitination and that this has regulatory consequences has led to considerable interest in other E3s with potentially similar roles under distinct biological scenarios [32–34]. Numerous examples of E3s that may facilitate ubiquitin-dependent regulation of PTKs are emerging and the list is likely to grow as more comprehensive studies of the ubiquitome are undertaken. A feature that sets the E3s of the Cbl family apart from all other E3s linked to PTK regulation is that they exclusively target PTKs and selected PTK signaling components in a “PTK activation-dependent” manner. The basis of this activated PTK-dependence involves mechanisms that mediate the binding interactions of Cbl-family E3s as well as the regulation of their activity by PTKs, as will be discussed below. Thus, Cbl-family E3s serve a unique role as activation-induced feedback negative regulators of PTKs. The discussion below on the biological consequences of genetic ablation of Cbl-family E3s together with emerging evidence that their mutations lead to human disease provide functional evidence that ubiquitination-dependent negative feedback provides a layer of regulation of activated PTKs that is distinct from the roles of PTPs and other mechanisms, and yet is essential for physiological control of PTKs.
4. Cbl-family ubiquitin ligases
4.1. General structure of Cbl-family E3s
Cbl (Casitas B-lineage Lymphoma) protein is the founding member of an evolutionarily-conserved family that includes three distinct gene products (Cbl or c-Cbl; Cbl-b; and Cbl-c, Cbl-3 or Cbl-SL) in mammals (Fig. 2). Cbl was initially isolated as the transforming oncogene (viral Cbl or v-Cbl) of a murine leukemia virus [54]. Subsequent isolation of the cellular homologue of mouse Cbl demonstrated that v-Cbl represents the truncated N-terminal part of Cbl fused to viral Gag sequences; the Gag sequences, however, proved dispensable for transformation of rodent fibroblasts by v-Cbl [55].
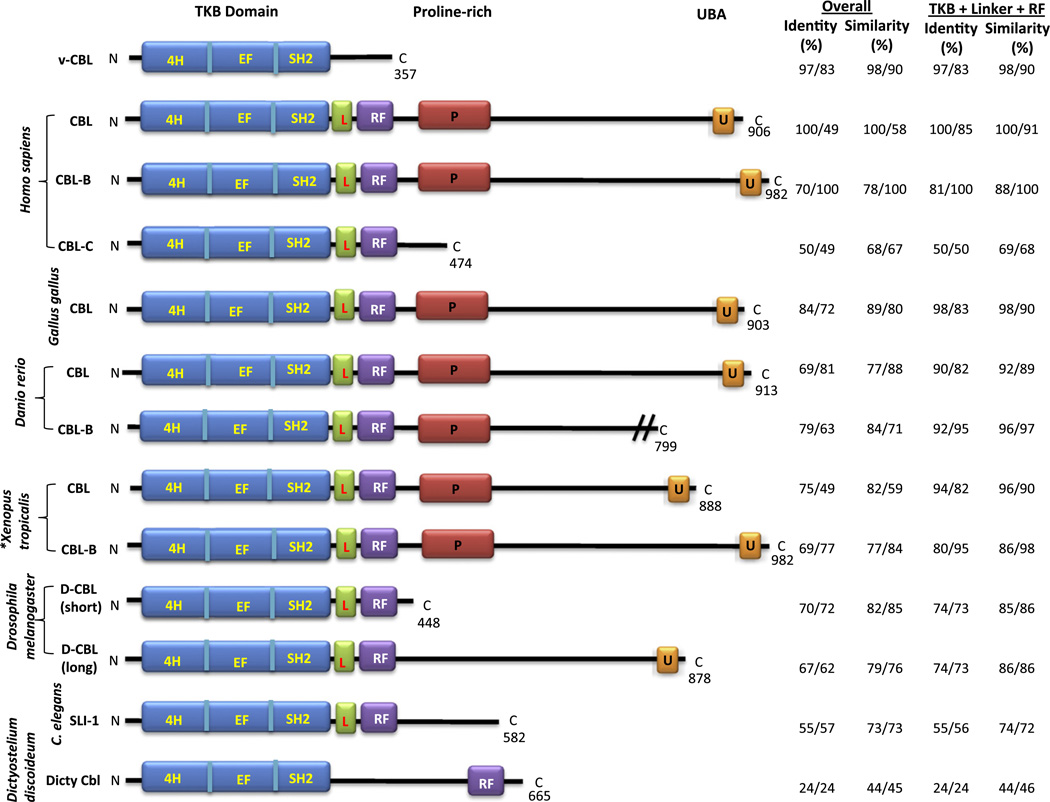
Fig. 2.
Evolutionary conservation of the primary structure and domain organization of Cbl proteins. The comparison includes: the three human (Homo sapiens) Cbl proteins (Cbl or c-Cbl; Cbl-b; and Cbl-c, Cbl-3 or Cbl- SL) as representative mammalian Cbl proteins; Chicken (Gallus gallus) Cbl; Zebra fish (Danio rerio) Cbl; Frog (Xenopus tropicalis) Cbl; Fly (Drosophila melanogaster) long and short Cbl; Worm (Caenorhabditis elegans) Cbl (SLI-1); and Dicty (Dictyostelium discoideum) Cbl (Cbl-A). * Xenopus tropicalis was used for comparison rather than Xenopus laevis, as Cbl sequences in the databases for the latter species were partial. Domain designations: TKB, Tyrosine Kinase-Binding; 4H, four-helical bundle; SH2, Src-Homology 2; RF, RING Finger; L, Linker helical region; P, Proline-rich region; U, Ubiquitin-associated (UBA) domain; The amino acid sequences were compared to human Cbl and Cbl-b and are shown as two values separated by “/” under % identity and similarity for whole protein (or available partial sequence; shown with // across C-terminal end) and for the N-terminal domains (TKB, Linker and RF). The latter emphasizes the higher evolutionary conservation of the N-terminal domains that constitute the core PTK-directed E3 activity of Cbl proteins. V-Cbl corresponds to amino acids 1–357 of mouse Cbl that are present in viral Cbl oncogene. Dicty Cbl 4H region (inferred in UniProt) was confirmed using the YASARA structure program (www.yasara.org); however, a linker helical region has not been identified in Dicty Cbl, making it an exception in the entire Cbl protein family. N and C refer to amino and carboxyl termini.
Mammalian Cbl proteins fall into two distinct categories (Fig. 2): the longer Cbl and Cbl-b gene products are highly related in their primary structure as well as their domain architecture. In contrast, Cbl-c represents a shorter version that lacks some of the key C-terminal domains and motifs. All three members of the Cbl family of proteins share a highly homologous N-terminal region that serves as the structural platform for direct binding to specific pY-containing peptide motifs in activated PTKs (Fig. 3) and is accordingly referred to as the Tyrosine Kinase-Binding (TKB) domain; this domain is in fact a unique assembly of a four-helical (4H) bundle, an EF hand domain and a variant SH2 domain, a tandem domain combination only found in Cbl-family proteins [56]. The TKB domain is followed by a highly conserved helical linker and a RING (Really Interesting New Gene) finger domain; these two regions together form the structural platform for binding to a ubiquitin conjugating enzyme (E2) [57] and this interaction is essential for the E3 activity of Cbl proteins. The three N-terminal domains (TKB, linker and RING finger) form the basic E3 module of Cbl proteins (Fig. 4) and these domains are necessary as well as sufficient for activated PTK-directed E3 activity of Cbl [58,59].
Fig. 3.
Phosphotyrosine-containing sequence motifs that mediate Cbl TKB domain binding to its targets. The consensus motifs and individual target protein sequence motifs with critical amino acids that contribute to binding affinity are indicated [208–210]. Φ indicates hydrophobic residue. The motifs and sequences are adapted from [106].
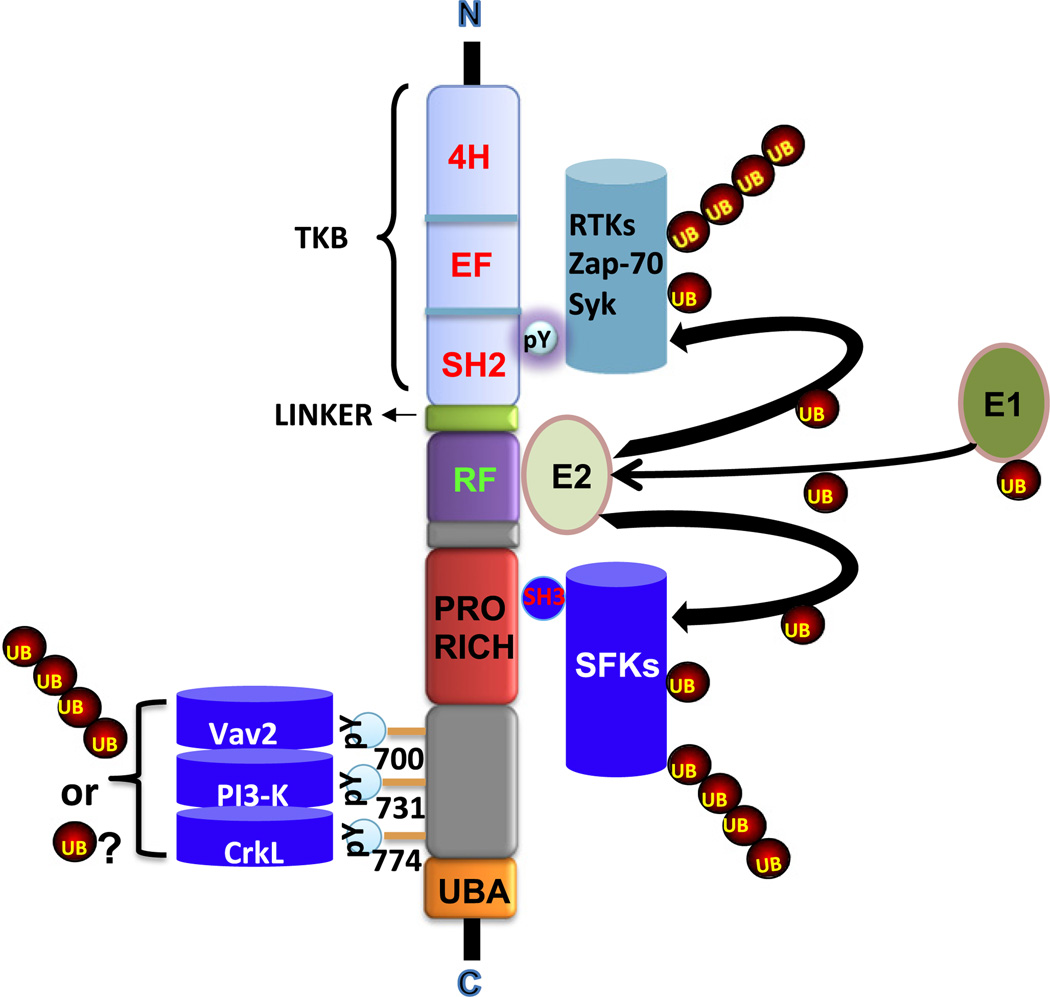
Fig. 4.
Domain architecture of Cbl proteins and major protein-protein interactions involving various domains/motifs. The N-terminal Tyrosine Kinase-Binding (TKB) domain binds to phosphotyrosine (pY)-containing sequence motifs in target proteins, that typically include activated receptor and non-receptor tyrosine kinases. The Linker region and the RING finger (RF) domain bind to ubiquitin conjugating enzymes (E2). The proline-rich motifs (Pro-rich) bind to SH3 domain containing signaling and endocytic proteins. Induced tyrosine phosphorylation sites (major sites at Y700, Y731 and Y774 are shown) recruit SH2 domain-containing signaling proteins. The Ubiquitin-associated (UBA) domain/leucine zipper near the C-terminus is involved in ubiquitin binding and dimerization. N and C refer to amino and carboxyl termini.
Cbl and Cbl-b proteins contain additional C-terminal motifs/domains known or presumed to mediate protein-protein interactions (Fig. 5) [60–65]. These include: a proline-rich region that mediates interactions of Cbl proteins with SH3 domain-containing proteins such as Src-family kinases, Grb2, CIN-85 and others; several tyrosine residues whose phosphorylation generates binding motifs for interaction with SH2 domain-containing signaling proteins that form key intermediates in PTK-dependent signaling networks, including Vav-family of guanine nucleotide exchange factors (GEFs) for Rho-family GTPases (bind to human Cbl Y700), p85 subunit of PI 3-kinase (binds to human Cbl Y731) and Crk-family of adaptor proteins that link Cbl proteins to C3G, a GEF for Ras-related small GTPase Rap1 (binds to human Cbl Y774). The sequences near the C-terminus of Cbl and Cbl-b encode a leucine zipper (LZ)-like motif/Ubiquitin-Associated (UBA) domain that is known to facilitate dimerization under certain experimental conditions [66]; this region also functions as a ubiquitin-associated (UBA) domain capable of binding to ubiquitin chains [67]. However, the functional roles of the LZ/UBA domain remain unclear although its ability to mediate ubiquitin-dependent dimerization has been suggested to provide a potential mechanism of activation of Cbl proteins [68].
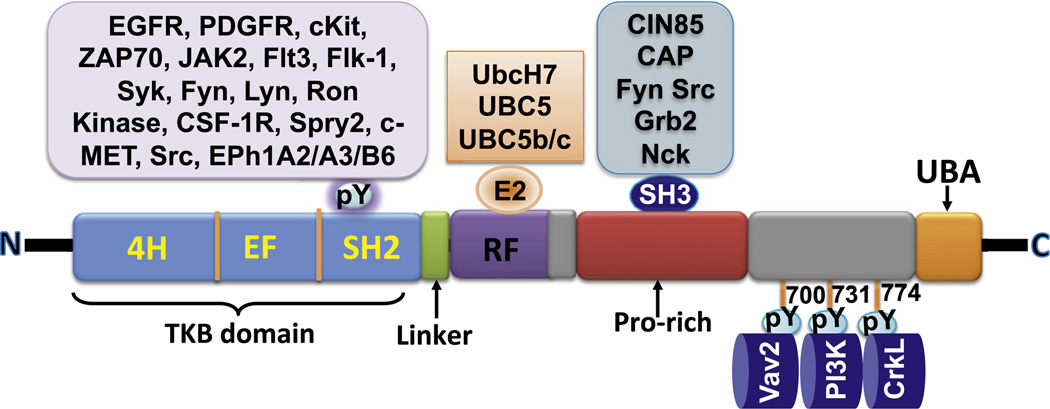
Fig. 5.
Schematic representation of the basic role of Cbl-family proteins as ubiquitin ligases (E3s) towards components of tyrosine kinase signaling pathways. Human Cbl is shown as a prototype of the family. The TKB domain, the proline-rich motifs and the induced tyrosine phosphorylation sites recruit targets for ubiquitin modification. The linker/RF-associated ubiquitin conjugating enzyme (E2) serves as an acceptor of activated ubiquitin from a ubiquitin-activating enzyme (E1) and transfers it to targets bound to various domain/motifs of Cbl to promote mono-ubiquitination (shown as a single UB subunit) or poly-ubiquitination (shown as four UB subunits).
In contrast to Cbl and Cbl-b, Cbl-c lacks most of the C-terminal regions except for a short proline-rich region for potential interactions with SH3 domain-containing proteins. Cbl-c also lacks a LZ/UBA domain as well as the region that harbors the tyrosine phosphorylation sites discussed above.
4.2. Evolutionary conservation of Cbl-family
Cbl proteins homologous to mammalian Cbl proteins have been identified in non-mammalian species including model genetic organisms such as C. elegans and Drosophila and even simple organisms such as Dictyostelium (Fig. 2) [60–65]. These Cbl orthologs exhibit a domain structure similar to that of mammalian Cbl-family proteins, with striking conservation of the domain architecture as well as the primary sequences of critical N-terminal functional domains. For example, the TKB, Linker and RING finger domains of the C. elegans Cbl ortholog SLI-1 together exhibit 55% identity and 74% similarity at the amino acid level with the corresponding regions of human Cbl (Fig. 2). The evolutionary comparison also shows that the equivalents of both the longer (Cbl and Cbl-b) and shorter (Cbl-c) Cbl-family proteins exist in simpler organisms: for instance, the C. elegans Cbl (SLI-1) and the shorter alternatively-spliced form of Cbl in Drosophila closely resemble mammalian Cbl-c in their domain organization while the domain structure of the longer form of Drosophila Cbl resembles that of mammalian Cbl and Cbl-b [69,70]. The Cbl protein family has a relatively ancient origin with an ortholog in Dictyostelium; the latter shows conservation of the N-terminal regions that mediate phospho-tyrosine sequence-dependent interaction of mammalian Cbl proteins with target proteins (TKB domain) and E2 (RING finger). Notably, however, Dicty-Cbl does not appear to have the helical linker region found between SH2 and RING finger domains of all other Cbl proteins, and functions as a positive regulator of signal transducer and activator of transcription (STAT) protein by binding to and down-regulating a phosphotyrosine phosphatase PTP3 [71]. The conservation of domain architecture as well as the primary structure strongly suggest a conservation of the biological functions as well the biochemical mechanisms by which Cbl proteins exert their functional effects, as discussed below.
4.3. Cbl-family proteins as “activated PTK-selective” E3s—evolution of the concept
A number of key early observations contributed to the current paradigm that Cbl-family E3s function as selective regulators of activated PTKs in higher organisms. Identification of the v-Cbl oncogene [54,72] implied that the N-terminal region of c-Cbl (which is incorporated in the oncogene) might interact with critical cellular proteins to dominantly deregulate their function and unleash an oncogenic program. However, the nature of cellular machinery targeted by Cbl remained unknown until multiple lines of evidence helped direct attention to PTKs. Several independent studies in the mid-1990s, investigating either Cbl or various PTKs, quickly converged to establish that Cbl is a direct binding partner and substrate of mammalian non-receptor and receptor type PTKs. Notably, Cbl was found to be tyrosine phosphorylated in cells oncogenically transformed with v-Abl indicating that Cbl is a substrate for a deregulated PTK [73]. This led to further analyses in which Cbl was found to be tyrosine phosphorylated in response to T-cell receptor engagement in cell lines, indicating that Cbl was a substrate of physiological PTK machinery [74]. Independent studies identified a novel 120 kD cellular protein (p120) that directly interacted with SH3 domains of T-cell receptor-associated Src-family kinases Fyn and Lck and was rapidly tyrosine phosphorylated upon T-cell receptor stimulation [75]. Subsequent purification of p120 identified it as Cbl [76]. Additional studies showed that stimulation through T-cell receptor as well as related B cell antigen receptor led to tyrosine phosphorylation of Cbl and its complex formation with downstream signaling proteins including Grb2, PI3-kinase p85 subunit, and Crk adaptor proteins [60,77–79]. Other studies quickly established tyrosine phosphorylation as well as signaling protein complexes of Cbl upon stimulation of EGFR in multiple cell systems [73–75,80–83]. In parallel, genetic screens in C. elegans identified the worm ortholog of Cbl (SLI-1) as a negative regulator of EGFR (LET23)-mediated vulva development and showed that the loss-of-function mutation in SLI-1 was a single amino acid missense mutation (G to E; equivalent to human Cbl G306) within the evolutionarily-conserved region preserved in v-Cbl [84]. Collectively, these advances suggested a critical role of the conserved N-terminal region of Cbl in its function with the possibility that this region may be related to the interaction of Cbl with PTKs.
Direct binding assays using purified recombinant Cbl fragments incorporating the N-terminal region of Cbl, corresponding to v-Cbl sequences (Cbl amino acids 1–357; also referred to as Cbl-N), allowed a test of this hypothesis. GST-Cbl-N selectively and directly bound to phosphorylated ZAP70 tyrosine kinase in an activated T cell line and to related PTK Syk in an activated B lymphocyte line [85,86]. From screening of oriented phosphopeptide libraries and mutagenesis, it was established that Cbl-N bound to the negative regulatory pY-containing peptide motifs within the linker region of ZAP70 and Syk [87] (Fig. 3). Importantly, a point mutation (human Cbl-G306E) corresponding to the loss of function allele of C. elegans SLI-1 completely abrogated the ability of the TKB domain to interact with pY-peptide motifs on activated PTKs thereby linking the genetically identified loss of Cbl's function as a negative regulator in C. elegans with PTK-binding activity in mammalian cells. Similar pY-containing motifs that allow direct binding on activated PTKs have been identified on other PTKs (Fig. 3) (hence the designation Tyrosine Kinase-Binding or TKB domain) [60,85].
The idea of a TKB domain-mediated regulatory interaction of Cbl with activated PTKs was further supported by observations in fibroblasts which showed that the overexpression of oncogenic mutants of Cbl hyper-activated while the overexpression of wild-type Cbl reduced the level of PDGFR signaling in a manner that required an intact TKB domain [82,88]. Studies of fibroblasts overexpressing human EGFR showed that transforming Cbl mutant expression was associated with increased EGFR phosphorylation [89]. Biochemical studies in fibroblasts with or without Cbl overexpression demonstrated that Cbl-dependent negative regulation of PDGF receptor was a consequence of its Cbl-dependent ubiquitination which was associated with an enhancement of the ligand-induced down-regulation of the receptor from the cell surface [88,90]. Parallel studies demonstrated the facilitation by Cbl of EGF-induced down-regulation of EGFR and further demonstrated this to be due to enhanced lysosomal traffic of the internalized EGFR [59]. These findings led to the idea that Cbl-dependent ubiquitination of RTKs, such as PDGFR and EGFR, facilitates their internalization and/or lysosomal sorting akin to a model based on genetic and biochemical studies of yeast G-protein coupled pheromone receptors [91,92]. Indeed, these analyses were extended to hepatocyte growth factor receptor c-MET and mutation of Cbl binding site of c-MET led to its acquiring oncogenic trait [93]. Furthermore, ubiquitination and downregulation of cell surface M-CSF receptor on bone marrow macrophages was reduced in Cbl-null mice [94]. [94,94,95] Further studies with a number of non-receptor PTKs such as Fyn, Lck, Src, ZAP-70 and Syk that are associated with other cell surface receptors [58,96–99] helped establish the generality of Cbl-dependent ubiquitination of activated PTKs.
The ability of Cbl to induce PTK ubiquitination and the presence of a highly conserved RING finger domain (shown in other proteins to bind to E2) adjacent to the TKB domain prompted in vitro biochemical analyses that established that Cbl functions as an E3 towards an activated PDGF receptor; the TKB domain was needed to contact the substrate and the RING finger was shown to recruit E2 [100]. Analyses of truncation and point mutants of Cbl in the context of Syk/ZAP70 and EGFR demonstrated that the TKB and RING finger domains were necessary as well as sufficient for PTK ubiquitination and degradation [58,59,99,101]. Subsequent studies established the E3 function of Cbl-b and Cbl-c and demonstrated that their TKB and RING finger domains were required for their E3 ligase activity [102–105]. Altogether, these studies helped establish that the core function of Cbl-family proteins is to link the ubiquitin machinery to negative regulation of activated PTKs.
4.4. Structural basis for activated PTK-selectivity of E3 function of Cbl proteins
The evolutionarily-conserved N-terminal region forms the core E3 module of Cbl-family proteins (Fig. 4). The TKB domain binds to specific phosphotyrosine-containing sequence motifs that are generated on PTKs upon their activation. Several flavors of these motifs have now been identified on known targets of Cbl proteins with different N and C-terminal residues relative to phosphotyrosine contributing to the affinity and specificity of binding (Fig. 5). A relatively common and high affinity motif is found on the Cbl TKB domain-binding site on ZAP70 ((D/N)XpY(S/T)XXP, initially identified based on the Cbl TKB domain motif selection in a phospho-peptide library screen and further refined through experimental studies [56,85–87,101,106]. A DpYR-based motif is found on c-MET and several other RTKs [107,108]. Finally, RAΦXNQpY(S/T)-related motifs are found in APS-related adaptor proteins that link Cbl to several cell surface receptor targets that lack direct TKB domain-binding motifs, such as the insulin receptor (Fig. 2) [109]. Furthermore certain RTKs such as c-kit use non-consensus TKB domain binding motifs to interact with Cbl [110]. It remains possible that other Cbl TKB domain-binding pY-bearing motifs will be uncovered as analyses of Cbl-interactome are expanded.
The second highly-conserved N-terminal domain that is an essential component of the basic E3 ubiquitin ligase function of Cbl proteins is the RING finger domain along with the helical linker region that connects it with the TKB domain. Together, these domains provide an interface for binding to E2s and thus help juxtapose the TKB domain-bound target PTKs with E2s, enabling their ubiquitin modification. Structural studies and mutational analyses clearly demonstrated that both the RING finger and helical region are required for E2 binding and ubiquitin ligase activity of Cbl towards PTKs [58,105,111–114]. Initial structural studies indicated that PTK targets (mimicked using the phospho-peptide motif from ZAP70) and E2 would lie on different sides of the TKB domain and hence did not clarify how the RING finger bound E2 would be positioned close to target PTKs to mediate ubiquitination [111]. Two lines of evidence however suggested a potential phosphorylation-dependent switch. First, the kinase activity of the target PTKs was found to be necessary for Cbl-dependent ubiquitination [115,116]. While the requirement of the kinase activity could reflect the need to generate a Cbl TKB domain-binding motif on RTKs or adaptor proteins, we showed that the Cbl TKB domain-mediated binding was dispensable yet the kinase activity was required for ubiquitination of the Src-family kinase Fyn [73,116]. Further studies revealed that target PTKs promote the phosphorylation of a conserved tyrosine residue in the helical linker of Cbl (human Cbl Y371) whose Y–F mutation leads to loss of E3 function [117]. Recent crystal structural analyses of the TKB, helical and RF domains in un-phosphorylated versus Y371 phosphorylated states provided evidence that phosphorylation of Y371 reorients the helical region resulting in increased E2 binding and positioning of E2 closer to the phospho-peptide binding pocket of the TKB domain [105,113,114]. Thus, binding to target PTKs is thought to promote a linker phosphorylation-dependent activation of Cbl E3 activity. While the recent structural studies investigated this in the context of a TKB domain-bound PTK target, it remains unclear if a similar mechanism might account for requirement of the kinase activity for ubiquitination of targets recruited via domains others than the TKB domain (as with SFKs and SH2 domain-containing proteins; see below). It is also not clear if Y371 in Cbl is directly phosphorylated by the TKB domain-associated PTKs or by secondarily-activated PTKs such as Src-family kinases. In this regard, it is notable that Cbl-mediated ubiquitination and down-regulation of PDGFR-α was impaired if Src-association sites on PDGFR were mutated or in cells lacking SFKs [118].
Once bound to target PTKs, Cbl and Cbl-b proteins also undergo phosphorylation on several tyrosine residues located within the sequences C-terminal to the RING finger domain and these generate binding sites for SH2 domain-containing signaling proteins (discussed above). It is noteworthy that the signaling proteins that form complexes with Cbl proteins through SH2–phosphopeptide interactions (hence PTK activity-dependent associations) also undergo Cbl-family E3-dependent ubiquitination; this has been clearly demonstrated for PI3-kinase, whose p85 subunit interacts with pY731 of Cbl [119], for Vav and Vav2 [120,121] which interacts with Cbl pY700, and Crk/CrkL-associated C3G protein, a guanine nucleotide exchanger for Rap GTPases that interacts with Cbl pY774 [122]. Since structural studies have not included the C-terminal region of Cbl or Cbl-b, it is at present unknown what mechanisms might juxtapose signaling proteins bound to these sequences with the RING finger domain-bound E2. The C-terminal sequences and their phosphorylation (and interactions with SH2 domain-containing proteins) however do not appear to be required for ubiquitination of TKB domain-bound targets as suggested by the functionality of mammalian Cbl-c and that of C. elegans SLI-1 and the shorter isoform of Cbl in Drosophila as well as studies with truncated Cbl proteins (discussed above).
4.5. Nature and consequences of Cbl E3-dependent ubiquitin modification of activated PTKs
In general, Cbl E3s facilitate mono- or poly-ubiquitination of the associated targets [112,123–128]. Cbl-dependent mono-ubiquitination and K63-linked poly-ubiquitination appears to be a feature of activated RTKs [112,124–128] while other targets, such as ZAP70/Syk and Src-family kinases, appear to undergo poly-ubiquitination [60,97–99]. Notably, in most cases, these distinct modifications dictate degradation of ubiquitinated targets of Cbl proteins but through distinct pathways: mono- or K63-linked poly-ubiquitination of activated cell surface receptors facilitates their sorting to the lysosomes, based on recognition of the ubiquitin chains by the Endosomal Sorting Complex for Transport (ESCRT) machinery which facilitates the entry of ubiquitin-tagged receptors into inner vesicles of the multi-vesicular body (MVB) compartment (see above) (Fig. 1); in contrast, many poly-ubiquitinated targets of Cbl proteins appear to undergo proteasomal degradation [35–39]. However, in some instances, cytoplasmic signaling proteins associated with activated RTK complexes may indeed be sorted together with RTKs into the MVB and thereby undergo lysosomal degradation [126,129]. Interestingly, degradation-independent negative regulation by Cbl proteins has been suggested in selected cases. For example, negative regulation of PI3-kinase in T cells by Cbl-b was associated with ubiquitination but not degradation, leading to the authors' suggestion that Cbl-family E3 dependent ubiquitination may mediate negative regulation of PI3-kinase and potentially other targets by regulating subcellular localization [119].
5. Functional roles of Cbl-family proteins
5.1. Functional roles of Cbl-family proteins based on evidence from in vitro model systems
A large body of literature now documents the functional consequences of the negative regulatory role of Cbl proteins using cellular systems. As these have been reviewed in detail over the past several years [63,77,78,130,131], this section will provide an overview of the message that has emerged out of these studies.
As PTKs generally serve as membrane proximal switches to convert extracellular signals into intracellular functional programs, the vast majority of in vitro studies have naturally focused on the role of Cbl proteins at the level of receptors themselves and at the level of proximal signaling events downstream of PTK-associated receptors. As stated above, the E3 activity of Cbl proteins plays a key role in the ubiquitination of RTKs as well as receptors (such as antigen receptors on immune cells) that are non-covalently linked to PTKs. There is general agreement that the overall consequence of Cbl-family protein-dependent ubiquitin modification of cell surface receptors is an enhancement of their lysosomal degradation via ESCRT protein-dependent facilitation of sorting at the MVB compartment [7,63,129,132]. Another proposed mechanism of the regulation of endocytic traffic of PTK-associated surface receptors by Cbl proteins, however, remains controversial. A series of investigations in the context of RTKs such as EGFR and c-MET, have indicated that Cbl proteins facilitate RTK internalization from the cell surface through a mechanism that may be independent of their E3 activity [133,134]. The model emanating from these studies suggests that Cbl proteins associate with activated RTKs via their TKB domains and recruit the Cbl proline-rich region-associated SH3 domain-containing proteins of the CIN85 family. CIN85 forms a complex with endophilin A, which is known to be important in receptor internalization. Thus, Cbl-family proteins are proposed to function as adapters to promote the internalization of activated RTKs from the cell surface into early endocytic vesicles (Fig. 1). Studies using mutant proteins in transfection studies support this model [133]. Studies in other systems, however, have suggested that Cbl proteins are dispensable for initial internalization of activated RTKs and are essential only for the later step of lysosomal sorting. A rather physiological approach using mouse embryonic fibroblasts with complete deletion of Cbl showed that initial internalization of activated EGFR was not materially impaired [135] and our initial findings using Cbl plus Cbl-b double knockout cells further support these conclusions (unpublished observations). Consistent with these findings, EGFR was shown to become internalized in a Cbl-independent manner in response to oxidative stress resulting in EGFR recycling rather than degradation [136]. In addition, EGFR mutants defective in Cbl binding as well as ubiquitin modification are still internalized [137,138]. Thus, while it is possible that Cbl proteins can facilitate receptor internalization this function may be easily compensated for in the absence of Cbl protein recruitment. However, Cbl proteins are essential for sorting of the internalized PTK-associated receptors to the lysosome (Fig. 1). Further analyses are needed to fully elucidate the relative importance of Cbl proteins working through an adapter mechanism versus E3 activity, at two distinct steps in the endocytic traffic of PTK-associated cell surface receptors.
The second aspect of the functional roles of Cbl-family proteins that has been elucidated through cellular studies relates to their importance in regulating early biochemical pathways activated by PTK-associated receptor signaling. Thus, in a range of cellular models ranging from transfection analyses, knockdown analyses or cells derived from mice with deletion of Cbl or Cbl-b studies have shown that Cbl proteins negatively regulate the signaling outputs from activated PTKs into major downstream pathways such as the Ras-Erk, PI3-kinase-AKT and STAT pathways [139–143] In general, loss of Cbl protein-dependent negative regulation kinetically prolongs the activation of these signaling pathways upon receptor stimulation. These effects are likely to be a result of the overall down-regulation of activated receptors through lysosomal degradation (which terminates their ability to continue to signal), targeting of associated PTKs (such as Src-family kinases, ZAP70/Syk) for proteasomal degradation, and the ubiquitination of signaling proteins themselves if these are able to interact with Cbl proteins directly. The latter mechanism appears to contribute to the negative regulation of PI3-kinase-AKT (via p85 subunit interaction with pY731 of Cbl), Erk (via interaction of Rap-GEF C3G with Cbl via Crk adaptors; possibly also via interactions with sprouty proteins), and Rho GTPase family (via interaction with GEFs of the Vav family) [97,99,115,120,121,140,144–147]. Other signaling pathways are often impacted in specific cell types.
How the regulation of PTK-associated receptor traffic and immediate biochemical pathways translate into specific biological responses is however poorly understood. Perhaps the best links in this regard have been established in specific immune system cellular studies. It is clear, for example, that Cbl proteins regulate the actin cytoskeletal remodeling in T-cells that in turn is required for the stability of the immunological synapse; as a result, Cbl-b deficient T cells have longer lived immunological synapses [148]. Similarly, Cbl-b deficient T cells are resistant to induction of anergy [149]. Loss of Cbl-b or Cbl plus Cbl-b in mice renders T-cells hypersensitive to T cell receptor activation for cell proliferation and cytokine secretion and removes the requirement for co-stimulatory signals normally delivered through CD28 or related proteins [150]. However, the linkage of these functional effects with specific biochemical pathways and regulation of T cell receptor endocytic trafficking remains poorly explored. How receptor proximal events regulated by Cbl proteins translate into distal functional consequences of PTK-associated receptor activation in other cell systems is even less clear. For example, even though RTKs such as EGFR, PDGFR and c-MET have been extensively analyzed with respect to receptor trafficking events and early biochemical responses regulated by Cbl proteins, how the physiological responses mediated by these receptors are regulated by Cbl proteins remains to be fully examined. One trait often altered in cells lacking Cbl proteins is cell migration response to growth factors such as EGF or HGF; for example, EGF or HGF-induced migration was increased in human mammary epithelial cells with Cbl plus Cbl-b knockdown yet their EGF-driven proliferation was unaffected [121]. In contrast, cytokine-induced proliferation of Cbl-null or Cbl plus Cblb-null hematopoietic stem and progenitor cells is markedly enhanced [151–153]. Little is known about how Cbl proteins might influence distal transcriptional events, cell proliferation and differentiation responses. These analyses are of fundamental importance to our understanding of the basis of physiological and pathological roles of Cbl proteins emerging from animal models and human diseases (see below).
While the focus of the current review is on the role(s) of Cbl-family E3s, it will be important for future studies to consider their roles in the context of other negative regulatory pathways that cells utilize to attenuate PTK-coupled receptor signaling. Apart from Cbl-family E3s other ubiquitin ligases have emerged as negative regulators of PTK-coupled receptor by impinging on pathways similar to those controlled by Cbl-family proteins; for example, Nedd proteins, ITCH, Nrdp1, CHIP, Cullin-5 and SIAH proteins have been shown to play a role in the ubiquitination and downregulation of PTK-coupled receptors [63,154–156]. Consistent with findings from earlier genetic studies in model organisms, recent analyses in mammalian systems have also revealed an important additional layer of negative feedback regulators that are induced upon receptor activation: examples of such inducible feedback inhibitors include LRIG1 (leucine rich repeats and immunoglobulin-like domain 1), suppressor of cytokine signaling 4 and 5 (SOCS4 and SOCS5) and receptor-associated late transducer (RALT, also known as MIG6 and ERRF11) [155]. LRIG1, a cell surface trans-membrane protein containing leucine-rich repeat domain and three Ig-like modules interacts with extracellular region of EGFR in a ligand independent fashion [157]. LRIG1 accelerates ligand induced ubiquitination and degradation of EGFR. RALT, a cytosolic protein with ErbB binding region (EBR) is recruited to the EGFR-kinase domain in a ligand-dependent fashion. Recruitment of RALT results in catalytic inactivation of the kinase domain and abrogation of downstream signaling [158]. RALT also binds to AP-2 and intersectins and promotes clathrin-dependent endocytosis [159]. Notably, RALT regulates receptors at multiple points during their endocytic traffic [160]. MIG6/RALT was recently implicated in promoting loss of cell survival under conditions of growth factor deprivation [158]. While SOCS proteins were initially characterized as inducible negative regulators of cytokine receptors as elongin-dependent E3s, recent studies show that SOCS4 and 5 bind EGFR in ligand-independent fashion through their N-terminal and SH2 domains [161,162] and mediate an together with elongin BC mediate EGFR downregulation [162].
These examples highlight the need for systematic studies to examine how Cbl-family E3s and other negative regulatory pathways function together to coordinate spatio-temporal regulation of PTK-coupled receptor signaling that is critical for specification of the nature and extent of cellular responses. Similarly, since Cbl-family proteins interact with their target PTKs in a phosphotyrosine-dependent manner, the potential role of phosphotyrosine phosphatases (PTPs) in regulating Cbl-family E3-dependent negative regulation of PTKs needs to be clarified. For example, a Cbl/SHP2 complex has been implicated in the ubiquitination and lysosomal degradation of gp130 receptor chain upon IL-6 stimulation [163]. Conversely, an siRNA screen of PTPs for dephosphorylation of EGFR identified density-enhanced phosphatase-1 (DEP-1) and tumor suppressor as an EGFR-targeting PTP; DEP-1-dependent dephosphorylation led to stabilization of EGFR by hampering its ability to associate with the CBL–GRB2 complex [164].
Negative regulation by Cbl-family E3s will undoubtedly function in the context of other negatively regulators of PTK-associated receptors that may function by mechanisms distinct from ubiquitin-dependent regulation. One family of such regulators that has received considerable attention is that mediated by Sprouty proteins as these proteins have been shown to directly associate with Cbl with dramatic functional consequences that appear to vary with the experimental systems [165]. Sprouty proteins are inducible feedback negative regulators of PTK-coupled receptor signaling that appear to primarily impinge on the Ras-Raf-Erk1/2 pathway [166,167]. Sprouty proteins have been shown to directly interact with Cbl proteins via two potentially cooperative interactions. The N-terminal region corresponding to residues 11–53 of human Sprouty 2 directly binds to RF domain of Cbl [168]. Stimulation of RTKs, such as EGFR or FGFR, also promotes tyrosine phosphorylation of Sprouty proteins on a tyrosine residue (tyrosine 55 on Sprouty 2; this residue is conserved in all family members and in other species) that serves as a binding site for Cbl TKB domain [145]. Analyses of RTKs such as EGFR, using in vitro cell culture systems, have shown that Cbl-Sprouty association sequesters Cbl from activated RTKs resulting in their inefficient ubiquitination and downregulation with a consequent increase in downstream signaling. Thus, this complex interaction appears to utilize a known negative regulator of RTKs (Sprouty) to abrogate the effects of another negative regulator (Cbl) effectively converting Sprouty into a positive regulator of RTK signaling [145]. Further analyses indicate that Cbl TKB-Sprouty 2 pY55 interaction is required for the negative regulatory function of Sprouty 2 towards Ras-Erk1/2 signaling [145,166]. In addition, this interaction has been shown to promote the ubiquitination and degradation of Sprouty 2 [169]. Finally, other studies have noted a biphasic regulation with positive as well as negative regulatory roles of Cbl-Sprouty complex that may influence RTK signaling in a spatio-temporal manner [170,171]. Notably, induction of Sprouty 1 upon activation of CD4+ T cells resulted in inhibition or enhancement of receptor signals depending on the differentiation state of cells, suggesting that Cbl and Sprouty proteins may function as a context-dependent regulatory complex for more precise and versatile regulation of receptor signals during development and tissue homeostasis [172]. Importantly, further studies, including in vivo analyses, of the relative importance of Cbl proteins versus other negative regulators and their cross-talk should provide fruitful avenues to elucidate the regulatory mechanisms that control biological processes mediated by PTK-associated receptor signaling.
5.2. Functional roles of Cbl-family proteins based on evidence from in vivo studies
5.2.1. Studies in model organisms
An early indication of a negative regulatory role of a Cbl-family protein for a PTK-associated receptor came from studies in C. elegans when a genetically identified negative regulator of the EGFR homolog LET23 was identified as the worm Cbl (SLI-1) [84] (Table 1). Drosophila Cbl homolog has similarly emerged as a negative regulator of EGFR signaling during photoreceptor development [173]. Drosophila Cbl is also required for dorso-ventral patterning and germ cell migration and functions by regulating RTK endocytic traffic [174]. More recently, a Cbl ortholog in zebrafish was shown to negatively regulate FGF8 signaling during embryonic development [175]. The mechanism and consequences of the regulatory role of Cbl identified in these studies are quite interesting with potentially important implications for other systems. FGF8 morphogen gradient is critical for shaping events during embryogenesis in Zebrafish. While loss of Cbl did not alter the morphogen gradient, Cbl-deficient tissues showed an expansion of the range of FGF8 target genes that were expressed apparently correlating with reduced lysosomal targeting of active FGFR1 receptor signaling complexes [175]. Thus, Cbl appears to regulate how a critical developmental morphogen gradient is interpreted by target tissues and this could have a bearing for the role of mammalian Cbl proteins as well. It is notable that combined deletion of Cbl and Cbl-b in mice results in early embryonic lethality through unknown mechanisms [150]. The elegant genetic studies in model organisms provide strong evidence that Cbl proteins play a critical role in physiological functions mediated by RTKs and provide a rationale for more directed studies in mammals.
Table 1.
In vivo roles of Cbl family genes revealed by genetic mutation and deletion approaches.
| Organism (Gene) | Mutation /Knockout (KO) | Phenotype | References |
|---|---|---|---|
| C. elegans (Sli-1) | Loss of function mutation | No phenotype in wildtype worms; Restoration of vulva development defects in worms expressing a hypomorphic EGFR (let-23) allele | [84] |
| Drosophila (D-Cbl) | Transgene overexpression | No phenotype in wildtype flies; expression in a sensitized background (expressing a disabled Sevenless kinase, SevE4) eliminated photoreceptor R7 differentiation. | [173] |
| Loss of function mutation of D-Cbl | Defective dorso-ventral patterning during oogenesis, ovarian development and embryonic development. Disruption of the wing development. | [174] | |
| Ring finger mutant of D-Cbl (mutation of Cys-369, corresponds to Cys-381 in human Cbl): affects both D-CblL and D-CblS | Abrogated border cell migration during oogenesis. | [211] | |
| Deletion mutation (identical to loss of function mutant of Schupbach group) and Ring finger mutation of D-Cbl (mutation at Arg-406): affects both D-CblL and D-CblS | Abnormal eye development, increased omatidial spacing and larger head. | [212] | |
| Zebra fish (Cbl) | Y->F mutation in linker region Tyr-352 and 355 (corresponding to Tyr-368 and 381 of human Cbl) | Mutant fish show patterning defects during embryonic development due to alterations of fgf8 (fibroblast growth factor 8) morphogen gradient. | [175] |
| Mouse (Cbl) | Complete (germline) Knockout (Cbltm1Ddlb) | Hyperplasia of lymphoid organs, splenomegaly, increased mammary gland branching. | [176] |
| Cbl−/− mice exhibited upregulated glucose metabolism, lean muscles and low fat deposition in the body despite hyperphagia, increased whole-body insulin action and energy expenditure. | [179] | ||
| Complete (germline) knockout (Cbltm1.1Hua) | Mice are born healthy and fertile. Mild splenomegaly due to erythroid hyperplasia. Loss of Cbl in these mice altered thymic positive selection by enhancing CD4 lineage Tcells. | [177] | |
| Cbl knockout male mice are hypofertile. Absence of Cbl in the testicular gonadal cells results in failure to induce apoptosis during spermatogenesis. | [178] | ||
| Cbl-deficient mice exhibit expansion of hematopoietic stem/progenitor (HSC) pool in the bone marrow. | [151] | ||
| Mouse Cbl-b | Knockout (Cblbtm1Pngr) | Born at expected Mendelian ratios and fertile. At the age of 6 month display huge mass of salivary gland due to accumulation of T cells and B-cells, and lymphoid neoorganogenesis in most vital organs of the body. Generalized spontaneous autoimmune disorder around 6 months of age due to hyperactivation of autoreactive T and B-cells. Older mice developed progressive changes in facial features and enhanced osteoclastogenesis. | [180]. |
| CD40-induced B-cell proliferation is enhanced in mutant mice. Furthermore, Cbl-b−/− mice display enhanced thymus-dependent antibody responses and germinal center formation. | [213] | ||
| Knockout (Cblbtm1Hua) | Mutant mice are fertile and grossly normal levels of T-, B-and Natural Killer cells in peripheral and central lymphoid organs. Mice more susceptible to induced experimental autoimmune encephalomyelitis. Deficiency of Cbl-b leads to loss of requirement for CD28 co-stimulation for T-cell activation. | [181] | |
| Mice showed symptoms of Type-2 diabetes by manifesting glucose intolerance and insulin resistance around 20 weeks of age. Infiltration of activated macrophages seen in white adipose tissue. | [214] | ||
| Cbl-b-deficient mice are resistant to unloading-induced muscle atrophy and loss of muscle function as a result of reduced IRS-1 (insulin receptor substrate-1) degradation. | [182] | ||
| Mouse Cbl-c | Knockout Cblctm1Pngr | No gross or micro-anatomic phenotype | [183] |
| Mouse Cbl and Cbl-b | Complete double Knockout | Embryonic lethal by day E10. | [150] |
| Conditional deletion in T-Cells using Lck-Cre (on Cbl-b KO background) | Severe vascular lesions, lymphadenopathy, splenomegaly, extramedulary hematopoiesis and symptoms of autoimmune diseases. Impaired T-cell receptor down modulation. Mutant mice exhibited disease sign after 3 weeks. Females became moribund at 12 weeks, male mice by 16 weeks. | [150] | |
| Conditional deletion in B-cells using CD19-Cre (on Cbl-b KO background) | Mice develop spontaneous SLE-like autoimmune diseases. Impaired B-cell intrinsic tolerance induction, Bcell and induction of B-cell anergy. Half of the mutant mice became moribund by 10 months of age. | [185] | |
| Conditional deletion in HSC as a result of MMTV-Cre induced deletion of floxed Cbl (on Cbl-b KO background); expected Cbl deletion in mammary gland. | Mutant mice born at sub-Mendelian ratios. Mice develop severe myeloproliferative disorders (MPD) by 5 weeks of age with a median survival of 67 days. | [153] | |
| Mouse Cbl-c | MMTV-Transgenic overexpression of Cbl-c in mammary epithelium. | Retardation in mammary ductal branching. | [215] |
| Mouse Cbl (G304E) | Knock in Cbltm1Wlan | No gross abnormality in organs and tissues except for mild splenomegaly and dilated uterine horns and fallopian tubes in mice of 6 to 9 weeks old. | [216] |
| Mouse CblA/− (C379A) | Knock in Cbltm2Wlan | Homozygous mutant mice died in utero or during first 24 h after birth. CblA/− mice resembled Cbl-knock out animals. However, the CblA/− mice exhibit additional defects, notably progressive loss of thymus. | [217] |
| CblA/− mice display reduced adiposity despite greater food intake, reduced circulating insulin, leptin and triglycerides level. The mice are protected against high-fat diet-induced obesity and insulin resistance. | [179] | ||
| Mouse CblYF (Y737) | Knock in Cbltm1Dejs | Mice expressing mutant Cbl with inability to interact with the SH2 domain of phosphatidylinositol (PI) 3-Kinase p85 subunit exhibit increased bone mass and volume, and decreased osteoclast function. | [218] |
| Mouse Cbl-b (C373A) | Knock in Cblbtm1Wlan | RING finger domain mutant mice are healthy and produce homozygous offspring with expected frequency. Enhanced high-affinity IgE receptor (FcεRI) signaling in mutant mice is independent of E3 ligase activity. | [219] |
5.2.2. Studies in mammalian systems
Complementing the evidence from biochemical and mammalian cell biological analyses, mouse models (Table 1) have provided a number of novel insights into physiological pathways where negative regulation provided by Cbl proteins is critical. Germ-line deletion of Cbl results in a series of phenotypes during development and in adult animals. Cbl-null mice exhibit mild hyper-cellularity of a number of lymphoid organs, including the bone marrow and spleen with increased numbers of megakaryocytes [176], expansion of hematopoietic stem/progenitor cells in the bone marrow [151], mild increase in mammary ductal branching in virgin mice [176], altered positive selection of T-cells in the thymus [177], reduced male fertility [178], lean muscle mass due to elevated insulin action [179] and other phenotypes; however, Cbl-null mice appear to live for a relatively normal life span under typical laboratory housing conditions. Cbl-b-null mice have no apparent developmental abnormalities. These mice, however, show hyperactive T cell responsiveness to antigens, leading to autoimmune disease [180,181], a failure to induce anergy in T cells [149], and reduced muscle atrophy in response to unloading [182]. Nonetheless these mice too live for a relatively normal life span if infectious challenges are avoided. In contrast to the mild to moderate phenotypes of individual Cbl or Cbl-b knockout mice, combined deletion of Cbl and Cbl-b in the germ-line is associated with early embryonic lethality [150]. This finding is of particular interest since the embryonic lethality of Cbl/Cbl-b double knockout mice precedes the time when Cbl-c is first expressed in the mid to late gestation [183], thereby providing clear evidence that Cbl and Cbl-b play a redundant but essential functional role during embryogenesis. The targets of Cbl proteins that mediate their embryonic developmental roles remain to be identified.
Given the apparently redundant but essential roles of Cbl and Cbl-b during embryonic development, it is of substantial interest to dissect out to what extent these two family members (with their closer biochemical similarity compared to Cbl-c) might function redundantly or in a specific manner. One mechanism for relatively specific functions might be provided by their relative expression levels. At the mRNA level, Cbl and Cbl-b appear to be widely expressed but with one or the other family member predominating in specific tissues, which appears to be concordant with the phenotypes of individual knockouts. For example, higher Cbl expression in the testes and thymus are consistent with male infertility and T cell developmental phenotypes of Cbl null mice while hypersensitivity of peripheral T cells in Cbl-b null mice coincides with higher Cbl-b expression relative to Cbl [150,178,184]. However, the overlapping expression patterns in most tissues suggest that either specific cell types within these tissues rely on one family member versus the other or that Cbl and Cbl-b function predominantly in a redundant manner in many tissues.
Mice with a conditional (floxed) allele of Cbl on a Cbl-b-null background have allowed tissue-selective KO of Cbl plus Cbl-b to assess the question of redundancy and to reveal the full functional impact of negative regulation by Cbl proteins. Combined deletion of Cbl plus Cbl-b in peripheral T lymphocytes (using Lck-Cre) indeed led to a substantially more severe and spontaneous systemic autoimmune/inflammatory disease and dramatic hyper-responsiveness of T cells to antigen stimulation [150]. Similarly, Cbl/Cbl-b double deletion in the B lymphoid compartment of mice led to loss of peripheral tolerance [185]. Finally, deletion of Cbl and Cbl-b in hematopoietic stem cells led to a myeloproliferative disease that was fatal within 8 weeks of birth [153]. The latter phenotype is replicated by a RING finger domain missense mutant knock-in on a Cbl-null background [152].
Collectively, the KO animal models suggest that Cbl and Cbl-b proteins serve selective physiological roles likely due to their relative levels of expression in various tissues. More importantly, these models have emphasized that the two Cbl-family members Cbl and Cbl-b function redundantly as essential regulators of both the embryonic development and adult tissue homeostasis in mammals. Given the success of conditional induction of Cbl deficiency in a Cbl-b-null background, further studies using this approach should help elucidate the roles of these widely expressed Cbl family members in mammals. In this regard, engineering of a Cbl-b floxed mouse model (ongoing in our laboratory) should provide a more facile and definitive tool to study functional impact of Cbl plus Cbl-b as redundant but critical negative regulators at various steps in development and in adult organ functions. Notably, deletion of the predominantly epithelial tissue-expressed Cbl-c gene is without a phenotype [183]; however, combined deletion of Cbl-c with Cbl or Cbl-b has not been examined; these studies are currently being carried out in our laboratory and should be of considerable general interest.
5.2.3. Evidence from human diseases
Given the critical roles of PTK signaling in animal development, homeostasis and diseases such as cancer, together with emerging evidence of the role of Cbl family proteins in animal models, there have been strong efforts to link aberrations of Cbl family proteins to human disease. Recent results have indeed linked Cbl mutations to a set of human malignancies providing unequivocal evidence of the critical role of Cbl family protein-dependent negative regulation of PTKs and establishing Cbl proteins as tumor suppressors. Since 2007, a number of investigations have identified mutations clustered in the helical linker and RING finger domain of Cbl (and occasionally of Cbl-b) in about 5% of myeloid leukemia patients with myelo-dysplastic syndromes/myeloproliferative disorders: these include the juvenile myelo-monocytic leukemia (JMML), chronic myelo-monocytic leukemia (CMML) and atypical chronic myeloid leukemia (aCML) subtypes [186–194]. The cancer-associated Cbl mutations predominantly involve residues that have been shown by mutational analyses and/or structural studies to be essential for the ubiquitin ligase activity of Cbl; those mutants that were examined in vitro were found to lack the E3 ligase activity [186,195,196]. Furthermore, a cancer-associated mutant introduced into mouse hematopoietic cells produced mastocytosis [197] and as stated above a RING finger knock-in mutant of Cbl in mice phenocopied the myeloproliferative disorder seen in human patients as also did the deficiency of Cbl and Cbl-b [152,153]. Findings of a direct link between Cbl-family E3mutations and human leukemia strongly support the critical role of Cbl protein-dependent negative regulation of PTK signaling. Identification of relevant target PTKs and a better understanding of the mechanisms of how mutant Cbl proteins promote oncogenesis represent exciting areas of ongoing and future research. Several recent reviews have summarized progress and emerging trends in this area [62,198–201]. It is somewhat intriguing that mutations of the linker and RING finger domains have thus far not surfaced in solid tumors. Whether this is a reflection of a lower frequency of mutations and need for larger studies or biological differences in the functional consequences of Cbl protein-dependent negative regulation in hematopoietic versus other tissues remains to be determined. Notably, recent studies of Cbl in lung cancer patients identified several mutations that are located in N- or C-terminal regions but not in the linker region and the RING finger domain [202]. As these mutations appear to moderately inhibit Cbl function in cell models, further studies with other solid tumors are warranted.
Given the growing evidence that human leukemias arise from hematopoietic stem/progenitor cells, linkage of Cbl mutations to leukemia also links these proteins to stem/progenitor cell homeostasis. Animal models with myeloproliferative disease as a result of the loss of Cbl protein function by gene deletion or knock-in of mutant Cbl [152,153] further support this role. In this regard, Cbl has also been shown to negatively regulate the PDGF-dependent survival of neural progenitor cells, and Cbl and Cbl-b were shown to regulate the asymmetric distribution of EGFR in neural stem cells as part of their self-renewal mechanisms [203,204]. Further studies should help clarify the role (general or limited) of Cbl proteins in stem cell maintenance and the potential target PTKs that they regulate in stem cell contexts. In this context, further analyses of the embryonic lethality of Cbl/Cbl-b double knockout mice should be quite revealing.
Prominent autoimmune phenotypes in mice with Cbl-b (or Cbl plus Cbl-b) deletion have prompted analyses of polymorphisms/mutations in human patients with autoimmune disease. While several smaller studies have failed to detect any associations, some recent studies suggest an association between particular polymorphisms of Cbl-b with systemic lupus erythematosus [205]. Studies in the Komeda diabetes prone rat model identified Cbl-b as a susceptibility gene but the mechanism by which the truncation mutation in a region past the RING finger (and hence unlikely to affect the E3 activity) might contribute to disease remains unclear [206,207]. Regardless of the paucity of evidence in this area, future studies are warranted especially in view of the clear evidence from mouse models that aberrant Cbl protein function can contribute to inflammatory and autoimmune phenomena.
6. Conclusions and future directions
Structural, biochemical and cell biological studies of Cbl proteins have established that members of the Cbl protein family function as activated PTK-selective E3 ubiquitin ligases. Cbl family protein-dependent ubiquitination targets activated PTKs for degradation either by facilitating their entry into lysosomes or by promoting their proteasomal degradation. Signaling intermediates associated with PTK signaling and/or associated with Cbl proteins also appear to be subjected to ubiquitination and negative regulation although the role of degradation needs to be clarified. Cellular and animal studies have established that at least two members (Cbl and Cbl-b) of the Cbl protein family play key physiological roles: prominent examples of these roles include protection against autoimmune disease and tumor suppressor function, the latter receiving validation from human studies that have linked mutations in Cbl to human leukemia. Cbl mutations in human leukemia and insights from animal models together support an important role of Cbl and Cbl-b in stem/progenitor cell maintenance. Thus, future studies of existing and emerging animal models and cellular models derived from these, together with studies of additional human pathologies should help fully realize the range of physiological functions regulated by the evolutionarily-conserved Cbl protein family.
While functions of the relatively widely-expressed Cbl-family members Cbl and Cbl-b have begun to be elucidated, the roles of the structurally distant family member Cbl-c remain unclear. As Cbl-c is selectively expressed in epithelial tissues, future studies of this protein by incorporating its deficiency in animals that lack Cbl, Cbl-b or both proteins should help clarify whether this protein functions redundantly with its family members or by itself, perhaps under specific physiological conditions. Concurrently, a better linkage of the roles of Cbl proteins in the early biochemical and cell biological events associated with activated PTKs and the cellular and organismic roles deciphered from genetic studies should help underscore the importance of the Cbl protein family and the need for its evolution into a multi-gene family in mammals. In this regard, systematic examination of protein-protein interaction networks of all Cbl proteins and their impact on transcriptional pathways and biological outputs of PTK signaling under different physiological conditions of cells and tissues should add to our understanding of how Cbl proteins perform their diverse functional roles.
Studies directed at understanding how Cbl mutants associated with hematological disorders promote oncogenesis should provide further avenues to explore the physiological functions of Cbl proteins as well as provide potential avenues to develop agents that might prove useful in mutant Cbl-driven human cancer.
Acknowledgements
This work was supported by the NIH grants CA87986, CA105489, CA99163, CA116552 and NCI 5U01CA151806-02 and Department of Defense grant W81WH-11-1-0167 to H.B; the NIH grants CA96844 and CA144027 and Department of Defense grants W81XWH-07-1-0351 and W81XWH-11-1-0171 to V.B; and the NCI Core Support Grant to UNMC-Eppley Cancer Center. B.M. is a recipient of a UNMC graduate studies fellowship; S.N. and TAB were pre-doctoral and postdoctoral trainees, respectively, under the NCI Institutional Cancer Biology Training Grant (CA009476). L.D. was an exchange scholar of the China Scholarship Council. D.F. was recipient of a scholarship from the China Medical University Shenyang, China. TAB was a postdoctoral fellowship recipient from the Susan G. Komen Foundation and is a UNMC Diversity Fund recipient.
Abbreviations
- TKB
Tyrosine kinase binding
- PTK
Protein tyrosine kinase
- EGFR
Epidermal growth factor receptor
- ErbB2
v-erb B2 erythroblastic leukemia viral oncogene 2 or EGFR-2
- E1
Ubiquitin activating enzyme
- E2
Ubiquitin conjugating enzyme (UBC)
- E3
Ubiquitin ligase
- LZ/UBA
Leucine zipper-like motif/ubiquitin associated domain
- DUB
deubiquitinase
- ESCRT
Endosomal sorting enzyme complex required for transport
- JAK
Janus kinase
- TGFα
Transforming growth factor alpha
- Grb2
Growth factor receptor bound protein-2
- Shc
Src homology and collagen protein
- Trk
Tropomyosin-receptor kinase
- NGF
Nerve growth factor
- FGF8
Fibroblast growth factor 8
- Erk
Extracellular signal-regulated kinase
- PLCγ1
Phospholipase C, gamma 1
- MAPK
Mitogen activated protein kinase
- ER
Endoplasmic reticulum
- HECT-domain
Homologous to the E6-AP carboxy terminus
- PDGFR
Platelet derived growth factor receptor
- CIN-85
Cbl-interacting protein of 85 kD
- SFKs
Src family kinases
- Lck
lymphocyte specific protein tyrosine kinase
- ZAP-70
Zeta chain associated protein kinase 70
- Syk
Spleen tyrosine kinase
- Lck-Cre
T-cell specific Cre recombinase
- PI3-K
Phosphatidylinositol-3-Kinase
- STAT
Signal transducer and activator of transcription
- KO
Knockout
- Cbl
Italicized Cbl represents Cbl gene
References
- 1.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss A. The right team at the right time to go for a home run: tyrosine kinase activation by the TCR. Nat. Immunol. 2010;11:101–104. doi: 10.1038/ni0210-101. [DOI] [PubMed] [Google Scholar]
- 3.Ghoreschi K, Laurence A, O'Shea JJ. Janus kinases in immune cell signaling. Immunol. Rev. 2009;228:273–287. doi: 10.1111/j.1600-065X.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendriks WJ, Elson A, Harroch S, Stoker AW. Protein tyrosine phosphatases: functional inferences from mouse models and human diseases. FEBS J. 2008;275:816–830. doi: 10.1111/j.1742-4658.2008.06249.x. [DOI] [PubMed] [Google Scholar]
- 5.Julien SG, Dube N, Hardy S, Tremblay ML. Inside the human cancer tyrosine phosphatome. Nat. Rev. Cancer. 2011;11:35–49. doi: 10.1038/nrc2980. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter G. The EGF receptor: a nexus for trafficking and signaling. Bioessays. 2000;22:697–707. doi: 10.1002/1521-1878(200008)22:8<697::AID-BIES3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 7.Waterman H, Yarden Y. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 2001;490:142–152. doi: 10.1016/s0014-5793(01)02117-2. [DOI] [PubMed] [Google Scholar]
- 8.Wells A, Welsh JB, Lazar CS, Wiley HS, Gill GN, Rosenfeld MG. Ligand-induced transformation by a noninternalizing epidermal growth factor receptor. Science. 1990;247:962–964. doi: 10.1126/science.2305263. [DOI] [PubMed] [Google Scholar]
- 9.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 10.Harari D, Yarden Y. Molecular mechanisms underlying ErbB2/HER2 action in breast cancer. Oncogene. 2000;19:6102–6114. doi: 10.1038/sj.onc.1203973. [DOI] [PubMed] [Google Scholar]
- 11.McClintock JL, Ceresa BP. Transforming growth factor-{alpha} enhances corneal epithelial cell migration by promoting EGFR recycling. Invest. Ophthalmol. Vis. Sci. 2010;51:3455–3461. doi: 10.1167/iovs.09-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouyang X, Gulliford T, Huang G, Epstein RJ. Transforming growth factor-alpha short-circuits downregulation of the epidermal growth factor receptor. J. Cell. Physiol. 1999;179:52–57. doi: 10.1002/(SICI)1097-4652(199904)179:1<52::AID-JCP7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 13.Roepstorff K, Grandal MV, Henriksen L, Knudsen SL, Lerdrup M, Grovdal L, Willumsen BM, van Deurs B. Differential effects of EGFR ligands on endocytic sorting of the receptor. Traffic. 2009;10:1115–1127. doi: 10.1111/j.1600-0854.2009.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sigismund S, Argenzio E, Tosoni D, Cavallaro E, Polo S, Di Fiore PP. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev. Cell. 2008;15:209–219. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 15.Di Fiore PP, De Camilli P. Endocytosis and signaling. An inseparable partnership. Cell. 2001;106:1–4. doi: 10.1016/s0092-8674(01)00428-7. [DOI] [PubMed] [Google Scholar]
- 16.Barker PA, Hussain NK, McPherson PS. Retrograde signaling by the neurotrophins follows a well-worn trk. Trends Neurosci. 2002;25:379–381. doi: 10.1016/s0166-2236(02)02199-9. [DOI] [PubMed] [Google Scholar]
- 17.McPherson PS, Kay BK, Hussain NK. Signaling on the endocytic pathway. Traffic. 2001;2:375–384. doi: 10.1034/j.1600-0854.2001.002006375.x. [DOI] [PubMed] [Google Scholar]
- 18.Seto ES, Bellen HJ, Lloyd TE. When cell biology meets development: endocytic regulation of signaling pathways. Genes Dev. 2002;16:1314–1336. doi: 10.1101/gad.989602. [DOI] [PubMed] [Google Scholar]
- 19.Burke P, Schooler K, Wiley HS. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol. Biol. Cell. 2001;12:1897–1910. doi: 10.1091/mbc.12.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qui MS, Green SH. PC12 cell neuronal differentiation is associated with prolonged p21ras activity and consequent prolonged ERK activity. Neuron. 1992;9:705–717. doi: 10.1016/0896-6273(92)90033-a. [DOI] [PubMed] [Google Scholar]
- 21.Pasic L, Eisinger-Mathason TS, Velayudhan BT, Moskaluk CA, Brenin DR, Macara IG, Lannigan DA. Sustained activation of the HER1-ERK1/2-RSK signaling pathway controls myoepithelial cell fate in human mammary tissue. Genes Dev. 2011;25:1641–1653. doi: 10.1101/gad.2025611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 23.Teis D, Wunderlich W, Huber LA. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell. 2002;3:803–814. doi: 10.1016/s1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- 24.Wunderlich W, Fialka I, Teis D, Alpi A, Pfeifer A, Parton RG, Lottspeich F, Huber LA. A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J. Cell Biol. 2001;152:765–776. doi: 10.1083/jcb.152.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, II, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 26.Bild AH, Turkson J, Jove R. Cytoplasmic transport of Stat3 by receptor-mediated endocytosis. EMBO J. 2002;21:3255–3263. doi: 10.1093/emboj/cdf351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panopoulou E, Gillooly DJ, Wrana JL, Zerial M, Stenmark H, Murphy C, Fotsis T. Early endosomal regulation of Smad-dependent signaling in endothelial cells. J. Biol. Chem. 2002;277:18046–18052. doi: 10.1074/jbc.M107983200. [DOI] [PubMed] [Google Scholar]
- 28.Pierce KL, Luttrell LM, Lefkowitz RJ. New mechanisms in heptahelical receptor signaling to mitogen activated protein kinase cascades. Oncogene. 2001;20:1532–1539. doi: 10.1038/sj.onc.1204184. [DOI] [PubMed] [Google Scholar]
- 29.Pennock S, Wang Z. Stimulation of cell proliferation by endosomal epidermal growth factor receptor as revealed through two distinct phases of signaling. Mol. Cell. Biol. 2003;23:5803–5815. doi: 10.1128/MCB.23.16.5803-5815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Pennock S, Chen X, Wang Z. Endosomal signaling of epidermal growth factor receptor stimulates signal transduction pathways leading to cell survival. Mol. Cell. Biol. 2002;22:7279–7290. doi: 10.1128/MCB.22.20.7279-7290.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiley HS, Herbst JJ, Walsh BJ, Lauffenburger DA, Rosenfeld MG, Gill GN. The role of tyrosine kinase activity in endocytosis, compartmentation, and down-regulation of the epidermal growth factor receptor. J. Biol. Chem. 1991;266:11083–11094. [PubMed] [Google Scholar]
- 32.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 33.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macgurn JA, Hsu PC, Emr SD. Ubiquitin and membrane protein turnover: from cradle to grave. Annu. Rev. Biochem. 2012;81:231–259. doi: 10.1146/annurev-biochem-060210-093619. [DOI] [PubMed] [Google Scholar]
- 35.Wegner CS, Rodahl LM, Stenmark H. ESCRT proteins and cell signalling. Traffic. 2011;12:1291–1297. doi: 10.1111/j.1600-0854.2011.01210.x. [DOI] [PubMed] [Google Scholar]
- 36.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 37.Babst M. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr. Opin. Cell Biol. 2011;23:452–457. doi: 10.1016/j.ceb.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rusten TE, Vaccari T, Stenmark H. Shaping development with ESCRTs. Nat. Cell Biol. 2011;14:38–45. doi: 10.1038/ncb2381. [DOI] [PubMed] [Google Scholar]
- 39.Shields SB, Piper RC. How ubiquitin functions with ESCRTs. Traffic. 2011;12:1306–1317. doi: 10.1111/j.1600-0854.2011.01242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol. Biol. Cell. 2005;16:5163–5174. doi: 10.1091/mbc.E05-06-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Row PE, Prior IA, McCullough J, Clague MJ, Urbe S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J. Biol. Chem. 2006;281:12618–12624. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- 42.Wright MH, Berlin I, Nash PD. Regulation of endocytic sorting by ESCRTDUB-mediated deubiquitination. Cell Biochem. Biophys. 2011;60:39–46. doi: 10.1007/s12013-011-9181-9. [DOI] [PubMed] [Google Scholar]
- 43.McCullough J, Row PE, Lorenzo O, Doherty M, Beynon R, Clague MJ, Urbe S. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr. Biol. 2006;16:160–165. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- 44.Ma YM, Boucrot E, Villen J, Affar el B, Gygi SP, Gottlinger HG, Kirchhausen T. Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J. Biol. Chem. 2007;282:9805–9812. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- 45.Row PE, Liu H, Hayes S, Welchman R, Charalabous P, Hofmann K, Clague MJ, Sanderson CM, Urbe S. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J. Biol. Chem. 2007;282:30929–30937. doi: 10.1074/jbc.M704009200. [DOI] [PubMed] [Google Scholar]
- 46.Wu X, Yen L, Irwin L, Sweeney C, Carraway KL., III Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol. Cell. Biol. 2004;24:7748–7757. doi: 10.1128/MCB.24.17.7748-7757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meenhuis A, Verwijmeren C, Roovers O, Touw IP. The deubiquitinating enzyme DUB2A enhances CSF3 signalling by attenuating lysosomal routing of the CSF3 receptor. Biochem. J. 2011;434:343–351. doi: 10.1042/BJ20101628. [DOI] [PubMed] [Google Scholar]
- 48.Berlin I, Schwartz H, Nash PD. Regulation of epidermal growth factor receptor ubiquitination and trafficking by the USP8.STAM complex. J. Biol. Chem. 2010;285:34909–34921. doi: 10.1074/jbc.M109.016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z, Zanata SM, Kim J, Peterson MA, Di Vizio D, Chirieac LR, Pyne S, Agostini M, Freeman MR, Loda M. The ubiquitin-specific protease USP2a prevents endocytosis-mediated EGFR degradation. Oncogene. doi: 10.1038/onc.2012.188. (in press) (Epub ahead of print, Jun 18 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pareja F, Ferraro DA, Rubin C, Cohen-Dvashi H, Zhang F, Aulmann S, Ben-Chetrit N, Pines G, Navon R, Crosetto N, Kostler W, Carvalho S, Lavi S, Schmitt F, Dikic I, Yakhini Z, Sinn P, Mills GB, Yarden Y. Deubiquitination of EGFR by Cezanne-1 contributes to cancer progression. Oncogene. 2012;31:4599–4608. doi: 10.1038/onc.2011.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harhaj EW, Dixit VM. Regulation of NF-kappaB by deubiquitinases. Immunol. Rev. 2012;246:107–124. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Z, Hunter T. Degradation of activated protein kinases by ubiquitination. Annu. Rev. Biochem. 2009;78:435–475. doi: 10.1146/annurev.biochem.013008.092711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mori S, Heldin CH, Claesson-Welsh L. Ligand-induced polyubiquitination of the platelet-derived growth factor beta-receptor. J. Biol. Chem. 1992;267:6429–6434. [PubMed] [Google Scholar]
- 54.Langdon WY, Hartley JW, Klinken SP, Ruscetti SK, Morse HC., III v-cbl, an oncogene from a dual-recombinant murine retrovirus that induces early B-lineage lymphomas. Proc. Natl. Acad. Sci. U.S.A. 1989;86:1168–1172. doi: 10.1073/pnas.86.4.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blake TJ, Heath KG, Langdon WY. The truncation that generated the v-cbl oncogene reveals an ability for nuclear transport, DNA binding and acute transformation. EMBO J. 1993;12:2017–2026. doi: 10.1002/j.1460-2075.1993.tb05851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meng W, Sawasdikosol S, Burakoff SJ, Eck MJ. Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature. 1999;398:84–90. doi: 10.1038/18050. [DOI] [PubMed] [Google Scholar]
- 57.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl–UbcH7 complex: RING domain function in ubiquitin–protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 58.Ota S, Hazeki K, Rao N, Lupher ML, Jr, Andoniou CE, Druker B, Band H. The RING finger domain of Cbl is essential for negative regulation of the Syk tyrosine kinase. J. Biol. Chem. 2000;275:414–422. doi: 10.1074/jbc.275.1.414. [DOI] [PubMed] [Google Scholar]
- 59.Lill NL, Douillard P, Awwad RA, Ota S, Lupher ML, Jr, Miyake S, Meissner-Lula N, Hsu VW, Band H. The evolutionarily conserved N-terminal region of Cbl is sufficient to enhance down-regulation of the epidermal growth factor receptor. J. Biol. Chem. 2000;275:367–377. doi: 10.1074/jbc.275.1.367. [DOI] [PubMed] [Google Scholar]
- 60.Lupher ML, Jr, Rao N, Eck MJ, Band H. The Cbl protooncoprotein: a negative regulator of immune receptor signal transduction. Immunol. Today. 1999;20:375–382. doi: 10.1016/s0167-5699(99)01484-x. [DOI] [PubMed] [Google Scholar]
- 61.Naramura M, Band V, Band H. Indispensable roles of mammalian Cbl family proteins as negative regulators of protein tyrosine kinase signaling: insights from in vivo models. Commun. Integr. Biol. 2011;4:159–162. doi: 10.4161/cib.4.2.14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naramura M, Nadeau S, Mohapatra B, Ahmad G, Mukhopadhyay C, Sattler M, Raja SM, Natarajan A, Band V, Band H. Mutant Cbl proteins as oncogenic drivers in myeloproliferative disorders. Oncotarget. 2011;2:245–250. doi: 10.18632/oncotarget.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duan L, Reddi AL, Ghosh A, Dimri M, Band H. The Cbl family and other ubiquitin ligases: destructive forces in control of antigen receptor signaling. Immunity. 2004;21:7–17. doi: 10.1016/j.immuni.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 64.Thien CB, Langdon WY. c-Cbl and Cbl-b ubiquitin ligases: substrate diversity and the negative regulation of signalling responses. Biochem. J. 2005;391:153–166. doi: 10.1042/BJ20050892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer. 2011;11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bartkiewicz M, Houghton A, Baron R. Leucine zipper-mediated homodimerization of the adaptor protein c-Cbl. A role in c-Cbl's tyrosine phosphorylation and its association with epidermal growth factor receptor. J. Biol. Chem. 1999;274:30887–30895. doi: 10.1074/jbc.274.43.30887. [DOI] [PubMed] [Google Scholar]
- 67.Davies GC, Ettenberg SA, Coats AO, Mussante M, Ravichandran S, Collins J, Nau MM, Lipkowitz S. Cbl-b interacts with ubiquitinated proteins; differential functions of the UBA domains of c-Cbl and Cbl-b. Oncogene. 2004;23:7104–7115. doi: 10.1038/sj.onc.1207952. [DOI] [PubMed] [Google Scholar]
- 68.Peschard P, Kozlov G, Lin T, Mirza IA, Berghuis AM, Lipkowitz S, Park M, Gehring K. Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol. Cell. 2007;27:474–485. doi: 10.1016/j.molcel.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 69.Nau MM, Lipkowitz S. Comparative genomic organization of the cbl genes. Gene. 2003;308:103–113. doi: 10.1016/s0378-1119(03)00471-2. [DOI] [PubMed] [Google Scholar]
- 70.Keane MM, Ettenberg SA, Nau MM, Banerjee P, Cuello M, Penninger J, Lipkowitz S. Cbl-3: a new mammalian Cbl family protein. Oncogene. 1999;18:3365–3375. doi: 10.1038/sj.onc.1202753. [DOI] [PubMed] [Google Scholar]
- 71.Langenick J, Araki T, Yamada Y, Williams JG. A Dictyostelium homologue of the metazoan Cbl proteins regulates STAT signalling. J. Cell Sci. 2008;121:3524–3530. doi: 10.1242/jcs.036798. [DOI] [PubMed] [Google Scholar]
- 72.Blake TJ, Shapiro M, Morse HC, III, Langdon WY. The sequences of the human and mouse c-cbl proto-oncogenes show v-cbl was generated by a large truncation encompassing a proline-rich domain and a leucine zipper-like motif. Oncogene. 1991;6:653–657. [PubMed] [Google Scholar]
- 73.Andoniou CE, Thien CB, Langdon WY. Tumour induction by activated abl involves tyrosine phosphorylation of the product of the cbl oncogene. EMBO J. 1994;13:4515–4523. doi: 10.1002/j.1460-2075.1994.tb06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Donovan JA, Wange RL, Langdon WY, Samelson LE. The protein product of the c-cbl protooncogene is the 120-kDa tyrosine-phosphorylated protein in Jurkat cells activated via the T cell antigen receptor. J. Biol. Chem. 1994;269:22921–22924. [PubMed] [Google Scholar]
- 75.Reedquist KA, Fukazawa T, Druker B, Panchamoorthy G, Shoelson SE, Band H. Rapid T-cell receptor-mediated tyrosine phosphorylation of p120, an Fyn/Lck Src homology 3 domain-binding protein. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4135–4139. doi: 10.1073/pnas.91.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fukazawa T, Reedquist KA, Trub T, Soltoff S, Panchamoorthy G, Druker B, Cantley L, Shoelson SE, Band H. The SH3 domain-binding T cell tyrosyl phosphoprotein p120. Demonstration of its identity with the c-cbl protooncogene product and in vivo complexes with Fyn, Grb2, and phosphatidylinositol 3-kinase. J. Biol. Chem. 1995;270:19141–19150. doi: 10.1074/jbc.270.32.19141. [DOI] [PubMed] [Google Scholar]
- 77.Rao N, Dodge I, Band H. The Cbl family of ubiquitin ligases: critical negative regulators of tyrosine kinase signaling in the immune system. J. Leukoc. Biol. 2002;71:753–763. [PubMed] [Google Scholar]
- 78.Thien CB, Langdon WY. Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 79.Tsygankov AY, Teckchandani AM, Feshchenko EA, Swaminathan G. Beyond the RING: CBL proteins as multivalent adapters. Oncogene. 2001;20:6382–6402. doi: 10.1038/sj.onc.1204781. [DOI] [PubMed] [Google Scholar]
- 80.Fukazawa T, Miyake S, Band V, Band H. Tyrosine phosphorylation of Cbl upon epidermal growth factor (EGF) stimulation and its association with EGF receptor and downstream signaling proteins. J. Biol. Chem. 1996;271:14554–14559. doi: 10.1074/jbc.271.24.14554. [DOI] [PubMed] [Google Scholar]
- 81.Panchamoorthy G, Fukazawa T, Miyake S, Soltoff S, Reedquist K, Druker B, Shoelson S, Cantley L, Band H. p120cbl is a major substrate of tyrosine phosphorylation upon B cell antigen receptor stimulation and interacts in vivo with Fyn and Syk tyrosine kinases, Grb2 and Shc adaptors, and the p85 subunit of phosphatidylinositol 3-kinase. J. Biol. Chem. 1996;271:3187–3194. doi: 10.1074/jbc.271.6.3187. [DOI] [PubMed] [Google Scholar]
- 82.Bonita DP, Miyake S, Lupher ML, Jr, Langdon WY, Band H. Phosphotyrosine binding domain-dependent upregulation of the platelet-derived growth factor receptor alpha signaling cascade by transforming mutants of Cbl: implications for Cbl's function and oncogenicity. Mol. Cell. Biol. 1997;17:4597–4610. doi: 10.1128/mcb.17.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meisner H, Conway BR, Hartley D, Czech MP. Interactions of Cbl with Grb2 and phosphatidylinositol 3′-kinase in activated Jurkat cells. Mol. Cell. Biol. 1995;15:3571–3578. doi: 10.1128/mcb.15.7.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yoon CH, Lee J, Jongeward GD, Sternberg PW. Similarity of sli-1, a regulator of vulval development in C. elegans, to the mammalian proto-oncogene c-cbl. Science. 1995;269:1102–1105. doi: 10.1126/science.7652556. [DOI] [PubMed] [Google Scholar]
- 85.Lupher ML, Jr, Reedquist KA, Miyake S, Langdon WY, Band H. A novel phosphotyrosine-binding domain in the N-terminal transforming region of Cbl interacts directly and selectively with ZAP-70 in T cells. J. Biol. Chem. 1996;271:24063–24068. doi: 10.1074/jbc.271.39.24063. [DOI] [PubMed] [Google Scholar]
- 86.Lupher ML, Jr, Rao N, Lill NL, Andoniou CE, Miyake S, Clark EA, Druker B, Band H. Cbl-mediated negative regulation of the Syk tyrosine kinase. A critical role for Cbl phosphotyrosine-binding domain binding to Syk phosphotyrosine 323. J. Biol. Chem. 1998;273:35273–35281. doi: 10.1074/jbc.273.52.35273. [DOI] [PubMed] [Google Scholar]
- 87.Lupher ML, Jr, Songyang Z, Shoelson SE, Cantley LC, Band H. The Cbl phosphotyrosine-binding domain selects a D(N/D)XpY motif and binds to the Tyr292 negative regulatory phosphorylation site of ZAP-70. J. Biol. Chem. 1997;272:33140–33144. doi: 10.1074/jbc.272.52.33140. [DOI] [PubMed] [Google Scholar]
- 88.Miyake S, Mullane-Robinson KP, Lill NL, Douillard P, Band H. Cbl-mediated negative regulation of platelet-derived growth factor receptor-dependent cell proliferation. A critical role for Cbl tyrosine kinase-binding domain. J. Biol. Chem. 1999;274:16619–16628. doi: 10.1074/jbc.274.23.16619. [DOI] [PubMed] [Google Scholar]
- 89.Thien CB, Langdon WY. Tyrosine kinase activity of the EGF receptor is enhanced by the expression of oncogenic 70Z-Cbl. Oncogene. 1997;15:2909–2919. doi: 10.1038/sj.onc.1201468. [DOI] [PubMed] [Google Scholar]
- 90.Miyake S, Lupher ML, Jr, Druker B, Band H. The tyrosine kinase regulator Cbl enhances the ubiquitination and degradation of the platelet-derived growth factor receptor alpha. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7927–7932. doi: 10.1073/pnas.95.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 92.Terrell J, Shih S, Dunn R, Hicke L. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol. Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- 93.Abella JV, Peschard P, Naujokas MA, Lin T, Saucier C, Urbe S, Park M. Met/Hepatocyte growth factor receptor ubiquitination suppresses transformation and is required for Hrs phosphorylation. Mol. Cell. Biol. 2005;25:9632–9645. doi: 10.1128/MCB.25.21.9632-9645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee PS, Wang Y, Dominguez MG, Yeung YG, Murphy MA, Bowtell DD, Stanley ER. The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J. 1999;18:3616–3628. doi: 10.1093/emboj/18.13.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peschard P, Fournier TM, Lamorte L, Naujokas MA, Band H, Langdon WY, Park M. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol. Cell. 2001;8:995–1004. doi: 10.1016/s1097-2765(01)00378-1. [DOI] [PubMed] [Google Scholar]
- 96.Rao N, Ghosh AK, Douillard P, Andoniou CE, Zhou P, Band H. An essential role of ubiquitination in Cbl-mediated negative regulation of the Src-family kinase Fyn. Signal Transduct. 2002;2:29–39. doi: 10.1002/1615-4061(200205)2:1/2<29::AID-SITA29>3.0.CO;2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rao N, Ghosh AK, Ota S, Zhou P, Reddi AL, Hakezi K, Druker BK, Wu J, Band H. The non-receptor tyrosine kinase Syk is a target of Cbl-mediated ubiquitylation upon B-cell receptor stimulation. EMBO J. 2001;20:7085–7095. doi: 10.1093/emboj/20.24.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rao N, Miyake S, Reddi AL, Douillard P, Ghosh AK, Dodge IL, Zhou P, Fernandes ND, Band H. Negative regulation of Lck by Cbl ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3794–3799. doi: 10.1073/pnas.062055999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rao N, Lupher ML, Jr, Ota S, Reedquist KA, Druker BJ, Band H. The linker phosphorylation site Tyr292 mediates the negative regulatory effect of Cbl on ZAP-70 in T cells. J. Immunol. 2000;164:4616–4626. doi: 10.4049/jimmunol.164.9.4616. [DOI] [PubMed] [Google Scholar]
- 100.Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin–protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 101.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 102.Ettenberg SA, Magnifico A, Cuello M, Nau MM, Rubinstein YR, Yarden Y, Weissman AM, Lipkowitz S. Cbl-b-dependent coordinated degradation of the epidermal growth factor receptor signaling complex. J. Biol. Chem. 2001;276:27677–27684. doi: 10.1074/jbc.M102641200. [DOI] [PubMed] [Google Scholar]
- 103.Kim M, Tezuka T, Tanaka K, Yamamoto T. Cbl-c suppresses v-Src-induced transformation through ubiquitin-dependent protein degradation. Oncogene. 2004;23:1645–1655. doi: 10.1038/sj.onc.1207298. [DOI] [PubMed] [Google Scholar]
- 104.Ryan PE, Sivadasan-Nair N, Nau MM, Nicholas S, Lipkowitz S. The N terminus of Cbl-c regulates ubiquitin ligase activity by modulating affinity for the ubiquitin-conjugating enzyme. J. Biol. Chem. 2010;285:23687–23698. doi: 10.1074/jbc.M109.091157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kales SC, Ryan PE, Lipkowitz S. Cbl exposes its RING finger. Nat. Struct. Mol. Biol. 2012;19:131–133. doi: 10.1038/nsmb.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ng C, Jackson RA, Buschdorf JP, Sun Q, Guy GR, Sivaraman J. Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates. EMBO J. 2008;27:804–816. doi: 10.1038/emboj.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Penengo L, Rubin C, Yarden Y, Gaudino G. c-Cbl is a critical modulator of the Ron tyrosine kinase receptor. Oncogene. 2003;22:3669–3679. doi: 10.1038/sj.onc.1206585. [DOI] [PubMed] [Google Scholar]
- 108.Peschard P, Ishiyama N, Lin T, Lipkowitz S, Park M. A conserved DpYR motif in the juxtamembrane domain of the Met receptor family forms an atypical c-Cbl/Cbl-b tyrosine kinase binding domain binding site required for suppression of oncogenic activation. J. Biol. Chem. 2004;279:29565–29571. doi: 10.1074/jbc.M403954200. [DOI] [PubMed] [Google Scholar]
- 109.Hu J, Hubbard SR. Structural characterization of a novel Cbl phosphotyrosine recognition motif in the APS family of adapter proteins. J. Biol. Chem. 2005;280:18943–18949. doi: 10.1074/jbc.M414157200. [DOI] [PubMed] [Google Scholar]
- 110.Masson K, Heiss E, Band H, Ronnstrand L. Direct binding of Cbl to Tyr568 and Tyr936 of the stem cell factor receptor/c-Kit is required for ligand-induced ubiquitination, internalization and degradation. Biochem. J. 2006;399:59–67. doi: 10.1042/BJ20060464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol. Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 112.Thien CB, Walker F, Langdon WY. RING finger mutations that abolish c-Cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol. Cell. 2001;7:355–365. doi: 10.1016/s1097-2765(01)00183-6. [DOI] [PubMed] [Google Scholar]
- 113.Kobashigawa Y, Tomitaka A, Kumeta H, Noda NN, Yamaguchi M, Inagaki F. Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20579–20584. doi: 10.1073/pnas.1110712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat. Struct. Mol. Biol. 2012;19:184–192. doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schmidt MH, Furnari FB, Cavenee WK, Bogler O. Epidermal growth factor receptor signaling intensity determines intracellular protein interactions, ubiquitination, and internalization. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6505–6510. doi: 10.1073/pnas.1031790100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ghosh AK, Reddi AL, Rao NL, Duan L, Band V, Band H. Biochemical basis for the requirement of kinase activity for Cbl-dependent ubiquitinylation and degradation of a target tyrosine kinase. J. Biol. Chem. 2004;279:36132–36141. doi: 10.1074/jbc.M404189200. [DOI] [PubMed] [Google Scholar]
- 117.Kassenbrock CK, Anderson SM. Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. J. Biol. Chem. 2004;279:28017–28027. doi: 10.1074/jbc.M404114200. [DOI] [PubMed] [Google Scholar]
- 118.Rosenkranz S, Ikuno Y, Leong FL, Klinghoffer RA, Miyake S, Band H, Kazlauskas A. Src family kinases negatively regulate platelet-derived growth factor alpha receptor-dependent signaling and disease progression. J. Biol. Chem. 2000;275:9620–9627. doi: 10.1074/jbc.275.13.9620. [DOI] [PubMed] [Google Scholar]
- 119.Liu YC, Gu H. Cbl and Cbl-b in T-cell regulation. Trends Immunol. 2002;23:140–143. doi: 10.1016/s1471-4906(01)02157-3. [DOI] [PubMed] [Google Scholar]
- 120.Miura-Shimura Y, Duan L, Rao NL, Reddi AL, Shimura H, Rottapel R, Druker BJ, Tsygankov A, Band V, Band H. Cbl-mediated ubiquitinylation and negative regulation of Vav. J. Biol. Chem. 2003;278:38495–38504. doi: 10.1074/jbc.M305656200. [DOI] [PubMed] [Google Scholar]
- 121.Duan L, Raja SM, Chen G, Virmani S, Williams SH, Clubb RJ, Mukhopadhyay C, Rainey MA, Ying G, Dimri M, Chen J, Reddi AL, Naramura M, Band V, Band H. Negative regulation of EGFR-Vav2 signaling axis by Cbl ubiquitin ligase controls EGF receptor-mediated epithelial cell adherens junction dynamics and cell migration. J. Biol. Chem. 2011;286:620–633. doi: 10.1074/jbc.M110.188086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang W, Shao Y, Fang D, Huang J, Jeon MS, Liu YC. Negative regulation of T cell antigen receptor-mediated Crk-L-C3G signaling and cell adhesion by Cbl-b. J. Biol. Chem. 2003;278:23978–23983. doi: 10.1074/jbc.M212671200. [DOI] [PubMed] [Google Scholar]
- 123.d'Azzo A, Bongiovanni A, Nastasi T. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic. 2005;6:429–441. doi: 10.1111/j.1600-0854.2005.00294.x. [DOI] [PubMed] [Google Scholar]
- 124.Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 125.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Umebayashi K, Stenmark H, Yoshimori T. Ubc4/5 and c-Cbl continue to ubiquitinate EGF receptor after internalization to facilitate polyubiquitination and degradation. Mol. Biol. Cell. 2008;19:3454–3462. doi: 10.1091/mbc.E07-10-0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fasen K, Cerretti DP, Huynh-Do U. Ligand binding induces Cbl-dependent EphB1 receptor degradation through the lysosomal pathway. Traffic. 2008;9:251–266. doi: 10.1111/j.1600-0854.2007.00679.x. [DOI] [PubMed] [Google Scholar]
- 128.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 129.Longva KE, Blystad FD, Stang E, Larsen AM, Johannessen LE, Madshus IH. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 2002;156:843–854. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huang C. Roles of E3 ubiquitin ligases in cell adhesion and migration. Cell Adh. Migr. 2010;4:10–18. doi: 10.4161/cam.4.1.9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ryan PE, Davies GC, Nau MM, Lipkowitz S. Regulating the regulator: negative regulation of Cbl ubiquitin ligases. Trends Biochem. Sci. 2006;31:79–88. doi: 10.1016/j.tibs.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 132.Reddi Alagarsamy L, Duan Lei, Rainey Mark A, Ortega-Cava Cesar, Clubb Robert, Chung Byung Min, Dimri Manjari, Tu Chun, George Manju, Raja Srikumar, Naramura Mayumi. Cbl-family E3 ubiquitin ligases as regulators of non-receptor tyrosine kinases. In: Tsygankov Alexander., editor. Cbl Proteins. Nova Science Publishers; 2008. pp. 53–74. [Google Scholar]
- 133.Soubeyran P, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I. Cbl-CIN85–endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature. 2002;416:183–187. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]
- 134.Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S. The endophilin–CIN85-Cbl complex mediates ligand-dependent downregulation of c-Met. Nature. 2002;416:187–190. doi: 10.1038/416187a. [DOI] [PubMed] [Google Scholar]
- 135.Duan L, Miura Y, Dimri M, Majumder B, Dodge IL, Reddi AL, Ghosh A, Fernandes N, Zhou P, Mullane-Robinson K, Rao N, Donoghue S, Rogers RA, Bowtell D, Naramura M, Gu H, Band V, Band H. Cbl-mediated ubiquitinylation is required for lysosomal sorting of epidermal growth factor receptor but is dispensable for endocytosis. J. Biol. Chem. 2003;278:28950–28960. doi: 10.1074/jbc.M304474200. [DOI] [PubMed] [Google Scholar]
- 136.Khan EM, Heidinger JM, Levy M, Lisanti MP, Ravid T, Goldkorn T. Epidermal growth factor receptor exposed to oxidative stress undergoes Src- and caveolin-1-dependent perinuclear trafficking. J. Biol. Chem. 2006;281:14486–14493. doi: 10.1074/jbc.M509332200. [DOI] [PubMed] [Google Scholar]
- 137.Huang F, Goh LK, Sorkin A. EGF receptor ubiquitination is not necessary for its internalization. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16904–16909. doi: 10.1073/pnas.0707416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Goh LK, Huang F, Kim W, Gygi S, Sorkin A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J. Cell Biol. 2010;189:871–883. doi: 10.1083/jcb.201001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Swaminathan G, Tsygankov AY. The Cbl family proteins: ring leaders in regulation of cell signaling. J. Cell. Physiol. 2006;209:21–43. doi: 10.1002/jcp.20694. [DOI] [PubMed] [Google Scholar]
- 140.Miura A, Sajan MP, Standaert ML, Bandyopadhyay G, Franklin DM, Lea-Currie R, Farese RV. Cbl PYXXM motifs activate the P85 subunit of phosphatidylinositol 3-kinase, Crk, atypical protein kinase C, and glucose transport during thiazolidinedione action in 3T3/L1 and human adipocytes. Biochemistry. 2003;42:14335–14341. doi: 10.1021/bi034808i. [DOI] [PubMed] [Google Scholar]
- 141.Standaert ML, Sajan MP, Miura A, Bandyopadhyay G, Farese RV. Requirements for pYXXM motifs in Cbl for binding to the p85 subunit of phosphatidylinositol 3-kinase and Crk, and activation of atypical protein kinase C and glucose transport during insulin action in 3T3/L1 adipocytes. Biochemistry. 2004;43:15494–15502. doi: 10.1021/bi049222q. [DOI] [PubMed] [Google Scholar]
- 142.Goh EL, Zhu T, Leong WY, Lobie PE. c-Cbl is a negative regulator of GH-stimulated STAT5-mediated transcription. Endocrinology. 2002;143:3590–3603. doi: 10.1210/en.2002-220374. [DOI] [PubMed] [Google Scholar]
- 143.Wang L, Rudert WA, Loutaev I, Roginskaya V, Corey SJ. Repression of c-Cbl leads to enhanced G-CSF Jak-STAT signaling without increased cell proliferation. Oncogene. 2002;21:5346–5355. doi: 10.1038/sj.onc.1205670. [DOI] [PubMed] [Google Scholar]
- 144.Timpson P, Lynch DK, Schramek D, Walker F, Daly RJ. Cortactin overexpression inhibits ligand-induced down-regulation of the epidermal growth factor receptor. Cancer Res. 2005;65:3273–3280. doi: 10.1158/0008-5472.CAN-04-2118. [DOI] [PubMed] [Google Scholar]
- 145.Fong CW, Leong HF, Wong ES, Lim J, Yusoff P, Guy GR. Tyrosine phosphorylation of Sprouty2 enhances its interaction with c-Cbl and is crucial for its function. J. Biol. Chem. 2003;278:33456–33464. doi: 10.1074/jbc.M301317200. [DOI] [PubMed] [Google Scholar]
- 146.Wong ES, Fong CW, Lim J, Yusoff P, Low BC, Langdon WY, Guy GR. Sprouty2 attenuates epidermal growth factor receptor ubiquitylation and endocytosis, and consequently enhances Ras/ERK signalling. EMBO J. 2002;21:4796–4808. doi: 10.1093/emboj/cdf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wong ES, Guy GR. Regulator of epidermal growth factor signaling: Sprouty. Methods Mol. Biol. 2006;327:61–83. doi: 10.1385/1-59745-012-X:61. [DOI] [PubMed] [Google Scholar]
- 148.Wiedemann A, Muller S, Favier B, Penna D, Guiraud M, Delmas C, Champagne E, Valitutti S. T-cell activation is accompanied by an ubiquitination process occurring at the immunological synapse. Immunol. Lett. 2005;98:57–61. doi: 10.1016/j.imlet.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 149.Jeon MS, Atfield A, Venuprasad K, Krawczyk C, Sarao R, Elly C, Yang C, Arya S, Bachmaier K, Su L, Bouchard D, Jones R, Gronski M, Ohashi P, Wada T, Bloom D, Fathman CG, Liu YC, Penninger JM. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity. 2004;21:167–177. doi: 10.1016/j.immuni.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 150.Naramura M, Jang IK, Kole H, Huang F, Haines D, Gu H. c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR down-modulation. Nat. Immunol. 2002;3:1192–1199. doi: 10.1038/ni855. [DOI] [PubMed] [Google Scholar]
- 151.Rathinam C, Thien CB, Langdon WY, Gu H, Flavell RA. The E3 ubiquitin ligase c-Cbl restricts development and functions of hematopoietic stem cells. Genes Dev. 2008;22:992–997. doi: 10.1101/gad.1651408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rathinam C, Thien CB, Flavell RA, Langdon WY. Myeloid leukemia development in c-Cbl RING finger mutant mice is dependent on FLT3 signaling. Cancer Cell. 2010;18:341–352. doi: 10.1016/j.ccr.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 153.Naramura M, Nandwani N, Gu H, Band V, Band H. Rapidly fatal myeloproliferative disorders in mice with deletion of Casitas B-cell lymphoma (Cbl) and Cbl-b in hematopoietic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16274–16279. doi: 10.1073/pnas.1007575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ledda F, Paratcha G. Negative regulation of receptor tyrosine kinase (RTK) signaling: a developing field. Biomark. Insights. 2007;2:45–58. [PMC free article] [PubMed] [Google Scholar]
- 155.Segatto O, Anastasi S, Alema S. Regulation of epidermal growth factor receptor signalling by inducible feedback inhibitors. J. Cell Sci. 2011;124:1785–1793. doi: 10.1242/jcs.083303. [DOI] [PubMed] [Google Scholar]
- 156.Ahmed AU, Schmidt RL, Park CH, Reed NR, Hesse SE, Thomas CF, Molina JR, Deschamps C, Yang P, Aubry MC, Tang AH. Effect of disrupting seven-in-absentia homolog 2 function on lung cancer cell growth. J. Natl. Cancer Inst. 2008;100:1606–1629. doi: 10.1093/jnci/djn365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, Amariglio N, Henriksson R, Rechavi G, Hedman H, Wides R, Yarden Y. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004;23:3270–3281. doi: 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–744. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Frosi Y, Anastasi S, Ballaro C, Varsano G, Castellani L, Maspero E, Polo S, Alema S, Segatto O. A two-tiered mechanism of EGFR inhibition by RALT/MIG6 via kinase suppression and receptor degradation. J. Cell Biol. 2010;189:557–571. doi: 10.1083/jcb.201002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schmid EM, McMahon HT. Integrating molecular and network biology to decode endocytosis. Nature. 2007;448:883–888. doi: 10.1038/nature06031. [DOI] [PubMed] [Google Scholar]
- 161.Nicholson SE, Metcalf D, Sprigg NS, Columbus R, Walker F, Silva A, Cary D, Willson TA, Zhang JG, Hilton DJ, Alexander WS, Nicola NA. Suppressor of cytokine signaling (SOCS)-5 is a potential negative regulator of epidermal growth factor signaling. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2328–2333. doi: 10.1073/pnas.0409675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kario E, Marmor MD, Adamsky K, Citri A, Amit I, Amariglio N, Rechavi G, Yarden Y. Suppressors of cytokine signaling 4 and 5 regulate epidermal growth factor receptor signaling. J. Biol. Chem. 2005;280:7038–7048. doi: 10.1074/jbc.M408575200. [DOI] [PubMed] [Google Scholar]
- 163.Tanaka Y, Tanaka N, Saeki Y, Tanaka K, Murakami M, Hirano T, Ishii N, Sugamura K. c-Cbl-dependent monoubiquitination and lysosomal degradation of gp130. Mol. Cell. Biol. 2008;28:4805–4818. doi: 10.1128/MCB.01784-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tarcic G, Boguslavsky SK, Wakim J, Kiuchi T, Liu A, Reinitz F, Nathanson D, Takahashi T, Mischel PS, Ng T, Yarden Y. An unbiased screen identifies DEP-1 tumor suppressor as a phosphatase controlling EGFR endocytosis. Curr. Biol. 2009;19:1788–1798. doi: 10.1016/j.cub.2009.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Guy GR, Jackson RA, Yusoff P, Chow SY. Sprouty proteins: modified modulators, matchmakers or missing links? J. Endocrinol. 2009;203:191–202. doi: 10.1677/JOE-09-0110. [DOI] [PubMed] [Google Scholar]
- 166.Hanafusa H, Torii S, Yasunaga T, Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat. Cell Biol. 2002;4:850–858. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- 167.Sasaki A, Taketomi T, Kato R, Saeki K, Nonami A, Sasaki M, Kuriyama M, Saito N, Shibuya M, Yoshimura A. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat. Cell Biol. 2003;5:427–432. doi: 10.1038/ncb978. [DOI] [PubMed] [Google Scholar]
- 168.Wong ES, Lim J, Low BC, Chen Q, Guy GR. Evidence for direct interaction between Sprouty and Cbl. J. Biol. Chem. 2001;276:5866–5875. doi: 10.1074/jbc.M006945200. [DOI] [PubMed] [Google Scholar]
- 169.Hall AB, Jura N, DaSilva J, Jang YJ, Gong D, Bar-Sagi D. hSpry2 is targeted to the ubiquitin-dependent proteasome pathway by c-Cbl. Curr. Biol. 2003;13:308–314. doi: 10.1016/s0960-9822(03)00086-1. [DOI] [PubMed] [Google Scholar]
- 170.Rubin C, Litvak V, Medvedovsky H, Zwang Y, Lev S, Yarden Y. Sprouty fine-tunes EGF signaling through interlinked positive and negative feedback loops. Curr. Biol. 2003;13:297–307. doi: 10.1016/s0960-9822(03)00053-8. [DOI] [PubMed] [Google Scholar]
- 171.Egan JE, Hall AB, Yatsula BA, Bar-Sagi D. The bimodal regulation of epidermal growth factor signaling by human Sprouty proteins. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6041–6046. doi: 10.1073/pnas.052090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Choi H, Cho SY, Schwartz RH, Choi K. Dual effects of Sprouty1 on TCR signaling depending on the differentiation state of the T cell. J. Immunol. 2006;176:6034–6045. doi: 10.4049/jimmunol.176.10.6034. [DOI] [PubMed] [Google Scholar]
- 173.Meisner H, Daga A, Buxton J, Fernandez B, Chawla A, Banerjee U, Czech MP. Interactions of Drosophila Cbl with epidermal growth factor receptors and role of Cbl in R7 photoreceptor cell development. Mol. Cell. Biol. 1997;17:2217–2225. doi: 10.1128/mcb.17.4.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pai LM, Barcelo G, Schupbach T. D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell. 2000;103:51–61. doi: 10.1016/s0092-8674(00)00104-5. [DOI] [PubMed] [Google Scholar]
- 175.Nowak M, Machate A, Yu SR, Gupta M, Brand M. Interpretation of the FGF8 morphogen gradient is regulated by endocytic trafficking. Nat. Cell Biol. 2011;13:153–158. doi: 10.1038/ncb2155. [DOI] [PubMed] [Google Scholar]
- 176.Murphy MA, Schnall RG, Venter DJ, Barnett L, Bertoncello I, Thien CB, Langdon WY, Bowtell DD. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol. Cell. Biol. 1998;18:4872–4882. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Naramura M, Kole HK, Hu RJ, Gu H. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15547–15552. doi: 10.1073/pnas.95.26.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.El Chami N, Ikhlef F, Kaszas K, Yakoub S, Tabone E, Siddeek B, Cunha S, Beaudoin C, Morel L, Benahmed M, Regnier DC. Androgen-dependent apoptosis in male germ cells is regulated through the proto-oncoprotein Cbl. J. Cell Biol. 2005;171:651–661. doi: 10.1083/jcb.200507076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Molero JC, Jensen TE, Withers PC, Couzens M, Herzog H, Thien CB, Langdon WY, Walder K, Murphy MA, Bowtell DD, James DE, Cooney GJ. c-Cbl-deficient mice have reduced adiposity, higher energy expenditure, and improved peripheral insulin action. J. Clin. Invest. 2004;114:1326–1333. doi: 10.1172/JCI21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bachmaier K, Krawczyk C, Kozieradzki I, Kong YY, Sasaki T, Oliveira-dos-Santos A, Mariathasan S, Bouchard D, Wakeham A, Itie A, Le J, Ohashi PS, Sarosi I, Nishina H, Lipkowitz S, Penninger JM. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 2000;403:211–216. doi: 10.1038/35003228. [DOI] [PubMed] [Google Scholar]
- 181.Chiang YJ, Kole HK, Brown K, Naramura M, Fukuhara S, Hu RJ, Jang IK, Gutkind JS, Shevach E, Gu H. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 2000;403:216–220. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- 182.Nakao R, Hirasaka K, Goto J, Ishidoh K, Yamada C, Ohno A, Okumura Y, Nonaka I, Yasutomo K, Baldwin KM, Kominami E, Higashibata A, Nagano K, Tanaka K, Yasui N, Mills EM, Takeda S, Nikawa T. Ubiquitin ligase Cbl-b is a negative regulator for insulin-like growth factor 1 signaling during muscle atrophy caused by unloading. Mol. Cell. Biol. 2009;29:4798–4811. doi: 10.1128/MCB.01347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Griffiths EK, Sanchez O, Mill P, Krawczyk C, Hojilla CV, Rubin E, Nau MM, Khokha R, Lipkowitz S, Hui CC, Penninger JM. Cbl-3-deficient mice exhibit normal epithelial development. Mol. Cell. Biol. 2003;23:7708–7718. doi: 10.1128/MCB.23.21.7708-7718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Langdon WY, Hyland CD, Grumont RJ, Morse HC., III The c-cbl proto-oncogene is preferentially expressed in thymus and testis tissue and encodes a nuclear protein. J. Virol. 1989;63:5420–5424. doi: 10.1128/jvi.63.12.5420-5424.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kitaura Y, Jang IK, Wang Y, Han YC, Inazu T, Cadera EJ, Schlissel M, Hardy RR, Gu H. Control of the B cell-intrinsic tolerance programs by ubiquitin ligases Cbl and Cbl-b. Immunity. 2007;26:567–578. doi: 10.1016/j.immuni.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sargin B, Choudhary C, Crosetto N, Schmidt MH, Grundler R, Rensinghoff M, Thiessen C, Tickenbrock L, Schwable J, Brandts C, August B, Koschmieder S, Bandi SR, Duyster J, Berdel WE, Muller-Tidow C, Dikic I, Serve H. Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood. 2007;110:1004–1012. doi: 10.1182/blood-2007-01-066076. [DOI] [PubMed] [Google Scholar]
- 187.Caligiuri MA, Briesewitz R, Yu J, Wang L, Wei M, Arnoczky KJ, Marburger TB, Wen J, Perrotti D, Bloomfield CD, Whitman SP. Novel c-CBL and CBL-b ubiquitin ligase mutations in human acute myeloid leukemia. Blood. 2007;110:1022–1024. doi: 10.1182/blood-2006-12-061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Makishima H, Cazzolli H, Szpurka H, Dunbar A, Tiu R, Huh J, Muramatsu H, O'Keefe C, Hsi E, Paquette RL, Kojima S, List AF, Sekeres MA, McDevitt MA, Maciejewski JP. Mutations of e3 ubiquitin ligase cbl family members constitute a novel common pathogenic lesion in myeloid malignancies. J. Clin. Oncol. 2009;27:6109–6116. doi: 10.1200/JCO.2009.23.7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Brecqueville M, Rey J, Bertucci F, Coppin E, Finetti P, Carbuccia N, Cervera N, Gelsi-Boyer V, Arnoulet C, Gisserot O, Verrot D, Slama B, Vey N, Mozziconacci MJ, Birnbaum D, Murati A. Mutation analysis of ASXL1, CBL, DNMT3A, IDH1, IDH2, JAK2, MPL, NF1, SF3B1, SUZ12, and TET2 in myeloproliferative neoplasms. Genes Chromosomes Cancer. 2012;51:743–755. doi: 10.1002/gcc.21960. [DOI] [PubMed] [Google Scholar]
- 190.Kao HW, Sanada M, Liang DC, Lai CL, Lee EH, Kuo MC, Lin TL, Shih YS, Wu JH, Huang CF, Ogawa S, Shih LY. A high occurrence of acquisition and/or expansion of C-CBL mutant clones in the progression of high-risk myelodysplastic syndrome to acute myeloid leukemia. Neoplasia. 2011;13:1035–1042. doi: 10.1593/neo.111192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nicholson L, Knight T, Matheson E, Minto L, Case M, Sanichar M, Bomken S, Vormoor J, Hall A, Irving J. Casitas B lymphoma mutations in childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2012;51:250–256. doi: 10.1002/gcc.20949. [DOI] [PubMed] [Google Scholar]
- 192.Saito Y, Aoki Y, Muramatsu H, Makishima H, Maciejewski JP, Imaizumi M, Rikiishi T, Sasahara Y, Kure S, Niihori T, Tsuchiya S, Kojima S, Matsubara Y. Casitas B-cell lymphoma mutation in childhood T-cell acute lymphoblastic leukemia. Leuk. Res. 2012;36:1009–1015. doi: 10.1016/j.leukres.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Schnittger S, Bacher U, Alpermann T, Reiter A, Ulke M, Dicker F, Eder C, Kohlmann A, Grossmann V, Kowarsch A, Kern W, Haferlach C, Haferlach T. Use of CBL exon 8 and 9 mutations in diagnosis of myeloproliferative neoplasms and myeloproliferative/myelodysplastic disorders: an analysis of 636 cases. Haematologica. 2012 doi: 10.3324/haematol.2012.065375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shiba N, Kato M, Park MJ, Sanada M, Ito E, Fukushima K, Sako M, Arakawa H, Ogawa S, Hayashi Y. CBL mutations in juvenile myelomonocytic leukemia and pediatric myelodysplastic syndrome. Leukemia. 2010;24:1090–1092. doi: 10.1038/leu.2010.49. [DOI] [PubMed] [Google Scholar]
- 195.Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S, Tamura A, Honda H, Sakata-Yanagimoto M, Kumano K, Oda H, Yamagata T, Takita J, Gotoh N, Nakazaki K, Kawamata N, Onodera M, Nobuyoshi M, Hayashi Y, Harada H, Kurokawa M, Chiba S, Mori H, Ozawa K, Omine M, Hirai H, Nakauchi H, Koeffler HP, Ogawa S. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460:904–908. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 196.Grand FH, Hidalgo-Curtis CE, Ernst T, Zoi K, Zoi C, McGuire C, Kreil S, Jones A, Score J, Metzgeroth G, Oscier D, Hall A, Brandts C, Serve H, Reiter A, Chase AJ, Cross NC. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood. 2009;113:6182–6192. doi: 10.1182/blood-2008-12-194548. [DOI] [PubMed] [Google Scholar]
- 197.Bandi SR, Brandts C, Rensinghoff M, Grundler R, Tickenbrock L, Kohler G, Duyster J, Berdel WE, Muller-Tidow C, Serve H, Sargin B. Study Alliance Leukemias, E3 ligase-defective Cbl mutants lead to a generalized mastocytosis and myeloproliferative disease. Blood. 2009;114:4197–4208. doi: 10.1182/blood-2008-12-190934. [DOI] [PubMed] [Google Scholar]
- 198.Liu X, Sabnis H, Bunting KD, Qu CK. Molecular targets for the treatment of juvenile myelomonocytic leukemia. Adv. Hematol. 2012;2012:308252. doi: 10.1155/2012/308252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mao JH, Sun XY, Liu JX, Zhang QY, Liu P, Huang QH, Li KK, Chen Q, Chen Z, Chen SJ. As4S4 targets RING-type E3 ligase c-CBL to induce degradation of BCR-ABL in chronic myelogenous leukemia. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21683–21688. doi: 10.1073/pnas.1016311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Makishima H, Sugimoto Y, Szpurka H, Clemente MJ, Ng KP, Muramatsu H, O'Keefe C, Saunthararajah Y, Maciejewski JP. CBL mutation-related patterns of phosphorylation and sensitivity to tyrosine kinase inhibitors. Leukemia. 2012;26:1547–1554. doi: 10.1038/leu.2012.7. [DOI] [PubMed] [Google Scholar]
- 201.Loh ML. Childhood myelodysplastic syndrome: focus on the approach to diagnosis and treatment of juvenile myelomonocytic leukemia. Hematol. Am. Soc. Hematol. Educ. Program. 2010;2010:357–362. doi: 10.1182/asheducation-2010.1.357. [DOI] [PubMed] [Google Scholar]
- 202.Tan YH, Krishnaswamy S, Nandi S, Kanteti R, Vora S, Onel K, Hasina R, Lo FY, El-Hashani E, Cervantes G, Robinson M, Hsu HS, Kales SC, Lipkowitz S, Karrison T, Sattler M, Vokes EE, Wang YC, Salgia R. CBL is frequently altered in lung cancers: its relationship to mutations in MET and EGFR tyrosine kinases. PLoS One. 2010;5:e8972. doi: 10.1371/journal.pone.0008972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Li Z, Dong T, Proschel C, Noble M. Chemically diverse toxicants converge on Fyn and c-Cbl to disrupt precursor cell function. PLoS Biol. 2007;5:e35. doi: 10.1371/journal.pbio.0050035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ferron SR, Pozo N, Laguna A, Aranda S, Porlan E, Moreno M, Fillat C, de la Luna S, Sanchez P, Arbones ML, Farinas I. Regulated segregation of kinase Dyrk1A during asymmetric neural stem cell division is critical for EGFR-mediated biased signaling. Cell Stem Cell. 2010;7:367–379. doi: 10.1016/j.stem.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 205.Doniz-Padilla L, Martinez-Jimenez V, Nino-Moreno P, Abud-Mendoza C, Hernandez-Castro B, Gonzalez-Amaro R, Layseca-Espinosa E, Baranda-Candido L. Expression and function of Cbl-b in T cells from patients with systemic lupus erythematosus, and detection of the 2126 A/G Cblb gene polymorphism in the Mexican mestizo population. Lupus. 2011;20:628–635. doi: 10.1177/0961203310394896. [DOI] [PubMed] [Google Scholar]
- 206.Yokoi N, Komeda K, Wang HY, Yano H, Kitada K, Saitoh Y, Seino Y, Yasuda K, Serikawa T, Seino S. Cblb is a major susceptibility gene for rat type 1 diabetes mellitus. Nat. Genet. 2002;31:391–394. doi: 10.1038/ng927. [DOI] [PubMed] [Google Scholar]
- 207.Yokoi N, Hayashi C, Fujiwara Y, Wang HY, Seino S. Genetic reconstitution of autoimmune type 1 diabetes with two major susceptibility genes in the rat. Diabetes. 2007;56:506–512. doi: 10.2337/db06-1027. [DOI] [PubMed] [Google Scholar]
- 208.Singh AJ, Meyer RD, Navruzbekov G, Shelke R, Duan L, Band H, Leeman SE, Rahimi N. A critical role for the E3-ligase activity of c-Cbl in VEGFR-2-mediated PLCgamma1 activation and angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5413–5418. doi: 10.1073/pnas.0700809104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sanjay A, Houghton A, Neff L, DiDomenico E, Bardelay C, Antoine E, Levy J, Gailit J, Bowtell D, Horne WC, Baron R. Cbl associates with Pyk2 and Src to regulate Src kinase activity, alpha(v)beta(3) integrin-mediated signaling, cell adhesion, and osteoclast motility. J. Cell Biol. 2001;152:181–195. doi: 10.1083/jcb.152.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wilhelmsen K, Burkhalter S, van der Geer P. C-Cbl binds the CSF-1 receptor at tyrosine 973, a novel phosphorylation site in the receptor's carboxy-terminus. Oncogene. 2002;21:1079–1089. doi: 10.1038/sj.onc.1205166. [DOI] [PubMed] [Google Scholar]
- 211.Jekely G, Sung HH, Luque CM, Rorth P. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev. Cell. 2005;9:197–207. doi: 10.1016/j.devcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 212.Wang Y, Werz C, Xu D, Chen Z, Li Y, Hafen E, Bergmann A. Drosophila cbl is essential for control of cell death and cell differentiation during eye development. PLoS One. 2008;3:e1447. doi: 10.1371/journal.pone.0001447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Qiao G, Li Z, Molinero L, Alegre ML, Ying H, Sun Z, Penninger JM, Zhang J. T-cell receptor-induced NF-kappaB activation is negatively regulated by E3 ubiquitin ligase Cbl-b. Mol. Cell. Biol. 2008;28:2470–2480. doi: 10.1128/MCB.01505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hirasaka K, Kohno S, Goto J, Furochi H, Mawatari K, Harada N, Hosaka T, Nakaya Y, Ishidoh K, Obata T, Ebina Y, Gu H, Takeda S, Kishi K, Nikawa T. Deficiency of Cbl-b gene enhances infiltration and activation of macrophages in adipose tissue and causes peripheral insulin resistance in mice. Diabetes. 2007;56:2511–2522. doi: 10.2337/db06-1768. [DOI] [PubMed] [Google Scholar]
- 215.Fiore F, Estebe B, Gibier P, Orsoni JC, Courbard JR, Chodosh LA, Birnbaum D, Delapeyriere O. Abnormal mammary gland development in MMTV-CBLC transgenic mouse. In Vivo. 2009;23:225–228. [PubMed] [Google Scholar]
- 216.Thien CB, Scaife RM, Papadimitriou JM, Murphy MA, Bowtell DD, Langdon WY. A mouse with a loss-of-function mutation in the c-Cbl TKB domain shows perturbed thymocyte signaling without enhancing the activity of the ZAP-70 tyrosine kinase. J. Exp. Med. 2003;197:503–513. doi: 10.1084/jem.20021498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Thien CB, Blystad FD, Zhan Y, Lew AM, Voigt V, Andoniou CE, Langdon WY. Loss of c-Cbl RING finger function results in high-intensity TCR signaling and thymic deletion. EMBO J. 2005;24:3807–3819. doi: 10.1038/sj.emboj.7600841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Adapala NS, Barbe MF, Langdon WY, Nakamura MC, Tsygankov AY, Sanjay A. The loss of Cbl-phosphatidylinositol 3-kinase interaction perturbs RANKL-mediated signaling, inhibiting bone resorption and promoting osteoclast survival. J. Biol. Chem. 2010;285:36745–36758. doi: 10.1074/jbc.M110.124628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Oksvold MP, Dagger SA, Thien CB, Langdon WY. The Cbl-b RING finger domain has a limited role in regulating inflammatory cytokine production by IgE-activated mast cells. Mol. Immunol. 2008;45:925–936. doi: 10.1016/j.molimm.2007.08.002. [DOI] [PubMed] [Google Scholar]