Abstract
The variation in several of the risk factors for osteoporotic fracture, including bone mineral density (BMD), has been shown to be strongly influenced by genetic differences. However, the genetic architecture of BMD is complex in both humans and in model organisms. We previously reported quantitative trait locus (QTL) results for BMD from a genome screen of 828 F2 progeny of Copenhagen and dark agouti rats. These progeny also provide an excellent opportunity to search for epistatic effects, or interaction between genetic loci, that contribute to fracture risk. Microsatellite marker data from a 20-cM genome screen was analyzed along with weight-adjusted bone density (DXA and pQCT) phenotypic data using the R/qtl software package. Genotype and phenotype data were permuted to determine genome-wide significance thresholds for the full model and epistasis (interaction) LOD scores corresponding to an alpha level of 0.01. A novel locus on chromosome 15 and a previously reported chromosome 14 QTL demonstrated a strong epistatic effect on BMD at the femur by DXA (LOD = 5.4). Two novel QTLs on chromosomes 2 and 12 were found to interact to affect total BMD at the femur midshaft by pQCT (LOD = 5.0). These results provide new information regarding the mode of action of previously identified QTL in the rat, as well as identifying novel loci that act in combination with known QTL or with other novel loci to contribute to BMD variation.
Introduction
Osteoporotic fractures at the hip and spine represent a major public health problem in developed countries (Anon 2000). Reduced bone mineral density (BMD) at one or more skeletal sites is the most widely used clinical indicator of fracture risk. BMD in humans and mammalian models follows a pattern of increase during growth and puberty, followed by a steady state after skeletal maturity (‘‘peak BMD’’), and then decline in later life. In humans, peak BMD is the primary determinant of osteoporotic fracture risk among older individuals, with high peak BMD levels providing protection against osteoporosis later on in life. Peak BMD is quite variable among individuals in the normal human population (Marshall et al. 1996) and is strongly influenced by heritable factors. However, Mendelian or major gene effects underlying this complex phenotype have not yet been identified (Peacock et al. 2002).
We previously reported results from a microsatellite genome screen for peak bone density phenotypes in the laboratory rat (Rattus norvegicus) using an F2 experimental design and employing two distinct crosses. The first cross involved the inbred Lewis (LEW) and Fischer 344 (F344) strains (Koller et al. 2005). More recently, we conducted an F2 quantitative trait locus (QTL) screen for BMD phenotypes in the Copenhagen (COP) and dark agouti (DA) rat strains (Koller et al. 2008b), and identified highly significant BMD QTL that appear to be unique to the COP/DA cross in addition to several QTLs shared in common with the F344/LEW F2 population.
In both the F344/LEW and COP/DA crosses, however, the sum of the QTL effects fell far short of the total heritable BMD variation observed among the F2 offspring. This led us to test whether additional factors such as gene-by-gene interaction, or epistasis, could be contributing to the observed complexity in the genetic architecture of peak BMD in the rat model. We demonstrated that the F344/LEW F2 population could be employed to detect pairs of genetic loci displaying evidence of epistasis and reported that a substantial portion of the variation in BMD phenotypes in the F344/LEW cross is due to epistatic interactions that we were able to detect in a QTL screen for epistasis (Koller et al. 2008a). The epistatic interactions detected in the F344/LEW cross involved both previously reported QTLs as well as novel loci that were detected only when epistasis was considered.
We have now undertaken to test the hypothesis that epistatic interactions also explain genetic variability in the COP/DA F2 animals that is not accounted for by the main-effect QTLs we have already reported. We expected that the additional genetic information regarding BMD variability would allow us to identify epistatic interactions involving both known and previously reported BMD QTLs in the COP/DA cross and that a subset of these interactions would overlap with the epistatic QTL pairs identified in the F344/LEW F2 cross.
Materials and methods
Animal breeding
Reciprocal mating of 12 breeding pairs of COP rats with DA rats was performed to first create an F1 population; then the 190 F1 rats were intercrossed to create 828 F2 offspring (405 males and 423 females). The rats were allowed to grow to 26 weeks of age, thereby attaining peak bone mineral density, before they were euthanized. Rat identities were recorded on data chips implanted subcutaneously and were verified using a scanner from Biomedic Data System (Seaford, DE). The rats were housed at Indiana University’s Laboratory Animal Resource Center (LARC), two rats per cage, and provided standard rat chow (NIH-31 Mouse/Rat diet 7017, Harlan Teklad, Madison, WI) and water ad libitum. After euthanasia, rat left femora and L3–5 vertebrae were removed and stripped of muscle and transferred to 70% ethyl alcohol at 4°C for densitometry analyses. The excised spleens were immediately stored in liquid nitrogen before transferring to −80°C. The procedures performed throughout the experiment followed the guidelines of the Indiana University Animal Care and Use Committee (IACUC).
Phenotypic measurements
In this study we employed computed tomography (CT) for measurement of true volumetric density (vBMD), as well as areal BMD (aBMD) obtained by DXA density measurement for direct comparison with the many human studies on which it has been used. To maintain maximal clinical relevance, we continued to focus on the same skeletal sites as in our previous reports, i.e., whole femur and lumbar (L3–5) spine for the aBMD measures, and midfemur, distal femur, and lumbar vertebrae for the vBMD measurements via CT.
Peripheral quantitative computed tomography (pQCT)
The left femurs were placed in plastic tubes filled with 70% ethyl alcohol and centered in the gantry of a Norland Stratec XCT Research SA + pQCT (Stratec Electronics, Pforzheim, Germany). Single slice measurements of 0.26-mm thickness and a voxel size of 0.07 mm were taken perpendicularly through the midfemoral shaft, distal femur, and the L5 vertebral body. For each slice, the X-ray source was rotated through 180° of projection. Total volumetric BMD (vBMD; mg/cm3) was measured using the XCT Research SA Plus software ver. 5.40. This is the bone mineral content divided by the total volume of the bone cross-section, including the marrow. The accuracy and repeatability of the pQCT measurements was confirmed using a method described previously (Koller et al. 2005).
Dual-energy X-ray absorptiometry (DXA)
The femur and L3–5 lumbar vertebrae were scanned using a fan-beam Hologic QDR 4500A DXA machine (Hologic, Bedford, MA) equipped with Hologic ver. 11.2:3 software. The machine was calibrated daily with an anthropomorphic spine phantom, as described previously. Repeatability of the aBMD measurements was assessed as described previously for pQCT (Koller et al. 2005).
DNA isolation and microsatellite marker genotyping
Genomic DNA was isolated from the individual rat spleen using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN, USA) according to the manufacturer’s protocol. Genotyping for each animal was accomplished using PCR with microsatellite markers (Research Genetics, Birmingham, AL, USA) previously shown to be polymorphic for COP and DA rats. The entire genome-wide screen (chromosomes 1–20, X) included 93 markers at an average interval of 20 cM and were analyzed using automated fluorescent microsatellite analysis. PCR products were sized on an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) using the Genotyper program ver. 3.6. All genotyping data were confirmed by two independent readers. Marker maps were generated using MAPMARKER/EXP with genotype data from all F2 animals (Lander et al. 1987). Marker order and distances were compared with those previously published Rat Genome Database (RGD) maps. This marker map was then used by the program R/qtl for epistasis analysis, with body weight and sex included as covariates for all QTL analyses (Broman et al. 2003).
Statistical analysis
A two-dimensional analysis of the genome screen marker data for epistatic interactions using the R/qtl software package (Broman et al. 2003) was performed to identify epistatic interactions contributing to variation in the six bone density measures: spine and femur aBMD by DXA, spine vBMD by CT, and midfemur, femur neck, and distal femur vBMD by CT. Each chromosomal region (rat chromosomes 1–19) was considered jointly with all other chromosomal regions throughout the genome, resulting in a genome-by-genome matrix of results for each BMD phenotype considered.
The statistical analysis of epistasis as implemented in the R/qtl software package consists of nested linear model-fitting for each pair of loci tested for an epistatic interaction, as described previously (Koller et al. 2008a). Briefly, two linear models are fitted to the F2 genotype and phenotype data. A full model (Eq. 1) contains both QTL main effects and a QTL interaction effect.
| (1) |
The full model is then compared with a model containing both QTLs but lacking the interaction term. This reduced model (Eq. 2) is
| (2) |
The statistical significance of each pair of loci is then evaluated via a likelihood ratio testing framework. Models (1) and (2) are fitted for each pair of QTLs for each BMD phenotype, and the logarithm of the odds (LOD) score for interaction is the difference in base-10 log-likelihood values between the two models. A large increase in log-likelihood for model (1) as opposed to model (2) indicates strong statistical evidence for an interaction effect.
To obtain appropriate genome-wide significance thresholds for the epistasis results and properly account for the large number of tests considered in the genome-by-genome scan, permutation tests (Doerge and Churchill 1996) were performed. Specifically, the five traits measured on each animal were randomly reassigned as a group across the 828 animals resulting in a permuted data set. By keeping all phenotypic data together, the underlying phenotypic correlations were preserved. For 5000 such permuted data sets, the epistatis analysis was then performed across the whole genome and the resulting maximum LOD scores for linkage for each phenotype were recorded. In this manner the LOD significance threshold for the 99th percentile of the maximum genome-wide interaction LOD scores across all phenotypes was found to be 4.5, and for the full model (1) it was found to be 7.9. Interacting QTL pairs were reported only if both of these thresholds were exceeded.
Effect size or relative impact of an epistatic interaction detected via the methods described above was estimated as the partial r2 associated with the QTL-by-QTL interaction term in model (1). This value corresponds to the proportion of phenotypic variability accounted for by the interaction after the main effects of both individual QTLs have been taken into account.
Results
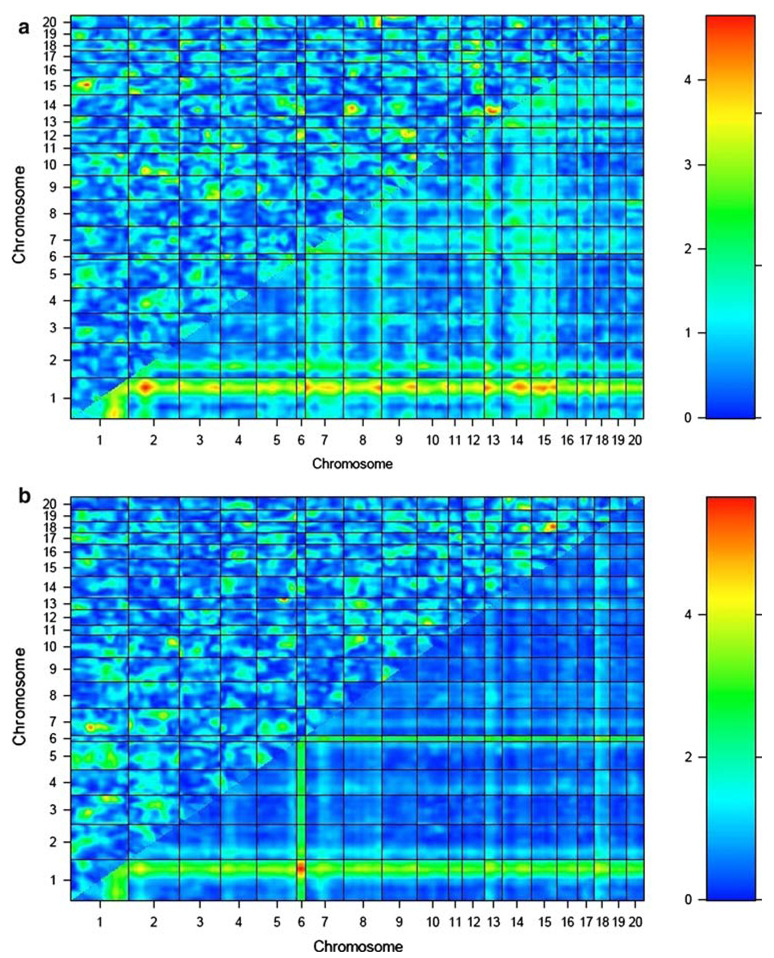
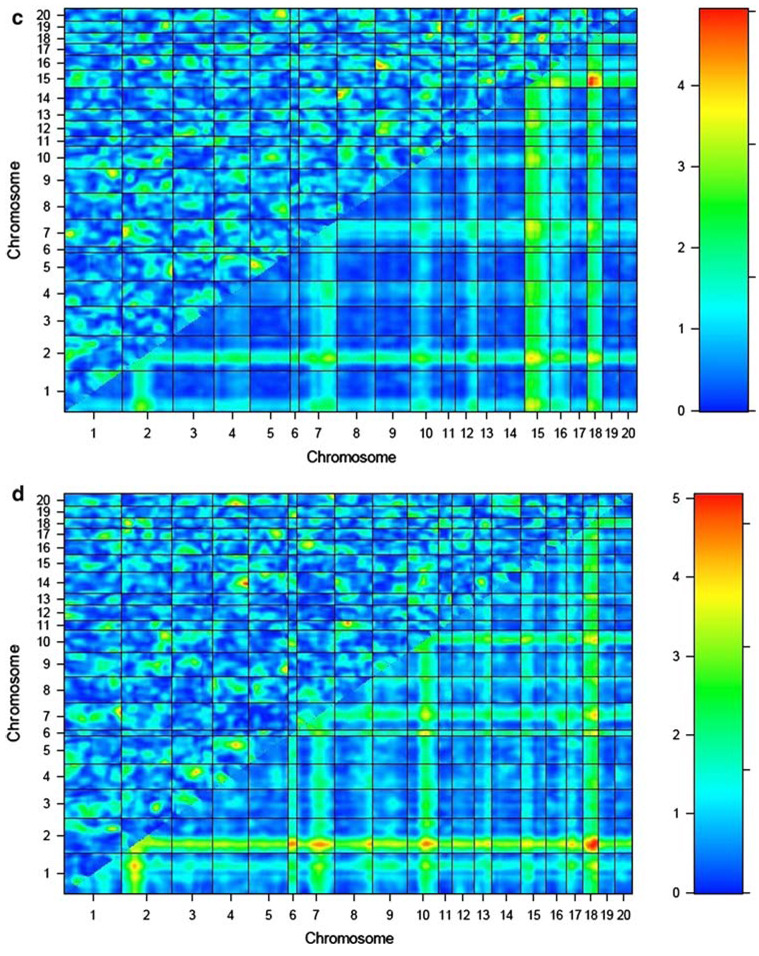
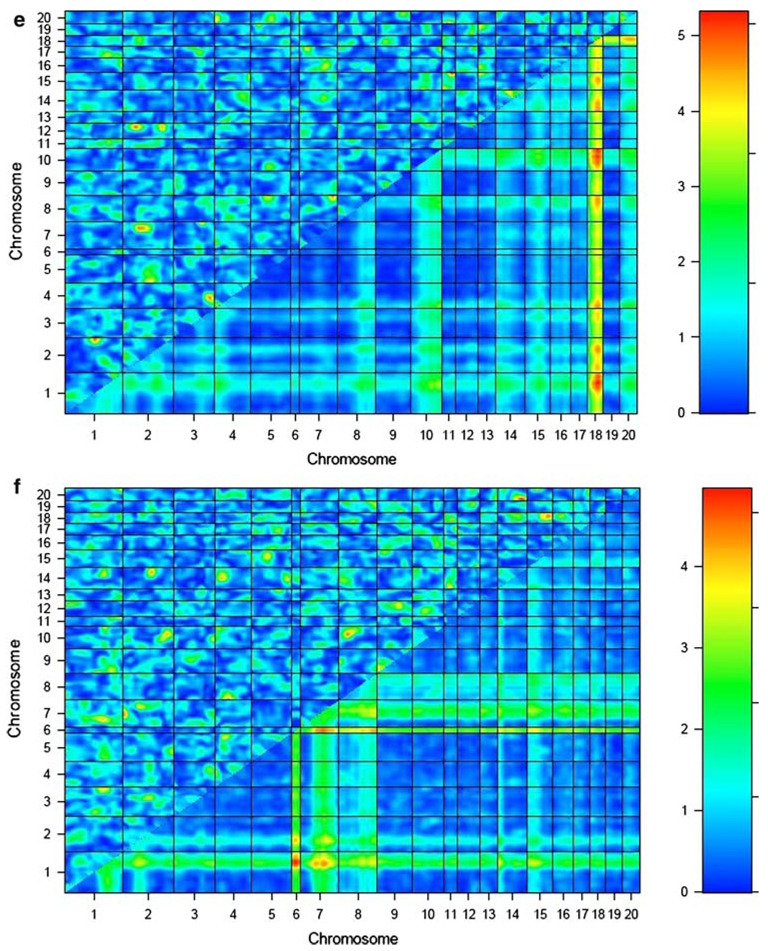
The results of the genome-by-genome epistasis analysis for each BMD phenotype in the COP/DA F2 rats are presented in Table 1 and Fig. 1a–f. This analysis revealed novel loci on chromosomes 2 and 12 interacting to influence vBMD at the femur midshaft (LODint = 5.0). This was the most significant interaction between novel QTLs identified over all analyzed BMD phenotypes. A distinct, previously reported chromosome 2 QTL also demonstrated evidence of epistasis with a previously detected QTL locus on chromosome 1 to impact vBMD variation at the femur midshaft (LODint = 5.0).
Table 1.
Interaction LOD scores for chromosome pairs and phenotypes significant at the genome-wide 0.01 level
| Phenotype | Chr | Position (cM) | Chr | Position (cM) | Full model | Additive model | Interaction LOD |
|---|---|---|---|---|---|---|---|
| Midshaft vBMD | 1 | 80.0 | 2 | 106.1 | 8.8 | 3.7 | 5.0 |
| Midshaft vBMD | 2 | 30.1 | 12 | 43.6 | 8.3 | 3.3 | 5.0 |
| Midshaft vBMD | 3 | 82.5 | 4 | 57.9 | 8.1 | 3.3 | 4.7 |
| Midshaft vBMD | 7 | 75.5 | 8 | 81.0 | 9.2 | 4.6 | 4.6 |
| Femur aBMD | 15 | 50.9 | 18 | 25.9 | 9.0 | 3.6 | 5.4 |
| Lumbar vBMD | 3 | 4.5 | 15 | 57.9 | 9.4 | 4.6 | 4.9 |
Fig. 1.
Epistasis plots for BMD phenotypes for COP/DA rat F2 offspring. Interaction effect is shown in the upper-left triangle in each plot, with color intensity corresponding to the LODint as indicated in the scale to the right of each plot. BMD phenotypes are as follows: a lumbar spine aBMD, b whole femur aBMD, c lumbar vBMD, d femur neck vBMD, e femur midshaft vBMD, and f distal femur vBMD
Strong evidence for epistatic interactions in the COP/DA cross is obtained with both novel QTLs and with loci detected in the main-effect QTL screen. A novel locus on chromosome 15 interacts with a previously reported COP/DA QTL for femur aBMD on chromosome 18 to influence the same phenotype (LODint = 5.4). This was the highest level of statistical significance obtained in our epistasis screen.
Several of the locus pairs below our threshold for significance also demonstrate other effects on BMD at other skeletal sites, suggesting pleiotropic effects of some of the interactions. The pair of QTLs on chromosomes 14 and 20 contributing to vBMD at the distal femur (LODint = 5.0) also had a significant effect on aBMD at the spine (LODint = 4.6) and vBMD at the spine (LODint = 4.9). The pair of loci on chromosomes 11 and 20 influencing lumbar vBMD also exerted an effect on vBMD at the femoral midshaft (LODint = 3.5). The pair of loci on chromosomes 4 and 14 influencing neck vBMD (LODint = 4.8) also exerted pleiotropic effects on vBMD at two other skeletal sites, lumbar aBMD (LODint = 3.6) and femur midshaft (LODint = 3.5).
Effect sizes of the epistatic interactions reported here were estimated as the partial r2 associated with the interaction term in model (1). The effect sizes for the interactions in Table 1 ranged from 0.019 (1.9% of total phenotypic variability), for the epistatic effect of chromosomes 2 and 12 on femur midshaft vBMD, to 0.046 (4.6%) for the interaction of chromosomes 15 and 18 impacting femur aBMD. The summed proportion of total phenotypic variability explained by the interactions reported in Table 1 ranges from 0.060 (6.0%) for lumbar aBMD to 0.150 (15.0%) for femur aBMD.
Discussion
We have performed a genome-wide epistasis screen in the COP/DA rat cross for BMD phenotypes and obtained evidence for multiple highly significant QTL pairs. Several of these QTL pairs involve loci that were previously detected in our published genome screen of COP/DA and F344/LEW F2 crosses, but many of the loci did not demonstrate significant effects on their own and thus were not detected previously. Some of the QTLs appear to have pleiotropic effects on BMD at both the spine and the femur. A minority of the pairs of epistatic loci detected in the COP/DA F2 cross, notably the interaction between QTLs on chromosomes 1 and 2 impacting variation in midfemur BMD, were also detected in the F344/LEW cross.
Several of the QTLs identified in our epistasis screen are located in rat chromosomal regions that are in fact syntenic to human chromosomal regions containing BMD QTLs that have been reported previously by our group and others. The novel distal chromosome 15 QTL that was found to act with chromosome 18 loci to influence whole-femur aBMD in female rats shares homology with human chromosome 1p, which has been implicated in BMD variation by other investigators (Karasik et al. 2002; Wilson et al. 2003). These findings exemplify an advantage of interaction analyses in animal models of complex phenotypes such as BMD, namely, that genes which are not detectable in the primary screen are able to reach significance in the epistasis screen, implicating human genomic regions via synteny that otherwise would likely have been passed over.
We have identified several novel pairs of QTLs that interact to contribute to variation in BMD in the COP/DA rat F2 cross. Identification of epistatic pairs of loci contribution to BMD variation in the rat represents a step forward in the delineation of the genetic architecture of these phenotypes in the rat species and provides a powerful approach to identify novel candidate genes and chromosomal regions for further pursuit in human studies. Our results, however, also illustrate the degree of complexity of the genetic architecture of these phenotypes. Together with the main effects of the QTLs identified previously (Koller et al. 2008b), the epistatic interactions reported here explain only 50–60% of the phenotypic variability in the BMD phenotypes that we have studied, all of which have estimated heritabilities substantially greater than what we have accounted for thus far. The remaining genetic variability is likely due to higher-order epistatic effects (interaction between three or more loci) and interactions between genes and environmental factors, which will require larger samples, sophisticated study designs, and additional phenotyping to detect.
Acknowledgments
This work was supported by the U.S. National Institutes of Health through grants R01AR047822 (CHT) and P01AG018397 (CHT, DLK, TF, MJE).
Contributor Information
Daniel L. Koller, Email: dkoller@iupui.edu, Department of Medical and Molecular Genetics, Indiana University Purdue University Indianapolis, Indianapolis, IN 46202, USA; Department of Medical and Molecular Genetics, Indiana University School of Medicine, 410 W. 10th St., HS 4023, Indianapolis, IN 46202, USA.
Lixiang Liu, Department of Medical and Molecular Genetics, Indiana University Purdue University Indianapolis, Indianapolis, IN 46202, USA.
Imranul Alam, Department of Biomedical Engineering, Indiana University Purdue University Indianapolis, Indianapolis, IN 46202, USA.
Qiwei Sun, Department of Biomedical Engineering, Indiana University Purdue University Indianapolis, Indianapolis, IN 46202, USA.
Michael J. Econs, Department of Medical and Molecular Genetics, Indiana University Purdue University Indianapolis, Indianapolis, IN 46202, USA Department of Medicine, Indiana University Purdue University Indianapolis, Indianapolis, IN 46202, USA.
Tatiana Foroud, Department of Medical and Molecular Genetics, Indiana University Purdue University Indianapolis, Indianapolis, IN 46202, USA.
Charles H. Turner, Department of Biomedical Engineering, Indiana University Purdue University Indianapolis, Indianapolis, IN 46202, USA Department of Orthopaedic Surgery, Indiana University Purdue University Indianapolis, Indianapolis, IN 46202, USA.
References
- Anon Osteoporosis prevention, diagnosis, and therapy. NIH Consensus Statement. 2000;17:1–36. [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasik D, Myers RH, Cupples LA, Hannan MT, Gagnon DR, et al. Genome screen for quantitative trait loci contributing to normal variation in bone mineral density: the Framingham Study. J Bone Miner Res. 2002;17:1718–1727. doi: 10.1359/jbmr.2002.17.9.1718. [DOI] [PubMed] [Google Scholar]
- Koller DL, Alam I, Sun Q, Liu L, Fishburn T, et al. Genome screen for bone mineral density phenotypes in Fisher 344 and Lewis rat strains. Mamm Genome. 2005;16:578–586. doi: 10.1007/s00335-004-2459-0. [DOI] [PubMed] [Google Scholar]
- Koller DL, Liu L, Alam I, Sun Q, Econs MJ, et al. Epistatic effects contribute to variation in BMD in Fischer 344 × Lewis F2 rats. J Bone Miner Res. 2008a;23:41–47. doi: 10.1359/JBMR.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller DL, Liu L, Alam I, Sun Q, Econs MJ, et al. Linkage screen for BMD phenotypes in male and female COP and DA rat strains. J Bone Miner Res. 2008b;23:1382–1388. doi: 10.1359/JBMR.080401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Marshall D, Johnell O, Wedel H. Meta-analysis of how well measures of bone mineral density predict occurrence of osteoporotic fractures. BMJ. 1996;312:1254–1259. doi: 10.1136/bmj.312.7041.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23:303–326. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- Wilson SG, Reed PW, Bansal A, Chiano M, Lindersson M, et al. Comparison of genome screens for two independent cohorts provides replication of suggestive linkage of bone mineral density to 3p21 and 1p36. Am J Hum Genet. 2003;72:144–155. doi: 10.1086/345819. [DOI] [PMC free article] [PubMed] [Google Scholar]