Abstract
The vast majority of Mycobacterium tuberculosis (M. tuberculosis) infected individuals are protected from developing tuberculosis and T cells are centrally involved in this process. MicroRNAs (miRNA) regulate T-cell functions and are biomarker candidates of disease susceptibility and treatment efficacy in M. tuberculosis infection. We determined the expression profile of 29 selected miRNAs in CD4+ T cells from tuberculosis patients and contacts with latent M. tuberculosis infection (LTBI). These analyses showed lower expression of miR-21, miR-26a, miR-29a, and miR-142-3p in CD4+ T cells from tuberculosis patients. Whole blood miRNA candidate analyses verified decreased expression of miR-26a, miR-29a, and miR-142-3p in children with tuberculosis as compared to healthy children with LTBI. Despite marked variances between individual donor samples, trends of increased miRNA candidate expression during treatment and recovery were observed. Functional in vitro analysis identified increased miR-21 and decreased miR-26a expression after re-stimulation of T cells. In vitro polarized Interleukin-17 positive T-cell clones showed activation-dependent miR-29a up-regulation. In order to characterize the role of miR-29a (a described suppressor of Interferon-γ in tuberculosis), we analyzed M. tuberculosis specific Interferon-γ expressing T cells in children with tuberculosis and healthy contacts but detected no correlation between miR-29a and Interferon-γ expression. Suppression of miR-29a in primary human T cells by antagomirs indicated no effect on Interferon-γ expression after in vitro activation. Finally, classification of miRNA targets revealed only a moderate overlap between the candidates. This may reflect differential roles of miR-21, miR-26a, miR-29a, and miR-142-3p in T-cell immunity against M. tuberculosis infection and disease.
Introduction
Approximately one third of humankind is infected with Mycobacterium tuberculosis (M. tuberculosis), the causative agent of human tuberculosis. Specific cellular immunity, especially CD4+ T cells, is capable of containing the pathogen in the vast majority of M. tuberculosis-infected individuals. These latently M. tuberculosis-infected individuals (LTBI) do not develop clinical symptoms of tuberculosis after infection, but remain at increased risk of developing active disease especially during the first years after M. tuberculosis infection. The exact mechanisms of how CD4+ T cells prevent the development of active disease remain elusive.
Animal models and human genetic studies point towards a crucial role of IFNγ expressing T helper type (TH) 1 cells in protective host immunity against tuberculosis [1]. Recent studies showed that ‘polyfunctional’ T cells characterized by the expression of multiple cytokine (i.e. IFNγ, TNFα, IL-2) were protective [2], [3] whereas single-cytokine (i.e. either TNFα ορ ΙΦΝγ) expressing TH1 cells were pathognomonic [4], [5]. Other T-cell subpopulations, like Interleukin-17 producing TH17 cells, may also influence disease susceptibility in tuberculosis, but previous studies revealed contrary results [6]–[11] Thus the quantity of a single T-cell cytokine, e.g. IFNγ, is not sufficient as a marker for protection against tuberculosis.
Little is known about the role of MicroRNAs (miRNAs) in containing M. tuberculosis infection and preventing disease progression. MiRNAs have been shown to fine-regulate target genes in eukaryotic cells, including immune cells, by modulating protein expression at the posttranscriptional level [12]. The maturation of miRNAs into functional regulators comprises three main steps: I) processing of the primary-miRNA transcripts (pri-miRNA) into the precursor miRNA (pre-miRNA) by the nuclear RNase III enzymes Drosha and DGRC8, II) export into the cytosol and subsequent processing into 22 bp duplexes by the cytosolic RNase III enzyme DICER and III) loading of the mature miRNA into the RNA-induced silencing complex (RISC), where it binds to the 3′-untranslated region (3′-UTR) of the target mRNA. MiRNAs exert their functions mainly by degradation of the target mRNA or inhibition of translation (reviewed in [13]). They play important roles in many physiological processes including the development and regulation of the immune system (reviewed in [14]). In T cells, miRNAs have already been shown to regulate several processes e.g. cytokine expression, polarization of distinct T-cell lineages, homeostasis of T cells, and T-cell receptor signaling [15]–[20]. Indications of immune pathological effects of miRNA-mediated regulation come from malignancies and autoimmune diseases (reviewed in [14]), but the role of miRNAs in chronic infectious diseases like tuberculosis is hardly defined. Recent studies identified miRNA (miR)-29 as a central non-redundant suppressor of IFNγ [21] and increased miR-29 expression promoted susceptibility against mycobacterial infections [22].
The aim of this study was to identify and characterize miRNAs involved in immunity against tuberculosis. For this purpose we initially compared expression of 29 pre-selected immune-related miRNAs in CD4+ T cells of tuberculosis patients, healthy LTBI, and non-M. tuberculosis infected individuals (PPDneg). A follow-up study was then performed to determine the expression of candidate miRNAs in whole blood of children with tuberculosis and healthy contacts.
Results
Identification of Differentially Expressed miRNAs in CD4+ T Cells of Tuberculosis Patients and LTBI
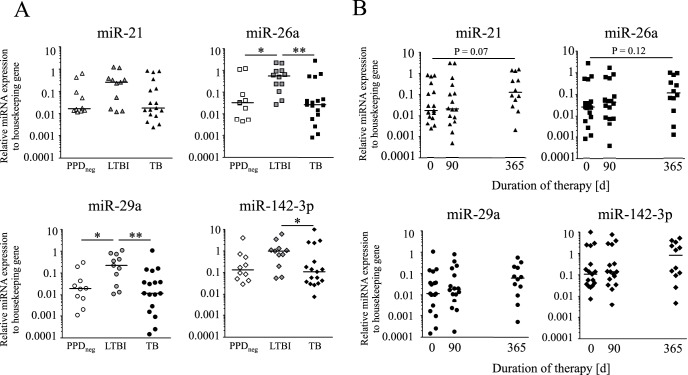
We compared the expression of 29 miRNAs in CD4+ T cells from peripheral blood of tuberculosis patients, LTBI, and non-infected control donors (PPDneg). MiRNAs were selected according to expression of T cells [23], [24]. Analyses were restricted to CD4+ T cells to avoid confounding influences of cellular heterogeneity on RNA expression [25]. For these analyses acute tuberculosis patients on diagnosis (i. e. prior to onset of chemotherapy) were included. 17 of 29 miRNAs were detectable according to defined criteria [for details see material and methods]. Median expression levels and standard deviations of all determined miRNAs are shown in Table 1. Four miRNAs, namely miR-21, miR-26a, miR-29a, and miR-142-3p, were differentially expressed between tuberculosis patients and LTBI (P = 0.035, P = 0.005, P = 0.008, and P = 0.002, respectively), whereas no differences were detected between LTBI and PPDneg (Table 1 and Figure 1A). Differentially expressed miRNAs showed decreased expression in CD4+ T cells from tuberculosis patients as compared to LTBI and PPDneg. Notably the level of differentially expressed miRNAs correlated markedly in T cells from individual donors (miR-21/miR-26a, P = 0.001; miR-21/miR-29a, P = 0.002; miR-21/miR-142-3p P = 0.001; miR-26a/miR-29a P<0.0001; miR-26a/miR-142-3p P<0.0001; miR-29a/miR-142-3p P<0.0001) (data not shown). This rendered a common cause of decreased miR-21, miR-26a, miR-29a, and miR-142-3p of CD4+ T cells from tuberculosis patients likely.
Table 1. Median Expression of miRNAs in PPDneg donors, LTBI and tuberculosis patients.
| PPDneg | LTBI | Tuberculosis | ||||
| Median | SD | Median | SD | Median | SD | |
| let-7a | 0,0008 | 0,0006 | nd | nd | nd | Nd |
| miR-15a | 0,0005 | 0,0003 | 0,0003 | 0,0013 | 0,0003 | 0,0008 |
| miR-17-3p | 0,1401 | 0,0311 | 0,1259 | 0,0403 | 0,071 | 0,02 |
| miR-21 | 0,0393 | 0,0161 | 0,0438 | 0,0252 | 0,0121 | 0,0047 |
| miR-23a | 0,00005 | 0,00005 | nd | nd | nd | Nd |
| miR-25 | 0,0032 | 0,0005 | 0,0027 | 0,0083 | 0,0015 | 0,0027 |
| miR-26a | 0,0808 | 0,0409 | 0,0928 | 0,0411 | 0,0148 | 0,0113 |
| miR-27a | 0,0016 | 0,001 | 0,0025 | 0,0054 | nd | Nd |
| miR-29a | 0,0687 | 0,0122 | 0,0815 | 0,0467 | 0,0233 | 0,0137 |
| miR-30a* | 0,728 | 0,0443 | 0,07 | 0,0294 | 0,036 | 0,0982 |
| miR-98 | nd | nd | nd | nd | nd | Nd |
| miR-99b | 0,0003 | 0,0001 | nd | nd | nd | Nd |
| miR-101 | 0,0008 | 0,0004 | 0,0014 | 0,001 | nd | Nd |
| miR-106a | 0,1478 | 0,0215 | 0,1185 | 0,0455 | 0,08 | 0,0222 |
| miR-106b | 0,0047 | 0,0011 | 0,003 | 0,0016 | 0,0018 | 0,0028 |
| miR-122 | nd | nd | nd | nd | nd | Nd |
| miR-126 | 0,0022 | 0,0007 | 0,0011 | 0,0013 | 0,0023 | 0,0055 |
| miR-142-3p | 0,8261 | 0,3628 | 0,6404 | 0,417 | 0,1 | 0,0909 |
| miR-143 | nd | nd | nd | nd | nd | Nd |
| miR-144* | nd | nd | nd | nd | nd | Nd |
| miR-146a | 0,3294 | 0,0417 | 0,3099 | 0,1966 | 0,2069 | 0,1075 |
| miR-150 | 2,459 | 0,7761 | 3,8087 | 1,6389 | 2,0281 | 1,3939 |
| miR-155* | nd | nd | nd | nd | nd | Nd |
| miR-181a-1 | 0,001 | 0,0003 | 0,0007 | 0,0009 | nd | Nd |
| miR-191 | 0,1224 | 0,366 | 0,0691 | 0,022 | 0,0558 | 0,0526 |
| miR-203 | nd | nd | nd | nd | nd | Nd |
| miR-218 | nd | nd | nd | nd | nd | Nd |
| miR-223 | 0,8505 | 0,7369 | 0,1425 | 0,3888 | 0,1249 | 0,8014 |
| miR-455-5p | nd | nd | nd | nd | nd | Nd |
| RNU481 | na | na | na | Na | ||
control; nd: miRNA not detectable in at least two donors.
Figure 1. Four miRNAs are differentially expressed in CD4+ T cells from tuberculosis patients and LTBI.

Expression of miR-21 (triangles), miR-26a (squares), miR-29a (circles), and miR-142-3p (diamonds) in CD4+ T cells from tuberculosis patients (TB) (black symbols, n = 6), LTBI (grey symbols, n = 7), and PPD negative healthy controls (PPDneg) (open symbols, n = 3). Median expression of candidate miRNAs relative to the housekeeping gene RNU48 is shown. Significant differences are indicated as asterisks (* for p<0.05; ** for p<0.01, Mann-Whitney U-test).
Differential Expression of Candidate miRNAs in Blood of Children with Tuberculosis, LTBI, and PPDneg
Limited donors numbers included for CD4+ T-cell miRNA expression analyses prompted us to perform a follow-up case/control study in children with tuberculosis, LTBI, and PPDneg contacts. Here isolation of CD4+ T cells was not feasible because of the limited sample volume. Therefore miRNA candidate expression in whole blood was determined. Although the expression of miRNAs was heterogeneous between individuals, we detected markedly lower expression of miR-26a (P<0.01), miR-29a (P<0.01), and miR-142-3p (P<0.05) in children with tuberculosis as compared to children with LTBI (Figure 2A). For miR-21 the same trend was identified without reaching significant levels. Notably, miR-21, miR-26a, miR-29a, and miR-142-3p expression levels were comparable between children with tuberculosis and PPDneg children. MiR-26a and miR-29a were lower (both P<0.05) in PPDneg as compared to children with LTBI (Figure 2A). Time course analyses of CD4+ T cells from selected tuberculosis patients revealed markedly increased miRNA candidate expression during the first months of chemotherapy (data not shown). Therefore we determined miRNA expression in children with tuberculosis also during treatment (3 months after onset of therapy) and after recovery (12 months after onset of therapy) (Figure 2B). We identified trends of increased miRNA candidates expression (P = 0.07 for miR-21; P = 0.12 for miR-26a) after recovery as compared to active disease (Figure 2B). We concluded that miRNA candidate expression in peripheral blood of children largely reflected differences between CD4+ T cells. Next we analyzed a possible role of miRNA candidates during T-cell activation and differentiation.
Figure 2. MiRNA candidate expression of peripheral blood cells from children with tuberculosis and LTBI.
Expression of miR-21 (triangles), miR-26a (squares), miR-29a (circles), and miR-142-3p (diamonds) in whole blood is shown for children with tuberculosis (TB, black symbols), healthy latently M. tuberculosis children (LTBI, grey symbols), and PPD negative contacts (PPDneg, open symbols) (A) as well as for children with tuberculosis under therapy and recovery (B). Each symbol indicates miRNA candidate expression for an individual donor relative to ‘housekeeping’ control RNU48. Significant differences are indicated as asterisks (* for P<0.05; ** for P<0.01, Mann-Whitney U-test). Exact P-values (Mann-Whitney U-test) are indicated for tendencies.
Expression of miR-21, miR-26a, miR-29a and miR-142-3p after in vitro Re-stimulation of Naïve T Cells
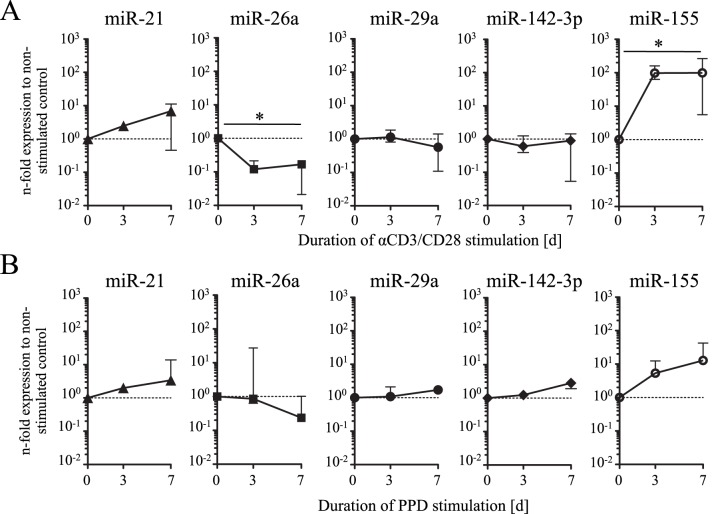
In an initial attempt to determine the role of candidate miRNAs in T-cell activation and differentiation, we determined expression of miR-21, 26a, 29a, 142-3p after in vitro restimulation. Naïve CD4+ T cells were stimulated with crosslinking αCD3/CD28 antibodies in vitro and expression of the miRNA candidates was determined. As a control we used miR-155 that is described to be up-regulated upon T-cell receptor-specific stimulation [26]. Following αCD3/CD28 stimulation expression of this control miRNA was increased up to 100-fold already on day three (P = 0.02) (Figure 3A). MiR-21 showed a tendency of increased expression on stimulation (up to 10-fold seven days post stimulation) (P = 0.1) (Figure 3A). MiR-26a expression decreased until day three post stimulation and remained relatively stable until day seven (P = 0.02) (Figure 3). MiR-29a and miR-142-3p showed only moderate expression differences with miR-142-3p being slightly decreased (Figure 3). Notably, similar changes in miR-21 expression were detected after PBMC stimulation from LTBI with M. tuberculosis-specific antigen PPD (Figure 3B). A trend of decreased expression was also detected for miR-26a on day seven (Figure 3B). We concluded that miR-21 expression increased upon activation of naïve CD4+ T cells in vitro whereas miR-26a decreased upon T-cell activation in vitro.
Figure 3. Candidate miRNA expression after activation of naïve T cells and PPD-specific PBMC stimulation.
MiRNA expression was analyzed three and seven days after in vitro activation of naïve T cells from healthy donors (n = 4) with αCD3/CD28 and for PBMCs of LTBI (n = 3) stimulated with M. tuberculosis purified protein derivative (PPD). Expression of the miRNAs was normalized to the housekeeping gene RNU48 and is shown relative to the respective non-stimulated control (dotted line). Median expression and range is shown. Significant differences are indicated as asterisks (* for P<0.05, Kruskal-Wallis test).
Expression of miR-21, miR-26a, miR-29a and miR-142-3p in Human TH1 and TH17 Clones
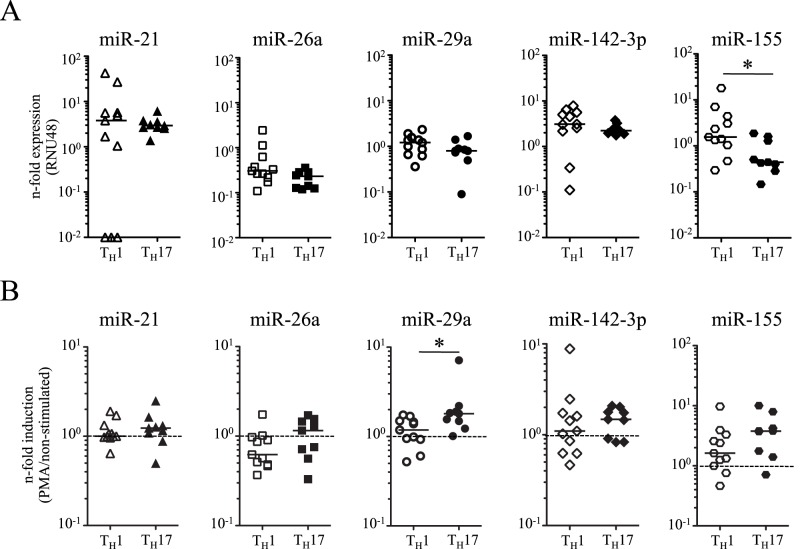
IFNγ-secreting TH1 cells are crucial for protective immunity against M. tuberculosis infection [27]–[29]. Polarization of T cells towards functional distinct TH-subsets (e.g. TH17) may affect susceptibility and protection against tuberculosis. Consequently we analyzed expression of candidate miRNAs in polarized T cell clones. T cell cloning (for details see Methods) resulted in IFNγ-expressing TH1 and IL-17-expressing TH17 clones. Comparison of miRNA-expression between resting TH1 and TH17 clones revealed no differences for candidate miRNAs (Figure 4A), whereas miR-155 expression was increased in TH1 clones (P = 0.03) (Figure 4A). Notably, activation of T-cell clones with PMA/Ionomycin increased miR-29a expression of TH17 clones (P = 0.01) but not of TH1 clones (Figure 4B). No differences were detected for miR-21, miR-26a, miR-142-3p and miR-155 upon activation (Figure 4B).
Figure 4. Expression of candidate miRNAs in TH1 and TH17 clones.
Expression of miR-21 (triangles), miR-26a (squares), miR-29a (circles), miR-142-3p (diamonds), and miR-155 (hexagons) was compared between IFNγ-expressing TH1 (open symbols, n = 11) and IL-17-expressing TH17 clones (black symbols, n = 9) of healthy donors prior to (A) and after in vitro activation (B). Expression of candidate miRNAs was normalized to the housekeeping gene RNU48. Median expression and range is shown. Significant differences are indicated as asterisks (* for P<0.05, Mann-Whitney-U test). (B) MiRNA expression after 12 h in vitro re-stimulation with PMA/Ionomycin. Relative expression to the respective non-activated control is shown (dotted line).
Similar IFNγ Expression in CD4+ T Cells from Children with Tuberculosis and LTBI and No Correlation between IFNγ and miRNA Candidate Expression Level
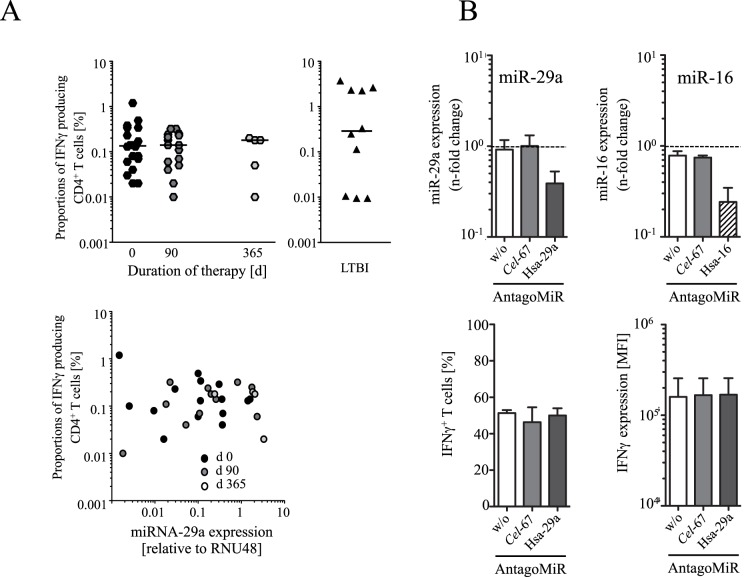
IFNγ has recently been identified as a target of miR-29a [21], [22]. Since miR-29a was differentially expressed between children with tuberculosis and LTBI, we compared proportions of IFNγ expressing T cells between these study groups and in children with tuberculosis during treatment and recovery. IFNγ expression was comparable between children with tuberculosis and LTBI after in vitro re-stimulation with M. tuberculosis-specific PPD (Figure 5A, upper graphs) and also for stimulation with staphylococcal superantigen SEB (data not shown). No differences were detected when comparing IFNγ expression between different time points of children with tuberculosis under chemotherapy and recovery (Figure 5A, upper left graph). Since IFNγ and miRNA expression varied between individuals we also compared expression of IFNγ and miR-29a in individual children with tuberculosis under chemotherapy and after recovery. No dependency between miR-29a and IFNγ expression was detected (R2 = 0.03; P = 0.36) (Figure 5A, lower graph).
Figure 5. IFNγ and miR-29a expression of infected children and ectopic miR-29a suppression in T cells.
(A) IFNγ expression after short-term in vitro re-stimulation of PBMC from children with tuberculosis (upper left graph) prior to chemotherapy (black symbols), three months under chemotherapy (grey symbols), after recovery (open symbols), as well as children with LTBI (upper right graph) with M. tuberculosis-specific PPD is shown. Proportions of IFNγ expressing CD4+ T cells (y-axis) for different time points after treatment onset of patients (x-axis) are depicted. Each symbol represents data from an individual patient with tuberculosis (hexagon) or LTBI (filled trigon). Ex vivo whole blood miR-29a expression (x-axis) in comparison to the proportions of IFNγ expressing CD4+ T cells (x-axis) for individual children with TB under therapy (circles) is shown in the lower graph. (B) Modulation of miR-29a expression (upper left graph) and control miR-16 (upper right graph) of CD4+ T cells by specific antagomirs (Homo sapiens (Hsa)-29a, Hsa-16, and Caenorhabditis elegans (Cel)-67) and effects on IFNγ expression (lower graphs) are depicted. IFNγ expression of activated CD4+ T cells after miR-29a or miR-16 suppression is shown as proportions of IFNγ+ cells (lower left graph) or as the IFNγ expression level per cell (indicated by median fluorescence intensities (MFI)) (lower right graph).
Modulation of miR-29a Expression in Primary Human CD4+ T Cells
Results of ex vivo miR-29a and in vitro IFNγ analyses may be confounded by several factors. Therefore, to validate these experiments, we modulated miR-29a expression in primary human T cells. Endogenous miR-29a expression was suppressed using specific antagomir assays (for details see methods). Antagomirs against human miR-16 and C. elegans miR-67 have been used as controls in these experiments. Both antagomirs against human miR-16 and miR-29a down-regulated respective targets (miR16, mean 70%; miR-29a, mean 58%) in CD4+ T cells, whereas negative control C. elegans miR-67 had no effects (Figure 5B, upper graphs). Activation of T cells using PMA/Ionomycin in vitro stimulation induced about 50% IFNγ producing T cells (Figure 5B, lower left graph). Notably decreased miR-29a expression did not affect the proportions of IFNγ producing T cells (Figure 5B, lower left graph). Also the cellular protein expression level of IFNγ (determined by single cell median fluorescence analysis) was not influenced by miR-29a suppression (Figure 5B, lower right graph). The same hold true for TNFα producing T cells in this assay (data not shown). We concluded that suppression of miR-29a did not affect IFNγ expression in our assay and therefore other target genes maybe regulated by differential miRNA expression. Especially common targets of the examined miRNAs maybe of interest in this context.
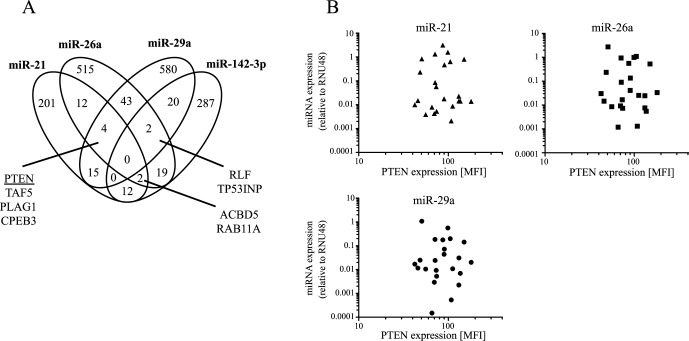
Common Immunological Targets of miRNAs and PTEN Protein Evaluation Compared to miR-21, 26a, and 29a Expression
Predicted miR-21, miR-26a, miR29a, and miR-142-3p targets from the miRGen Internet platform were determined for expression in immunological tissues using Unigene Internet database (for details see Materials and Methods). The predicted candidate miRNA targets expressed in immunological tissues (i.e. 201 genes for miR-21; 287 genes for miR-142-3p; 515 genes for miR-26a; and 580 genes for miR-29a) were then analyzed for overlapping target groups. Figure 6A shows a venn diagram of overlapping miRNA targets. The overall overlap of target genes between the candidate miRNAs was small and not a single gene was predicted target of all four miRNAs. One common target predicted for miR-21, miR-26a, and miR-29a was PTEN, a regulatory protein involved in T cell receptor signaling [30]. Therefore ex vivo PTEN protein expression in lymphocytes of children with tuberculosis and contacts was compared. We found no evidence for a correlation of miR-21 (Figure 6B; upper left graph), miR-26a (Figure 6B; upper right graph), or miR-29a (Figure 6B; lower graph) with PTEN protein levels. Ongoing studies aim at validating additional common targets of miRNA candidates to claryfy the role of miR-21, miR-26a, miR-29a, and miR-142-3p expression in tuberculosis.
Figure 6. Target genes and overlaps for miR-21, miR-26a, miR-29a and miR-142-3p.
(A) A Venn diagram indicates the overlap of target genes for miR-21, miR-26a, miR-29a and miR-142-3p. Common targets of at least three candidate miRNAs are listed by name. (B) Comparison between expression miR-21 (upper left graph), miR-26a (upper right graph), miR-29a (lower graph) and protein expression of the common target PTEN in lymphocytes from children with tuberculosis (n = 19) and LTBI (n = 3) is shown. We indicate relative miRNA expression to house keeping gene RNU48 by quantitative PCR and compare this to PTEN protein expression determined by flow cytometry using median fluorescence intensity (MFI) analysis. Each symbol represents data from an individual donor.
Discussion
MiRNAs are involved in the regulation of T-cell immunity and failure may lead to malignancies or autoimmune diseases. The present study provided evidence for a role of miR-21, miR-26a, miR-29a, and miR-142-3p in the immune response against human tuberculosis, a chronic infectious disease. The candidate miRNAs identified in this study were expressed at lower levels in T cells of tuberculosis patients and increased during chemotherapy in individual donors. Analyses of miRNA candidates in whole blood of children with tuberculosis and contacts verified lower miR-26, miR-29a, and miR-142-3p expression in children with tuberculosis. MiR-21 and miR-26a were shown to be regulated during T-cell activation. Initial experiments to modulate miR-29a expression in primary T cells did not suggest a suppressive role of miR-29a on IFNγ regulation.
Host immunity against M. tuberculosis is complex and different innate and adaptive immune components are crucial for protection against disease [1]. Because of the unambiguous crucial role of CD4+ T cells, we initially focused on this population for identification of candidate miRNAs in the present study. The expression of miR-21, miR-26a, miR-29a, and miR-142-3p was homogeneous within the study groups and already moderate numbers of tuberculosis patients, LTBI, and PPDneg were sufficient to detect significant differences. In addition, the level of differentially expressed miRNAs of tuberculosis patients and controls correlated significantly and therefore, common regulation for miR21, miR-26a, miR-29a, and miR-142 was likely. Previous studies identified key genes for microRNA regulation and a central factor was c-myc [31]. The oncogene c-myc is a transcription factor that regulates several genes and recently it became clear that c-myc also dramatically reprograms miRNA expression [31]. Besides regulating the important miR-17-92 cluster, c-myc has also been shown to suppress miR-26a and miR-29 family members [32], [33]. The related transcription factor n-myc has been identified as a suppressor of miR-21 [34]. Previously we showed that c-myc is up-regulated in T cells from tuberculosis patients prior to therapy [35]. It is tempting to speculate that c-myc is part of the mechanisms leading to concordant down-regulation of miR-26a, miR-29a (and probably miR-21). It remains elusive whether miR-142-3p is also regulated by c-myc. Notably the promotor of c-myc has been identified as a target of nucleoside diphosphate kinase secreted by M. tuberculosis [36]. Future studies will address the question if pathognomonic differences in host miRNA expression were caused by direct interaction of M. tuberculosis virulence factors with host genes.
MiR-21 and miR-26 showed differential expression after in vitro re-stimulation. Here miR-21 was up-regulated and miR-26a was down-regulated after TCR-specific in vitro stimulation of naïve T cells and M. tuberculosis specific T cells of LTBI. Recently a comprehensive study of miRNA expression in T cells detected decreased expression of miR-26a upon in vitro T-cell activation [37]. Our results confirm miR-26a down-regulation after T-cell receptor specific stimulation and likely reflect involvement in T-cell activation and development.
To examine possible roles of the identified candidate miRNAs we performed target gene analysis and focused on targets expressed in immunological tissues (i.e. blood, lymph node, lymph, spleen, thymus). The overlap between target genes was moderate and none of these were predicted for all four miRNAs. Only eight targets were predicted for three candidate miRNAs and classification did not reveal pathways dominantly targeted by miRNA candidates (data not shown). Hence we considered it likely that distinct immunologic processes are involved by differential miRNA expression. A crucial T-cell regulator identified as target of miR-21, miR-26a, and miR-29a is PTEN (phosphatase and tensin homolog deleted on chromosome 10). PTEN is a lipid phosphatase that negatively regulates antigen-activation of T cells by inhibiting the phosphoinositide 3-kinase (PI3K) signaling pathway [38]. Recently Huang et al. found that cell lines deficient for PTEN have an increased susceptibility to Mycobacterium bovis BCG infection [39]. Initial experiments have been performed to characterize PTEN protein expression in lymphocytes of children with tuberculosis and LTBI. The fact that we did not detect a correlation between miR-21, 26a, or 29a expression and PTEN protein levels did not exclude a possible role of PTEN regulation since other factors may confound analyses. In vitro modulation of miRNA candidate expression is currently performed to verify candidate miRNA targets.
A key factor for susceptibility against tuberculosis is IFNγ and the underlying TH1 immune response. It is a matter of discussion if protective TH1 immunity against tuberculosis may be impeded by other T helper subtypes (i.e. TH2, TH17) in a process called immune polarization. Analysis of miRNA candidate expression from ex vivo isolated TH17 cells was not feasible because of the low frequency of this subpopulation in peripheral blood [40]. Therefore we compared polarized TH1 and TH17 clones (TH2 clones could not be generated) for candidate miRNA expression. Differences were detected for miR-29a that was up-regulated after in vitro activation in TH17 but not in TH1 clones. This is in accordance with previous findings showing an IFNγ suppressive function of miR-29 [22]. In a recent study miR-29a reconstitution alone corrected aberrant IFNγ expression induced in the absence of miRNAs [21]. Furthermore, Ma et al. showed IFNγ itself is a target for miR-29a and that infection of mice with intracellular bacteria led to down-regulation of miR-29a expression in T cells.
In an attempt to characterize the effect of miR-29a expression we compared IFNγ expression in children with tuberculosis and LTBI as well as expression of both (miR-29a and predicted target IFNγ) in lymphocytes of individual donors but detected no dependency between these factors. In addition we determined the influence of miR-29a suppression on IFNγ production of primary human T cells using antagomir technology but did not detect differences of IFNγ expression upon inhibition of miR-29a expression (about 58% reduction). Several reasons may account for these seemingly contrary results. First, approximately 3-fold reduction may not be sufficient for induction of additional IFNγ producing T cells, taking into account that PMA/Ionomycin stimulation activated high proportions of IFNγ expression T cells (about 50% of all T cells). Second, given the dominant TH1 response – marked by IFNγ production – induced by in vitro stimulation, it is possible that ex vivo miR-29a expression levels in T cells were too low to inhibit IFNγ expression. In this case down-regulation would be without consequences but over-expression of miR-29a may lead to IFNγ-inhibition. Ongoing experiments in our laboratory address these questions. Finally, it is tempting to speculate that there are differences in the regulation of IFNγ by miR-29 between animal models and humans. E.g. there may be additional miRNAs that suppress IFNγ in humans and, hence, down-regulation of miR-29a was not sufficient.
We consider the present study an initial step to characterize the role of microRNAs in tuberculosis. Consistent pattern of differentially expressed miR-21, miR-26a, miR-29a, and miR-142-3p suggested a role of these candidates in T-cell immunity during tuberculosis disease and recovery. Further studies will have to be performed to elucidate the processes targeted by candidate miRNAs and to reveal if miRNAs are possible targets for disease intervention strategies in tuberculosis.
Materials and Methods
Study Design and Samples
A pilot study was conducted in order to compare miRNA expression among tuberculosis patients and healthy contacts recruited at the University hospital Hamburg-Eppendorf and at the Asklepios Center for Respiratory Medicine and Thoracic Surgery Munich-Gauting, Germany Altogether 16 donors were recruited and peripheral blood (20 ml) was obtained from each donor. Participants were categorized according to the following criteria:
Healthy contacts of tuberculosis patients with a positive T-cell response against purified protein derivative (PPD) of M. tuberculosis (Statens Serum Institute) (as described previously [41]) were termed latently M. tuberculosis infected (LTBI) (n = 6; median age 32 y; range 29–58 y).
Healthy donors without PPD-specific immunity were termed PPD negative (PPDneg; n = 3).
Diagnosis of active tuberculosis was based on patient history, chest X-ray, tuberculin skin test, QuantiFERON-TB-Gold® test, and mycobacterial culture (TB patients; n = 7; median age 35 y; range 22–52 y).
T-cell clones (TCCs) have been generated using buffy coat cells from anonymous donors. All donors gave written informed consent. The local ethics committee approved this study (Aerztekammer Hamburg, WF-07/09).
In the follow-up prospective cohort study, we recruited 22 children with tuberculosis (median age, 5 y; range 0–16 y), 14 children with LTBI (median age 8 y; range 3–16 y), and 19 non-M. tuberculosis infected (PPDneg) children (median age, 3 y; range, 0–11 y) at the Department of Pediatric Pneumology and Immunology, Charité in Berlin, Germany. All children received follow-up examinations three and twelve months after diagnosis. Peripheral blood (max. of 7 ml) was taken at all time points to perform IGRA tests and isolation of peripheral blood mononuclear cells (PBMCs). Of 22 children with tuberculosis initially recruited, 18 were included three months, and of these 13 were included 12 months after onset of therapy. Children were classified according to the following criteria:
Children with LTBI had reported contact to a contagious tuberculosis index case, had positive tuberculin skin test (TST), and Interferon gamma (IFNγ-release assay (IGRA, QuantiFERON®-TB Gold) but showed no clinical symptoms of tuberculosis (including chest X-ray imaging without pathological findings). Children with tuberculosis were diagnosed based primarily on patient history (especially reported contact to index case) and chest X-ray and diagnosis was supported by positive TST or IGRA in all included cases. Children with LTBI and children with tuberculosis were treated according to German guidelines. All legal guardians of included children gave written informed consent. This study was approved by the local ethics committee (EA4/019/09).
MiRNA Isolation, Reverse Transcription, and Analysis
MiRNAs were isolated from T cells (minimum of 5×106) or frozen blood pellets (of 500 µl heparanized blood) using the mirVana™ miRNA Isolation Kit (Ambion) following manufacturer’s instructions. Reverse transcription was performed with the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems) using two different methods: I) for plate assay analyses using the Megaplex™ RT Primers Human Pool A and B (500 ng RNA each) (Applied Biosystems), II) for single TaqMan® miRNA Assays containing RNA-specific RT primer (100 ng RNA) (Applied Biosystems) following manufacturer’s instructions. For quantitative PCR-based detection of miRNA either a custom-designed 96-well plate (Applied Biosystems) for detection of 29 candidate miRNAs (Table 1) or single TaqMan® miRNA assays (Applied Biosystems) were used. For the 96-well plates reverse transcription method I and for the single assays reverse transcription method II was used. The ‘housekeeping’ control gene RNU48 (official symbol: SNORD48) was applied. The real time PCR was performed in triplicates. Median expression of candidate miRNAs (Ct) relative to the housekeeping gene was calculated (ΔCt). MiRNAs that were detectable in less than 5 tuberculosis patients and 6 LTBI were excluded from further analysis and termed not detectable (nd). For in vitro stimulated T cells, ΔCt values of the respective non-stimulated sample were subtracted.
Enrichment of Total and Naive CD4+ T Cells
Peripheral blood mononuclear cells (PBMC) were isolated from 20 ml heparinized blood by density centrifugation (Biocoll, Gibco) and enrichment of CD4+ T cells was performed using the magnetic cell sorting system (IMag, Becton Dickinson) following manufacturer’s instructions. The purity of enriched T-cell populations was generally higher than 95% as determined by flow cytometry. Naïve CD4+ T cells for in vitro stimulation experiments were enriched using ‘untouched’ magnetic cell separation (MACS, Miltenyi) following manufacturer’s instructions and as described before [42].
CD4+ T-cell subpopulation enrichment for ex vivo miRNA analyses was performed by fluorescence-activated cell sorting (FACS) using the following antibodies: CD4 (APC-H7, clone RPA-T4, BD), CD45RA (FITC, clone HI100, BD), CD3 (PerCP, clone SK7, BD) and CCR7 (PE-Cy7, clone 3D12, BD). Cell sorting was performed using a FACS-ARIA III (BD). A minimum of 2.5×105 sorted cells was acquired and immediately frozen at −20°C for miRNA isolation.
In vitro Stimulation of T Cells
Naïve T cells (1.2×106) were stimulated with crosslinking αCD3/CD28 Dynabeads (Dynal) (2,5 µl/ml) in 96-well microtiter plates containing 200 µl X-Vivo15 medium (supplemented with 1% Penicillin/Streptomycin). PBMCs of LTBI were stimulated with PPD of M. tuberculosis. Samples were incubated for three or seven days at 37°C and 5% CO2. Afterwards cells were immediately frozen at −20°C for miRNA analyses. Short term (overnight) re-stimulation of T cells with M. tuberculosis specific PPD for analysis of IFNγ expressing T cells proportions was performed as described previously [41].
Intracellular PTEN Protein Expression
PBMCs were fixed in para-Formaldehyde (2%) for 10 min at 37°C and were then permeabilized using Phosflow buffer 3 (BD) following manufacturer’s instructions. After single staining with PTEN-specific PE-labeled antibodies (BD) (to avoid confounding effects of multi-color analyses), cells were wash twice and fluorescence intensities were determined using a FACS-Canto flow cytometer (BD). Median fluorescence intensities (MFI) have been shown.
Generation of TH1 and TH17 Clones
TH1 and TH17 clones were generated as described previously [40]. In brief, purified PBMCs (2×107) were stained with monoclonal antibodies (anti-CD4 APC-Cy7, BD and anti-CD161 FITC, Miltenyi Biotec), and CD4+ CD161+ T cells were sorted into 96-well round bottom plates (1, 2, 3, 5 or 10 cells/well) using a FACS Aria II (BD). Prior to sorting irradiated heterologous antigen presenting cells (1×105 cells/well) and αCD3/CD28 Dynabeads (0,1 µL/well) were added. On day 0, 7, 14, and 21 IL-2 (20 U/ml) was added. After 14 days expanded T-cells clones were counted for each plate. Only T-cells clones from plates with less than 37% positive wells were selected to ensure derivation from a single precursor cell. The cytokine profile of the TCC was determined after PMA/Ionomycin activation over night as described previously [40]. TCC with distinct IFNγ and IL-17 expression were analyzed miRNA candidate levels.
Modulation of MiRNA Expression Using Antagomirs
Transfection of PBMCs with antagomir-29a and control antagomirs for miR-16, and C. elegans cel-miR-67 (all Dharmacon) was performed following manufacturer’s instructions. Briefly, PBMCs (1.5×105 cells/well ) were seeded in 96-well round bottom plates containing X-Vivo15 w/o antibiotics (200 µL/well) and αCD3/CD28 Dynabeads (2 µL/well). After 16 h at 37°C and 5% CO2 transfection was done. Antagomirs (5 µM in siRNA buffer (Dharmacon)) were diluted 1∶10 with X-Vivo15 and transfection-solution was prepared mixing DharmaFECT4 (Dharmacon) with X-Vivo15 in a 0.5/10 Ratio. After incubation for 5 min at room temperature (RT) the antagomir-solution was added to the transfection-solution in the ratio of 1∶1 and was incubated for 20 min at RT. X-Vivo15 was added in a ratio of 1∶5. Stimulated cells were centrifuged and the supernatant was discarded. The prepared solution of antagomir, DharmaFECT4, and X-Vivo15 was added (100 µL/well) and the cells were incubated for 48 h at 37°C and 5% CO2. Subsequently cytokines (i.e IFNγ, TNFα, IL-2) and miRNA expression was analyzed by flow cytometry and real-time PCR, respectively.
Identification and Functional Annotation of miRNA Targets
We predicted miRNA targets on the miRGen internet platform (www.diana.pcbi.upenn.edu/cgi-bin/miRGen/v3/Targets.cgi) using PicTar, TargetScanS, miRanda (miRBase), DIANA-microT. Only targets predicted by a minimum of two software algorithms were selected for further analyses. The Unigene EST profile viewer was applied to constrain predicted miRNA candidate targets to those expressed in at least two of five immune tissues (i.e. blood, lymph node, lymph, spleen, thymus). Target annotations and clustering were done using the DAVID internet platform (http://david.abcc.ncifcrf.gov/) Medium classification stringency was chosen and p-values adjusted for multiple comparisons (according to Benjamini-Hochberg) are shown.
Statistical Analyses
Non-parametric tests have been applied throughout since moderate sample numbers were not sufficient to verify normal distributions. For comparison of miRNA expression between study groups and T-cell clones we applied the Mann-Whitney U-test. The Kruskal-Wallis test has been applied for comparison of miRNA expression in naïve T cells at different time points after in vitro stimulation. For correlation analyses of miRNA candidates the Spearman test was used.
Acknowledgments
We thank A. Neher, H. Henkel, G. Schulz, H. Hoffmann, and S. Schmiedel for their support concerning donor recruitment, J. Dunkel for screening of miRNA target gene expression pattern, and B. Schattling und M. Piédavent for their assistance in miRNA expression analyses. This study has been presented at the joint annual meeting of the German and Italian Societies of Immunology September 2011 in Riccione, Italy and is part of the dissertations of K. Heesch and S. Schattling.
In commemoration of Dr. K. Magdorf, initiator and mentor of the childhood tuberculosis study, who died in January 2013.
Funding Statement
This study was supported by the German Leprosy and TB relief association (DAHW), project: TB Biomarkers and by the Deutsche Forschungsgemeinschaft (DFG), project: Immunpolarization in childhood TB (JA 1479/3-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dorhoi A, Reece ST, Kaufmann SH (2011) For better or for worse: the immune response against Mycobacterium tuberculosis balances pathology and protection. Immunol Rev 240: 235–251. [DOI] [PubMed] [Google Scholar]
- 2. Lindenstrom T, Agger EM, Korsholm KS, Darrah PA, Aagaard C, et al. (2009) Tuberculosis subunit vaccination provides long-term protective immunity characterized by multifunctional CD4 memory T cells. J Immunol 182: 8047–8055. [DOI] [PubMed] [Google Scholar]
- 3. Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, et al. (2007) Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med 13: 843–850. [DOI] [PubMed] [Google Scholar]
- 4. Harari A, Rozot V, Enders FB, Perreau M, Stalder JM, et al. (2011) Dominant TNF-alpha+ Mycobacterium tuberculosis-specific CD4+ T cell responses discriminate between latent infection and active disease. Nat Med 17: 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Day CL, Abrahams DA, Lerumo L, Janse van Rensburg E, Stone L, et al. (2011) Functional capacity of Mycobacterium tuberculosis-specific T cell responses in humans is associated with mycobacterial load. J Immunol 187: 2222–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wozniak TM, Saunders BM, Ryan AA, Britton WJ (2010) Mycobacterium bovis BCG-specific Th17 cells confer partial protection against Mycobacterium tuberculosis infection in the absence of gamma interferon. Infect Immun 78: 4187–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen X, Zhang M, Liao M, Graner MW, Wu C, et al. (2010) Reduced Th17 response in patients with tuberculosis correlates with IL-6R expression on CD4+ T Cells. Am J Respir Crit Care Med 181: 734–742. [DOI] [PubMed] [Google Scholar]
- 8. Kumar NP, Anuradha R, Suresh R, Ganesh R, Shankar J, et al. (2011) Suppressed type 1, type 2, and type 17 cytokine responses in active tuberculosis in children. Clin Vaccine Immunol 18: 1856–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marin ND, Paris SC, Rojas M, Garcia LF (2012) Reduced frequency of memory T cells and increased Th17 responses in patients with active tuberculosis. Clin Vaccine Immunol 19: 1667–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jurado JO, Pasquinelli V, Alvarez IB, Pena D, Rovetta AI, et al. (2012) IL-17 and IFN-gamma expression in lymphocytes from patients with active tuberculosis correlates with the severity of the disease. J Leukoc Biol 91: 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Basile JI, Geffner LJ, Romero MM, Balboa L, Sabio YGC, et al. (2011) Outbreaks of mycobacterium tuberculosis MDR strains induce high IL-17 T-cell response in patients with MDR tuberculosis that is closely associated with high antigen load. J Infect Dis 204: 1054–1064. [DOI] [PubMed] [Google Scholar]
- 12. Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD (2008) MicroRNAs: new regulators of immune cell development and function. Nat Immunol 9: 839–845. [DOI] [PubMed] [Google Scholar]
- 13. Sun W, Julie Li YS, Huang HD, Shyy JY, Chien S (2010) microRNA: a master regulator of cellular processes for bioengineering systems. Annu Rev Biomed Eng 12: 1–27. [DOI] [PubMed] [Google Scholar]
- 14. O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D (2010) Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol 10: 111–122. [DOI] [PubMed] [Google Scholar]
- 15. Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, et al. (2009) Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 30: 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Curtale G, Citarella F, Carissimi C, Goldoni M, Carucci N, et al. (2010) An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood 115: 265–273. [DOI] [PubMed] [Google Scholar]
- 17. Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, et al. (2007) miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 129: 147–161. [DOI] [PubMed] [Google Scholar]
- 18. Mattes J, Collison A, Plank M, Phipps S, Foster PS (2009) Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci U S A 106: 18704–18709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei B, Pei G (2010) microRNAs: critical regulators in Th17 cells and players in diseases. Cell Mol Immunol 7: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, et al. MicroRNA-21 Limits In Vivo Immune Response-Mediated Activation of the IL-12/IFN-{gamma} Pathway, Th1 Polarization, and the Severity of Delayed-Type Hypersensitivity. J Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, et al. (2011) MicroRNA-29 Regulates T-Box Transcription Factors and Interferon-gamma Production in Helper T Cells. Immunity 35: 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma F, Xu S, Liu X, Zhang Q, Xu X, et al. (2011) The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol 12: 861–869. [DOI] [PubMed] [Google Scholar]
- 23. Tili E, Michaille JJ, Calin GA (2008) Expression and function of micro-RNAs in immune cells during normal or disease state. Int J Med Sci 5: 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, et al. (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jacobsen M, Repsilber D, Gutschmidt A, Neher A, Feldmann K, et al. (2006) Deconfounding microarray analysis - independent measurements of cell type proportions used in a regression model to resolve tissue heterogeneity bias. Methods Inf Med 45: 557–563. [PubMed] [Google Scholar]
- 26. Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, et al. (2002) T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol 217: 78–86. [DOI] [PubMed] [Google Scholar]
- 27. Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, et al. (1993) Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med 178: 2243–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, et al. (1993) An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med 178: 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ottenhoff TH, De Boer T, van Dissel JT, Verreck FA (2003) Human deficiencies in type-1 cytokine receptors reveal the essential role of type-1 cytokines in immunity to intracellular bacteria. Adv Exp Med Biol 531: 279–294. [DOI] [PubMed] [Google Scholar]
- 30. Buckler JL, Liu X, Turka LA (2008) Regulation of T-cell responses by PTEN. Immunol Rev 224: 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bui TV, Mendell JT (2010) Myc: Maestro of MicroRNAs. Genes Cancer 1: 568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, et al. (2008) Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mott JL, Kurita S, Cazanave SC, Bronk SF, Werneburg NW, et al. (2010) Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J Cell Biochem 110: 1155–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Buechner J, Henriksen JR, Haug BH, Tomte E, Flaegstad T, et al. (2011) Inhibition of mir-21, which is up-regulated during MYCN knockdown-mediated differentiation, does not prevent differentiation of neuroblastoma cells. Differentiation 81: 25–34. [DOI] [PubMed] [Google Scholar]
- 35. Jacobsen M, Repsilber D, Kleinsteuber K, Gutschmidt A, Schommer-Leitner S, et al. (2011) Suppressor of cytokine signaling-3 is affected in T-cells from tuberculosisTB patients. Clin Microbiol Infect 17: 1323–1331. [DOI] [PubMed] [Google Scholar]
- 36. Kumar P, Verma A, Saini AK, Chopra P, Chakraborti PK, et al. (2005) Nucleoside diphosphate kinase from Mycobacterium tuberculosis cleaves single strand DNA within the human c-myc promoter in an enzyme-catalyzed reaction. Nucleic Acids Res 33: 2707–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, et al. (2011) Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol 12: 796–803. [DOI] [PubMed] [Google Scholar]
- 38. Buckler JL, Walsh PT, Porrett PM, Choi Y, Turka LA (2006) Cutting edge: T cell requirement for CD28 costimulation is due to negative regulation of TCR signals by PTEN. J Immunol 177: 4262–4266. [DOI] [PubMed] [Google Scholar]
- 39. Huang G, Redelman-Sidi G, Rosen N, Glickman MS, Jiang X (2012) Inhibition of mycobacterial infection by the tumor suppressor PTEN. J Biol Chem 287: 23196–23202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kleinsteuber K, Heesch K, Schattling S, Sander-Juelch C, Mock U, et al. (2012) SOCS3 promotes interleukin-17 expression of human T cells. Blood 120: 4374–4382. [DOI] [PubMed] [Google Scholar]
- 41. Mueller H, Detjen AK, Schuck SD, Gutschmidt A, Wahn U, et al. (2008) Mycobacterium tuberculosis-specific CD4+, IFNgamma+, and TNFalpha+ multifunctional memory T cells coexpress GM-CSF. Cytokine 43: 143–148. [DOI] [PubMed] [Google Scholar]
- 42. Jacobsen M, Repsilber D, Gutschmidt A, Neher A, Feldmann K, et al. (2005) Ras-associated small GTPase 33A, a novel T cell factor, is down-regulated in patients with tuberculosis. J Infect Dis 192: 1211–1218. [DOI] [PubMed] [Google Scholar]