Abstract
Numerous studies have recently suggested that miRNAs contribute to the development of various types of human cancer as well as to their invasive and metastatic capacities. The aim of this study was to investigate the functional significance of miR-424 and to identify its possible target genes in osteosarcoma (OS) cells. Previously, inhibition of fatty acid synthase (FASN) has been shown to suppress OS cell proliferation, invasion and migration. The prediction was made using the microRNA.org and TargetScan.human6.0.database. The results showed that FASN is a promising target gene of miR-424. FASN may be a direct target of miR-424 as shown by the luciferase reporter assays. Furthermore, miR-424 expression was increased in osteosarcoma cells by transfection with has-miR-424. FASN mRNA and protein expression levels were measured by RT-PCR and western blot analysis. Cell migration and invasion was measured using Transwell migration and Transwell invasion assays. Expression levels of FASN mRNA and protein were greatly decreased in U2OS cells transfected with has-miR-424. The migration and invasion of cells was significantly decreased by the upregulation of miR-424. These findings suggested that miR-424 plays a key role in inhibiting OS cell migration and invasion through targeting FASN.
Keywords: miR-424, metastasis, osteosarcoma, fatty acid synthase
Introduction
Osteosarcoma (OS) is one of the most common primary malignant bone tumors in childhood and adolescence. It was not until the early 1970s that the introduction of doxorubicin and methotrexate with leucovorin rescue revealed the potential to improve survival. With the advent of effective chemotherapy, the 5-year survival rate of patients treated with intensive multidrug chemotherapy and aggressive local control have been reported as 55–80% (1–3). Despite the encouraging trend towards longer survival many patients still face a dismal outcome. Numerous articles have reported that the 5-year survival rate of patients with metastatic diseases is <20% (4–6). Clearly, the impact of identifying factors that govern metastasis is significant in the management of osteosarcoma.
Fatty acid metabolic pathways play an important role in carcinogenesis (7). Fatty acid synthase (FASN) is an enzyme crucial for endogenous lipogenesis in mammals responsible for catalyzing the synthesis of long-chain fatty acids. FASN is critical to sustain the biological features of cancer cells (8). FASN is expressed at high levels in a variety of human tumors (9–13) with low levels in normal tissues. Various reports have shown that inhibiting expression of FASN suppresses cancer cell proliferation in vitro and in vivo (14–19). Recent studies revealed that FASN may contribute to cancer cell metastasis (20–22). FASN is, thus, considered a novel promising target for anticancer therapy.
miRNAs are small endogenous RNAs averaging 20 to 24 nucleotides, transcribed from non-protein-coding genes or introns, which mediate translational suppression or cleavage of their target mRNAs by binding to complementary sites in their 3′UTR (23–25). A large number of miRNAs are located inside or close to fragile chromosomal sites that are frequently lost or amplified in cancer (26). miRNAs have been characterized as oncogenes, tumor suppressors or as components of regulatory pathways critical for tumorigenesis. miRNAs play an important role in tumorigenesis and metastasis.
The aim of this study was to investigate the functional significance of miR-424 and to identify its possible target genes in osteosarcoma (OS) cells.
Materials and methods
Cell culture and transfection
Human OS cell line U2OS (Shanghai Cell Bank, Chinese Academy of Sciences, China) was cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and incubated at 37°C in 5% CO2. U2OS cells were seeded in six-well plates at 30% confluence on the day before transfection. Transfection with has-miR-424 or negative miRNA was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Transfection complexes were prepared according to the manufacturer’s instructions. Our study was approved by the ethics committee of the First Affiliated Hospital of Nanchang University, Nanchang, China.
Quantitative real-time PCR (qRT-PCR)
Total RNA from cells treated with has-miR-424 or negative control miRNA was isolated using TRIzol reagent (Tiangen, Beijing, China) and reverse transcribed using a reverse transcription kit (Tiangen) according to the manufacturer’s instructions. Reactions were performed and analyzed using an ABI 7300 system (Applied Biosystems, Carlsbad, CA, USA). β-actin was used as the internal control to quantify initial cellular transcripts. Details of the primers and probes used in this study are summarized in Table I. All qRT-PCR were performed six times according to the manufacturer’s instructions. The relative expression level of FASN was normalized to that of β-actin by 2-ΔΔCtcycle threshold method. The ΔCt data were collected automatically. The average ΔCt of each group was calculated using the following formula: ΔCt = average miR-424 Ct - Δaverage of β-actin Ct. ΔΔCt was calculated by ΔΔCt=ΔCt of miR-424 group - ΔΔCt of the negative control group. The fold-change in FASN expression level was calculated using 2−ΔΔCt.
Table I.
Primers and probes used.
| Primers and probes | Sequences 5′-3′ |
|---|---|
| FASN | |
| Sense | 5′-AAGCAGGCACACACGATGG-3′ |
| Antisense | 5′-TCGGAGTGAATCTGGGTTGATG-3′ |
| Probe | 5′-CTGCGGCTGCTGCTGGAAGTCACC-3′ |
| β-actin | |
| Sense | 5′-TGCCCATCTACGAGGGGTATG-3′ |
| Antisense | 5′-CTCCTTAATGTCACGCACGATTTC-3′ |
| Probe | 5′-CCTGCGTCTGGACCTGGCTGGC-3′ |
FASN, fatty acid synthase.
Luciferase activity assay
Primers were designed in accordance with the Genbank query FASN gene mRNA (NM_004104.4) sequence. A fragment of the 3′-UTR of FASN was amplified from U2OS cells by PCR using the forward primers 5′-CCCCTCGAGCCTGCCACCGGAGGTCACT-3′ and the reverse primers 5′-CGGGCGGCCGCGTGGGAGGC TGAGAGCAGCA-3′. After digestion of the PCR product by XhoI and NotI, the FASN 3′-UTR was cloned in pSiCHECK2 (Promega, Madison, WI, USA) at the XhoI and NotI sites. All PCR products were verified by DNA sequencing. U2OS cells were cotransfected with the pSiCHECK2 vectors containing the 3′-UTR variants and has-miR-424 or negative miR-001. Luciferase activity was measured 48 h after transfection. The firefly luciferase activity was then normalized against the renilla luciferase activity.
Transwell invasion assay in vitro
Invasion assays were performed in triplicate using Transwell invasion chambers coated with Matrigel (50 μl per filter) (BD Biosciences, Franklin Lakes, NJ, USA) as described in the manufacturer’s protocol. U2OS cells were transfected with either has-miR-424 or negative control oligonucleotide, cultured for 48 h and transferred on the top of Matrigel-coated invasion chambers in a 1% fetal calf serum DMEM/F12 (2×104 cells/well). DMEM/F12 containing 10% fetal calf serum was added to the lower chambers. After incubation for 24 h at 37°C in an atmosphere containing 5% CO2, invaded cells on the lower surface were stained with crystal violet stain and counted under a light microscope. All experiments were repeated six times over multiple days.
Migration assay
Migration assays were performed using a 24-well Transwell chamber system (Costar 3422, Corning Inc., NY, USA). Cells were seeded in the upper chamber at 2×104 cells/ml in 0.1 ml serum-free DMEM/F12 media. Media supplemented with 10% fetal bovine serum was placed in the bottom well in a volume of 0.8 ml (used as a chemoattractant). After incubation for 24 h at 37°C in an atmosphere containing 5% CO2, migrated cells on the lower surface were stained with crystal violet stain and counted under a light microscope. All experiments were repeated six times over multiple days.
Western blot analysis
U2OS cells in the exponential growth phase were transfected with has-miR-424 for 48 h. Total proteins were isolated from U2OS cells. Protein concentrations were measured using a Micro BCA protein assay kit (Pierce, Biotechnology, Inc, Rockford, IL, USA). Proteins were resolved using 10% SDS-PAGE gel, transferred to the nitrocellulose membrane, blocked in 5% non-fat dry milk in Tris-buffered saline (pH 7.4) containing 0.05% Tween-20 and blotted with a rabbit polyclonal antibody against FASN (1:1,000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and goat anti-rabbit IgG (1:3,000; Santa Cruz Biotechnology, Inc.). GAPDH was used as a loading control. Signals were detected using secondary antibodies labeled with HRP. All western blot analyses were performed six times.
Statistical analysis
Data were expressed as means ± SD of at least six experiments. The independent samples test was used for statistical analysis. P<0.05 was considered to indicate a statistically significant result. All analyses were performed using SPSS Version 13.0 (Statistical Software for Social Sciences, Chicago, IL, USA).
Results
Upregulation of miR-424 inhibits cell migration in vitro
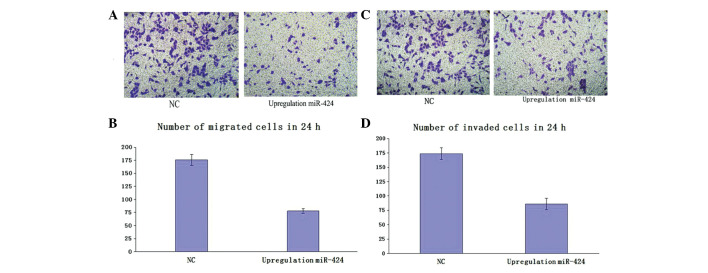
To corroborate the effect of the upregulation of miR-424 on U2OS cell migration, migration was measured by Transwell migration assay. U2OS cells were transfected with miR-424 or negative miRNA. The results showed that the migration of cells transfected with has-miR-424 was significantly inhibited when compared with cells transfected with negative miRNA (Fig. 1A and B, P<0.05). These results suggested that upregulation of miR-424 inhibited the migration of U2OS cells.
Figure 1.
Upregulation of miR-424 suppresses osteosarcoma (OS) cell migration and invasion. (A) and (C) A representative image of six experiments of Transwell migration and invasion assays is shown for each group (10×10), respectively. (B) Columns, mean (n=6); bars, SD. *P<0.05, vs. negative control group (NC). It indicated that upregulation of miR-424 expression in the U2OS cells suppressed cell migration. (D) Quantification of cell invasion expressed by cell counting. Columns, mean (n=6); bars, SD. *P<0.05, vs. negative control group (NC). It indicated that upregulation of miR-424 significantly inhibits cell invasion.
Upregulation of miR-424 inhibits cell invasion in vitro
To examine the effect of upregulation of miR-424 on U2OS cell migration, the Transwell invasion assay was performed. U2OS was transfected with has-miR-424 or negative miRNA. The results showed that the invasion of cells transfected with has-miR-424 was significantly inhibited when compared with cells transfected with negative miRNA. (Fig 1C and D; P<0.05). These results suggested that upregulation of miR-424 inhibits the invasion of U2OS cells.
FASN is a direct target of miR-424
To validate whether miR-424 regulates FASN directly through a putative binding site in U2OS cells, we cloned FASN 3′-UTR in the predicted miRNA binding site into the luciferase gene (pSiCHECK2; Promega). Following cotransfection with the pSiCHECK2 vectors and miR-424 or negative control miR-001, the upregulation of miR-424 in U2OS cells transfected with has-miR-424 resulted in a significant decrease in the luciferase activity of the wild-type FASN 3′-UTR (Fig. 2; P<0.05). The results indicate that FASN is a direct target of miR-424.
Figure 2.
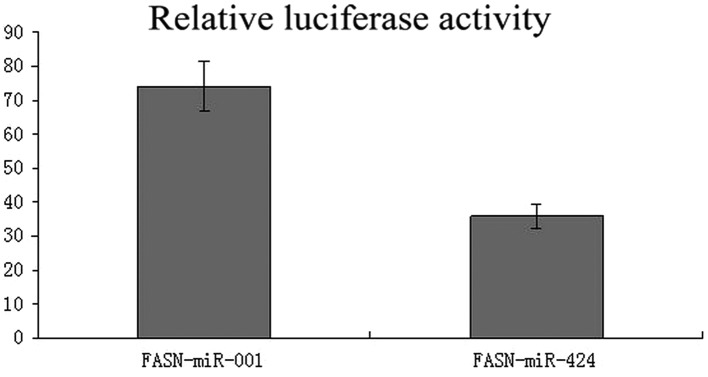
Analysis of the reporter activity in U2OS cells. The luciferase activity was normalized by renilla luciferase. Columns, mean (n=6); bars, SD. *P<0.05 vs. fatty acid synthase (FASN)-miR-001 group. It indicated that miR-424 inhibits reporter activity of gene expression by targeting FASN 3′-UTR in osteosarcoma (OS) cells.
miR-424 negatively regulates FASN mRNA expression in U2OS cells
To investigate the effect of upregulation of miR-424 on the expression of FASN mRNA, miR-424 was upregulated in U2OS cells by treatment with has-miR-424 for 48 h. The expression level of FASN mRNA was measured by qRT-PCR. The data (2−ΔΔCt=0.254±0.01157) showed that the FASN mRNA expression in cells transfected with negative control vector was four-fold that in cells transfected with has-miR-424. It indicated that miR-424 may negatively regulate the expression of FASN mRNA.
miR-424 inhibits FASN protein expression in U2OS cells
To investigate the effect of upregulation miR-424 on the expression of FASN protein, miR-424 was upregulated in U2OS cells by treatment with has-miR-424 for 48 h. The expression level of FASN protein was measured using western blot analysis. The results revealed that the upregulation of miR-424 in OS cells resulted in decreasing the expression of FASN protein (Fig 3A and B). It suggested that miR-424 may negatively regulate the expression of FASN protein.
Figure 3.
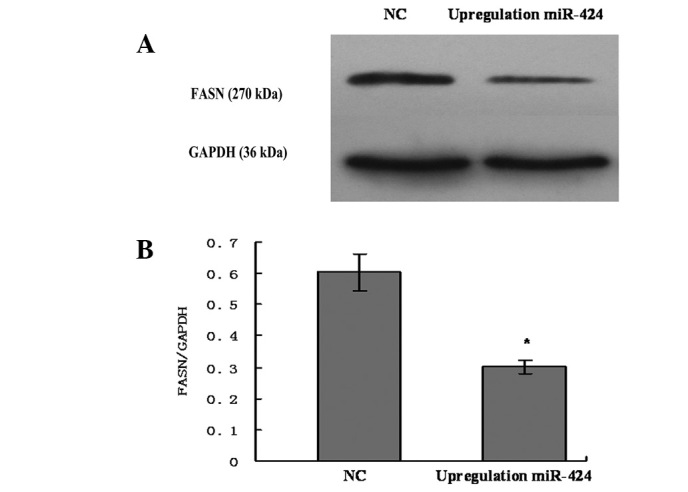
Fatty acid synthase (FASN) protein expression was inhibited by upregulation of miR-424 in U2OS cells. (A) A representative image of western blots showing the expression of FASN protein was suppressed by upregulation of miR-424. (B) Columns, mean (n=6); bars, SD. *P<0.05, vs. negative control group (NC). It indicated that miR-424 inhibits FASN protein expression in U2OS cells.
Discussion
miR-424, one of the miR-16/15/195/424/497 family members, induces muscle differentiation and promotes cell cycle quiescence and differentiation (27–29) and regulates cell-autonomous angiogenesis (30–32). Recent evidence demonstrated that miR-424 plays an important role in tumorigenesis (33–34). miR-424 has been reported to be downregulated in cervical cancer (33), senile hemangioma (30), tongue cancer (35), chronic myelogenous leukaemia (36) and acute myeloid leukemia (34). However, miR-424 is upregulated in human colorectal cancer (37–38) and atypical chronic myeloid leukemia (37). Deregulation of miR-424 may be different in different types of cancer and the roles of miR-424 in carcinogenesis and progression should not be assumed as a tumor suppressor or oncogene. The roles of miR-424 deregulation in cancer development remains to be further investigated. In the present study, miR-424 was downregulated in the human osteosarcoma cell line U2OS. The inhibiting effect on OS cell migration and invasion by upregulation of miR-424 was observed. It suggested that miR-424 plays a role as a tumor suppressor in inhibiting OS cell migration and invasion.
A previous study reported that inhibition of FASN causes a decrease in OS cell invasion and migration. The prediction was made using software (microRNA.org and TargetScan. human6.0). It revealed that miR-424 may be targeting FASN. In this study, to investigate the molecular mechanism that inhibits the migration and invasion by restoration of miR-424 in OS cells, RT-PCR and western blot analysis were performed to detect the expression level of FASN mRNA and protein in the OS cell line U2OS. The results showed that FASN expression was significantly inhibited in cells transfected with has-miR-424 compared with the control group (Fig. 4). This suggested that restoration of miR-424 expression could inhibit FASN expression in OS cells. Furthermore, to identify whether miR-424 regulates the expression of FASN, the FASN 3′-UTR was cloned into the pSiCHECK2, placing the 3′-UTR with most potential miRNA binding sites downstream of coding sequence of luciferase (pMiR-Report; Promega). OS cells were cotransfected with the pSiCHECK2 vector containing the 3′-UTR and miR-424 or negative miR-001. Overexpression of miR-424 significantly reduced luciferase activity from the reporter construct containing the FASN 3′-UTR. It indicated that FASN is a direct miR-424 target. There are hundreds of predicted targets of miR-424 in the TargetScan prediction and a single miRNA is demonstrated to target multiple mRNAs to regulate gene expression (39), therefore it is probable that other targets of miR-424 may also participate in OS migration and invasion. miR-424 may also target different molecules in different types of cancer. Additionally, the tumor micro-environment may influence tumor progression, invasion and migration. This study indicates that expression of FASN is negatively regulated by miR-424 through a special binding site of the FASN 3′-UTR. Moreover, miR-424 inhibits cell invasion and migration in OS cells in vitro. These results suggest that miR-424 plays a key role in inhibiting OS cell migration and invasion by targeting FASN. Further research is necessary to identify the entire roles of miR-424 in osteosarcoma metastasis.
Acknowledgments
This study was supported by grants from the health department of Jiangxi Province (NO.20121036) and the Nature Science of Jiangxi Province (NO.20114BAB205093).
References
- 1.Meyers PA, Schwartz CL, Krailo M, et al. Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J Clin Oncol. 2005;23:2004–2011. doi: 10.1200/JCO.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 2.Bacci G, Forni C, Longhi A, et al. Local recurrence and local control of non-metastatic osteosarcoma of the extremities: A 27-year experience in a single institution. J Surg Oncol. 2007;96:118–123. doi: 10.1002/jso.20628. [DOI] [PubMed] [Google Scholar]
- 3.Jawad MU, Cheung MC, Clarke J, et al. Osteosarcoma: Improvement in survival limited to high-grade patients only. J Cancer Res Clin Oncol. 2011;137:597–607. doi: 10.1007/s00432-010-0923-7. [DOI] [PubMed] [Google Scholar]
- 4.Mialou V, Philip T, Kalifa C, et al. Metastatic osteosarcoma at diagnosis: Prognostic factors and long-term outcome - the French pediatric experience. Cancer. 2005;104:1100–1109. doi: 10.1002/cncr.21263. [DOI] [PubMed] [Google Scholar]
- 5.Hegyi M, Semsei AF, Jakab Z, et al. Good prognosis of localized osteosarcoma in young patients treated with limb-salvage surgery and chemotherapy. Pediatr Blood Cancer. 2011;57:415–422. doi: 10.1002/pbc.23172. [DOI] [PubMed] [Google Scholar]
- 6.Stokkel MP, Linthorst MF, Borm JJ, et al. A reassessment of bone scintigraphy and commonly tested pretreatment biochemical parameters in newly diagnosed osteosarcoma. J Cancer Res Clin Oncol. 2002;128:393–399. doi: 10.1007/s00432-002-0350-5. [DOI] [PubMed] [Google Scholar]
- 7.Yeh CS, Wang JY, Cheng TL, et al. Fatty acid metabolism pathway play an important role in carcinogenesis of human colorectal cancers by microarray-bioinformatics analysis. Cancer Let. 2006;233:297–308. doi: 10.1016/j.canlet.2005.03.050. [DOI] [PubMed] [Google Scholar]
- 8.Hess D, Igal RA. Genistein downregulates de novo lipid synthesis and impairs cell proliferation in human lung cancer cells. Exp Biol Med (Maywood) 2011;236:707–713. doi: 10.1258/ebm.2011.010265. [DOI] [PubMed] [Google Scholar]
- 9.Alo PL, Amini M, Piro F, et al. Immunohistochemical expression and prognostic significance of fatty acid synthase in pancreatic carcinoma. Anticancer Res. 2007;27:2523–2527. [PubMed] [Google Scholar]
- 10.Walter K, Hong SM, Nyhan S, et al. Serum fatty acid synthase as a marker of pancreatic neoplasia. Cancer Epidemiol Biomarkers Prev. 2009;18:2380–2385. doi: 10.1158/1055-9965.EPI-09-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okawa Y, Hideshima T, Ikeda H, et al. Fatty acid synthase is a novel therapeutic target in multiple myeloma. Br J Haematol. 2008;141:659–671. doi: 10.1111/j.1365-2141.2008.07114.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Migita T, Ruiz S, Fornari A, et al. Fatty acid synthase: a metabolic enzyme and candidate oncogene in prostate cancer. J Natl Cancer Inst. 2009;101:519–532. doi: 10.1093/jnci/djp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu ZL, Wang G, Shu Y, et al. Enhanced antitumor activity of epirubicin combined with cerulenin in osteosarcoma. Mol Med Report. 2012;5:326–330. doi: 10.3892/mmr.2011.661. [DOI] [PubMed] [Google Scholar]
- 14.Orita H, Coulter J, Tully E, et al. Inhibiting fatty acid synthase for chemoprevention of chemically induced lung tumors. Clin Cancer Res. 2008;14:2458–2464. doi: 10.1158/1078-0432.CCR-07-4177. [DOI] [PubMed] [Google Scholar]
- 15.Coleman DT, Bigelow R, Cardelli JA. Inhibition of fatty acid synthase by luteolin post-transcriptionally down-regulates c-Met expression independent of proteosomal/lysosomal degradation. Mol Cancer Ther. 2009;8:214–224. doi: 10.1158/1535-7163.MCT-08-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saati GE, Archer MC. Inhibition of fatty acid synthase and Sp1 expression by 3,3′-diindolylmethane in human breast cancer cells. Nutr Cancer. 2011;63:790–794. doi: 10.1080/01635581.2011.570896. [DOI] [PubMed] [Google Scholar]
- 17.Notarnicola M, Pisanti S, Tutino V, et al. Effects of olive oil polyphenols on fatty acid synthase gene expression and activity in human colorectal cancer cells. Genes Nutr. 2011;6:63–69. doi: 10.1007/s12263-010-0177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Notarnicola M, Messa C, Refolo MG, et al. Polyunsaturated fatty acids reduce fatty acid synthase and hydroxy-methyl-glutaryl CoA-reductase gene expression and promote apoptosis in HepG2 cell line. Lipids Health Dis. 2011;10:10. doi: 10.1186/1476-511X-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zecchin KG, Rossato FA, Raposo HF, et al. Inhibition of fatty acid synthase in melanoma cells activates the intrinsic pathway of apoptosis. Lab Invest. 2011;91:232–240. doi: 10.1038/labinvest.2010.157. [DOI] [PubMed] [Google Scholar]
- 20.Carvalho MA, Zecchin KG, Seguin F, et al. Fatty acid synthase inhibition with Orlistat promotes apoptosis and reduces cell growth and lymph node metastasis in a mouse melanoma model. Int J Cancer. 2008;123:2557–2565. doi: 10.1002/ijc.23835. [DOI] [PubMed] [Google Scholar]
- 21.Murata S, Yanagisawa K, Fukunaga K, et al. Fatty acid synthase inhibitor cerulenin suppresses liver metastasis of colon cancer in mice. Cancer Sci. 2010;101:1861–1865. doi: 10.1111/j.1349-7006.2010.01596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu ZL, Zhou Y, Luo QF, et al. Inhibition fatty acid synthase suppress osteosarcoma cell invasion and migration. Indian J Pathol Microbiol. 2012;55:163–169. doi: 10.4103/0377-4929.97849. [DOI] [PubMed] [Google Scholar]
- 23.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 24.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 26.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkar S, Dey BK, Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell. 2010;21:2138–2149. doi: 10.1091/mbc.E10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forrest AR, Kanamori-Katayama M, Tomaru Y, et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia. 2010;24:460–466. doi: 10.1038/leu.2009.246. [DOI] [PubMed] [Google Scholar]
- 29.Rosa A, Ballarino M, Sorrentino A, et al. The interplay between the master transcription factor PU. 1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci USA. 2007;104:19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakashima T, Jinnin M, Etoh T, et al. Down-regulation of mir-424 contributes to the abnormal angiogenesis via MEK1 and cyclin E1 in senile hemangioma: Its implications to therapy. PLoS One. 2010;5:e14334. doi: 10.1371/journal.pone.0014334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chamorro-Jorganes A, Araldi E, Penalva LO, et al. MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. Arterioscler Thromb Vasc Biol. 2011;31:2595–2606. doi: 10.1161/ATVBAHA.111.236521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh G, Subramanian IV, Adhikari N, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J Clin Invest. 2010;120:4141–4154. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu J, Li Y, Wang F, et al. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. 2012 Apr 2; doi: 10.1038/onc.2012.121. (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 34.Faraoni I, Laterza S, Ardiri D, et al. MiR-424 and miR-155 deregulated expression in cytogenetically normal acute myeloid leukaemia: correlation with NPM1 and FLT3 mutation status. J Hematol Oncol. 2012;5:26. doi: 10.1186/1756-8722-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rentoft M, Fahlén J, Coates PJ, et al. miRNA analysis of formalin-fixed squamous cell carcinomas of the tongue is affected by age of the samples. Int J Oncol. 2011;38:61–69. [PubMed] [Google Scholar]
- 36.Hussein K, Büsche G, Muth M, et al. Expression of myelopoiesis associated microRNA in bone marrow cells of atypical chronic myeloid leukaemia and chronic myelomonocytic leukaemia. Ann Hematol. 2011;90:307–313. doi: 10.1007/s00277-010-1072-4. [DOI] [PubMed] [Google Scholar]
- 37.Guo ST, Jiang CC, Wang GP, et al. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene. 2012 Jun 18; doi: 10.1038/onc.2012.214. (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Wang J, Ma H, et al. Downregulation of miR-195 correlates with lymph node metastasis and poor prognosis in colorectal cancer. Med Oncol. 2012;29:919–927. doi: 10.1007/s12032-011-9880-5. [DOI] [PubMed] [Google Scholar]
- 39.Selbach M, Schwanhäusser B, Thierfelder N, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]