Abstract
Solid-state nanopores have emerged as versatile single-molecule sensors for applications including DNA sequencing, protein unfolding, micro-RNA detection, label-free detection of single nucleotide polymorphisms, and mapping of DNA-binding proteins involved in homologous recombination. While machining nanopores in dielectric membranes provides nanometer-scale precision, the rigid silicon support for the membrane contributes capacitive noise and limits integration with microfluidic networks for sample pre-processing. Herein, we demonstrate a technique to directly transfer solid-state nanopores machined in dielectric membranes from a silicon support into a microfluidic network. The resulting microfluidic-addressable nanopores can sense single DNA molecules at high bandwidths and with low noise, owing to significant reductions in membrane capacitance. This strategy will enable large-scale integration of solid-state nanopores with microfluidic upstream and downstream processing and permit new functions with nanopores such as complex manipulations for multidimensional analysis and parallel sensing in two and three-dimensional architectures.
Keywords: Integrated Nanopores, DNA, Noise, Translocation, Single Molecule
Solid-state nanopores are beginning to emerge as key elements in the study of single biomolecules such as nucleic acids and proteins. Owing to their small dimensions and ability to directly probe single molecules with high fidelity and throughput, a number of applications have been investigated using solid-state nanopores such as single molecule DNA sequencing 1, protein unfolding 2, micro-RNA detection3, label-free detection of single nucleotide polymorphisms4, and mapping of DNA-binding proteins involved in homologous recombination5. Solid-state nanopores with diameters <20 nm are typically constructed by drilling a nanometer-scale hole in a thin dielectric membrane using ion6 or electron beam irradiation7. The dielectric membrane, usually suspended on a rigid silicon substrate that acts as the support, separates two reservoirs filled with an electrolyte solution. Application of a voltage bias across the substrate drives an ionic current through the pore, and when a molecule translocates through, a measurable signal indicative of its structure and composition is obtained. Conventional fabrication techniques place several limitations on nanopore functionality and performance. First, the nanopores are situated between macroscale reservoirs, making it difficult to perform complex manipulations such as sorting of single molecules 8 based on electrical signals obtained during translocation through a nanopore. Second, fabrication in planar silicon architectures induces high noise, which limits the spatial and temporal resolution of nanopore-based analysis of nucleic acids. Limitations are also placed on the ability to arrange nanopores in simple networks with microfluidic devices that can potentially provide upstream and downstream processing of molecules.
Herein, we demonstrate fabrication of microfluidic-integrated nanopore devices using a modular strategy that relies on transfer printing. Whereas thick nanoporous polymeric membranes have previously been integrated into microfluidic devices9,10, the transfer printing technique presented here permits functional registration of single solid-state nanopores in thin (< 100 nm) dielectric membranes without a silicon support between polymeric microchannels. The resulting device architecture significantly reduces capacitive noise and permits sampling at high bandwidths, which is critical to precisely resolve single DNA molecule translocation events. Finally, we demonstrate functionality of the devices by high-fidelity sensing of single DNA molecules with the microfluidics-integrated nanopore devices and contrast the results with those obtained using conventional fabrication methods. These advances provide avenues to perform exciting new functions with nanopores such as integrated sample preparation and analysis, monitoring of dynamic molecular interactions and rearrangements, nanopore-enabled single-molecule sorting, and complex manipulations for multidimensional analysis in two and three-dimensional microfluidic architectures.
MATERIALS AND METHODS
Fabrication of Microfluidics-Integrated Nanopore Devices
Photolithography (SU-8, Shipley) was used to create a negative mold defining the microchannel system. The mold was chemically treated with 1H,1H,2H,2H-perflurododecyl trichlorosilane (Sigma Aldrich) for 6 h prior to use. Polydimethylsiloxane (PDMS, Sylgard 184) mixed in a 3:1 polymer to cross-linker ratio was poured over the SU-8 mold and cured for 8 h at 80°C. The PDMS component comprised larger access microchannels that tapered into smaller microchannels with widths ranging from 3-15 μm and a height of 10 μm. The width of the smaller microchannels determined the membrane area exposed to the electrolyte solution, which could be tuned to <10 μm2. Two PDMS components were fabricated for assembly of each device with one component acting as the top and the other component acting as the bottom. Devices used for DNA translocations were made from PDMS mixed in a 10:1 polymer to cross-linker ratio for the bottom piece, and 5:1 polymer to cross-linker ratio for the top piece, both being cured for 40 minutes at 80°C before transferring membrane and bonding the two PDMS substrates. Inlet/outlet ports were punched at the ends of each microchannel before the transfer process.
Commercially available 3-mm diameter transmission electron microscopy grids, (Ted Pella / TEM windows) with 50-nm thick LPCVD SiNx membranes and free-standing areas of 100 × 100 μm on a silicon substrate, were machined with a focused ion beam (FIB). Nanopores with diameters ranging from 25 to 50 nm were drilled in a square array (Figure 1a) with a Ga+ FIB (Helios Nanolabs 600) in conjunction with NanoPattern Generating System (JC Nabity Lithography Systems). The pitch of the nanopore array was set to equal the microchannel width (3-15 μm) to ensure a high probability that a single pore in the array was registered between two microchannels. Cuts around the membrane (visible in Figure 1a, 1b) were made with the Ga+ FIB directly after nanopore fabrication, with a milling time of roughly 2 min. These cuts left the membrane suspended on the silicon wafer at eight 5 μm wide segments (two on each side of each corner), and facilitated subsequent membrane transfer to the receiving PDMS substrate. 11, 12 The patterned grid was then spatially located over the PDMS microchannel using a custom-built aligner (see Supplementary Section S4) and brought into conformal contact (Figure 1b). The flexibility of the polymeric substrate allows for good contact with the TEM grid even in the presence of particulates or debris. The 3:1 PDMS ratio increased the Young’s Modulus of the PDMS, making it less flexible and facilitating manual alignment of the top PDMS piece over the bottom. The fabrication process, however, was verified to work for a PDMS polymer to cross-linker ratio of 10:1 for both PDMS substrates. Prior to contact, the patterned grid and the PDMS substrate were treated with plasma for 15 s at a pressure of 700 mTorr of air (atmospheric composition) at a power of 7.16 W. The time interval before bonding was controlled between 3 to 4 min (required for setting up the membrane for transfer) to ensure reproducible adhesion so that the TEM grid could be peeled away, depositing the SiNx membrane over the microchannel (Figure 1c). Another identical PDMS microchannel (rotated 90°) was plasma treated (29.6 W, 700 mTorr of air, 1 min), positioned over the membrane in an identical fashion and brought into conformal contact such that a single nanopore electrically and fluidically connected the two microfluidic channels (Figure 1d). Devices in which a 3:1 ratio was used for both the top and bottom PDMS substrates were baked for 3 d at 80°C prior to use to ensure a strong bond strength between them. Devices in which the partial curing method was used (10:1 ratio for the bottom substrate, 5:1 for the top substrate) were baked for 1 d at 80°C prior to use. Except for the polymer to pre-cursor ratio and the curing times, there were no other differences between the two PDMS bonding methods.
Figure 1.
Overview of fabrication process for microfluidic-addressable low-noise solid-state nanopore devices. (a) [top] 3-D rendering of TEM Grid with cross section illustrating Focused Ion Beam fabrication of square nanopore arrays in the 100 μm × 100 μm × 50 nm SiNx membrane. [bottom] SEM Image of nanopore array and cuts around the membrane. Membrane area inside cuts is 90 μm × 90 μm. 3D rendering [top], and optical micrograph [bottom] of: (b) SiNx membrane with Si wafer support being aligned over, and brought into contact with a microchannel on a PDMS substrate using a custom built aligner. Membrane area inside cuts is 90 μm × 90 μm (c) PDMS microchannel with detached SiNx membrane (d) Assembled device following bonding of a second PDMS substrate with perpendicular microchannel. Deposited membrane width in (c) and (d) is 90 μm.
Atomic Layer Deposition (ALD)
Thin conformal films of alumina (Al2O3) were deposited at 140°C from alternating exposures of a precursor, trimethylaluminum (Al(CH3)3, TMA), and water, at a growth rate of approximately 1.1 Å/cycle. Al2O3 depositions were performed directly on the membrane after nanopore fabrication, before depositing the membrane onto the PDMS. A decrease in the pore diameter estimated from current measurements and increase in surface charge (Figure 2) indicated successful deposition. SiO2 ALD depositions were performed after membrane transfer and prior to bonding of the second PDMS microchannel, by exposing the substrate to alternating exposure of TMA, water, and three consecutive silane pulses at 140°C. The deposited thickness of the Al2O3 and SiO2 ALD films was varied across devices, but in all cases was less than 20 nm. Many devices (> 30) have been made using this method, and device characterization data presented here are representative of the observations made over the course of device testing.
Figure 2.
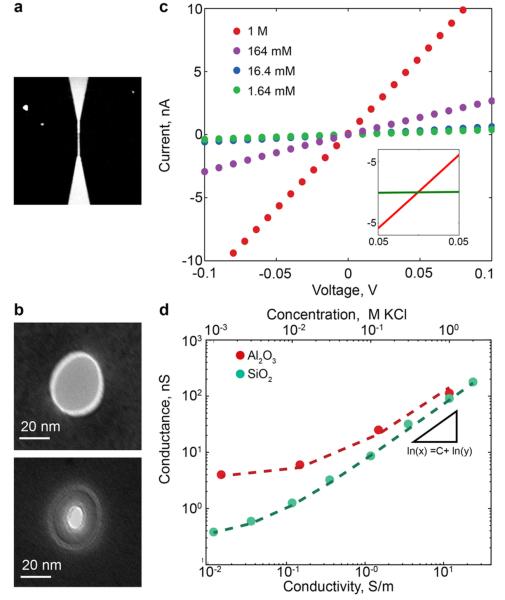
Characterization of microfluidic-addressable nanopore devices. (a) Epifluorescence image showing absence of fluorescent dye transport after diffusion for one hour followed by applying 1 V across the membrane for 1 min in a device without nanopores. Image size is 400 μm × 400 μm. (b) TEM images of a nanopore before and after ALD of Al2O3. (c) I-V curves for 30-nm effective diameter Al2O3 nanopore at four electrolyte concentrations (1 M, 164 mM, 16.4 mM, 1.64 mM KCl, at pH of 7.4). Inset shows I-V curve comparison with a device without nanopores at 1 M KCl. (d) Effect of ionic concentration on conductance for nanopores coated with SiO2 (pH 8) and Al2O3 (pH 7.4). Measured buffer conductivity and corresponding KCl concentration are shown on the X-axes. The estimated effective diameters were 20 nm and 19 nm, and surface charge was −8 mC/m2 and +40 mC/m2 for the SiO2 and Al2O3 nanopores, respectively.
Electrical Characterization
Current traces were measured using an Axopatch 200B patch clamp amplifier and digitized with a Digidata 1440 (Molecular Devices). A stock solution of 2 M KCl (Sigma Aldrich), 0.2X TBE (Tris Borate EDTA buffer, Sigma Alrich) solution at pH 8 was prepared using Nuclease/RNAase-free water. The salt concentration was adjusted by diluting the stock solution in Nuclease/RNAase-free water and the ratio of KCl to TBE was kept constant across different salt concentrations used for measuring nanopore conductance. Ag/AgCl electrodes (In Vivo Metric) were inserted into the microchannel access ports. I-V curve measurements were taken by ramping the voltage from the maximum voltage down to its negative value in 20 discrete steps. I-V traces were low-pass filtered at 2 kHz with the 8-pole Bessel filter in the Axopatch 200B, and sampled at a frequency of 20 kHz. RMS noise values, and Power Spectral Densities (PSD) were obtained by measuring 10 s of ionic current data, low-pass filtered at 100 kHz and sampled at 250 kHz with 0 V applied voltage. PSD estimation was performed using Matlab functions for spectrum estimation. For all noise and DNA translocation measurements, the Axopatch 200B Gain, β , was set to β = 1. This setting, corresponding to a gain of 1 V/nA, limits the maximum measurable current to ± 10 nA, and is the lowest noise setting for the amplifier.
Fluorescence Visualization of Membrane Integrity
A 0.1 mg/mL solution of Alexa Fluor 488 (Molecular Probes) was prepared in 100 mM KCl and 0.01X TBE. The fluorescent solution was introduced into only the top microchannel, with an identical electrolyte solution without Alexa Fluor 488 in the bottom. Alexa 488 was then allowed transport through all available accessible fluid paths connected to the top microchannel via diffusion for 1 h. A 1 V voltage bias was then applied for one minute to drive the dye across the membrane before image acquisition. The optical discharge of a mercury lamp, bandpass filtered at 465 – 495 nm, illuminated the device. The device was imaged with a Nikon Eclipse TE-2000U optical microscope, with an emission bandpass filter of 515 – 555 nm.
Conventional Nanopore Device Fabrication for Noise Comparisons
A punched aperture (70 μm) in a thin (< 1 mm thick) gasket was aligned over a 100 μm × 100 μm freestanding SiNx membrane in a TEM grid to control the exposed membrane area. The TEM grid with the thin gasket was sandwiched between identical 2 mm thick, 3.5 mm diameter silicone elastomer gaskets, and mounted into a custom flow cell. Large fluidic channels of width 1 mm and length of 2 mm in each of the silicone gaskets provided a fluid pathway for cross-flow between two external reservoirs on both sides of the membrane (see Supplementary Section S7). The flow cell was rinsed with ethanol, flushed with deionized water, and then with 1 M KCl, 0.1X TBE solution. Electrodes were placed in reservoirs on either side of the SiNx membrane to measure noise across the exposed membrane area. A 500 μm × 500 μm free-standing SiNx membrane was used instead of the 100 μm × 100 μm SiNx membrane to measure noise across membrane areas larger than (100 μm)2.
DNA Translocations
A solution (1 μg/mL) of homogenous DNA molecules (48,502 bp λ-DNA, Fermentas) in 1 M KCl, 0.1X TBE buffer was loaded on one side of the nanopore, while the other side was loaded with the same buffer without DNA. Ag/AgCl electrodes were inserted into the microchannel access ports, and a 100 mV voltage bias was applied across the nanopore with the negative electrode on the DNA side (voltage drop in the microchannels was negligible). The ionic current was sampled at 250 kHz and low-pass filtered at 100 kHz. The amplitude and duration of DNA translocations were measured off-line and statistics were compiled. 3 μm-wide microchannels were used when measuring DNA translocation in the microfluidics-integrated nanopore device. Identical measurement conditions and concentrations were used when measuring DNA translocations in the custom flow cell, where the exposed membrane area was restricted to (70 μm)2.
RESULTS AND DISCUSSION
The fabricated devices comprised microchannels defined in polydimethylsiloxane (PDMS) that sandwiched a suspended thin dielectric membrane with an array of nanopores (Figure 1). When the 3 layers – two PDMS substrates and the dielectric membrane – are assembled and aligned by appropriately matching the nanopore array pitch with the microchannel width, a single nanopore electrically and fluidically connects the two adjacent microfluidic channels.
Verification of Device Integrity
The device integrity was first examined with a transferred membrane without nanopores using fluorescence leakage experiments. When a fluorescent dye solution was introduced into only one microchannel, the thin dielectric membrane acted as a barrier to fluid flow. No fluorescence was detected in the other microchannel after an hour of diffusion and even after application of a voltage bias across the membrane (Figure 2a), demonstrating that the dielectric membrane was able to isolate the solutions in the neighboring microchannels. I-V curve measurements across a membrane without any nanopores confirmed that the suspended membrane was able to form a high-resistance seal with the polymeric support with a leakage resistance of ~0.7 GΩ for a 1 M KCl, 0.1X TBE solution. There was no observed difference in electrical or fluidic leakage measurements between plasma bonding of the two PDMS substrates (where both substrates were made with polymer to cross-linker ratio of 3:1), and the partial curing method (where the bottom substrate was made with a 10:1 ratio, and top substrate with a 5:1 ratio).
Next, we measured the I-V characteristics of devices with nanopores machined in 50-nm thick dielectric SiNx membranes at different electrolyte concentrations. For these devices, the FIB machining with Ga+ is expected to produce conical nanopores13 with an angle of roughly 14° and typical diameters14 of 25-50 nm. We illustrate the flexibility of the fabrication approach by further tuning the nanopore diameter and surface charge using ALD of alumina (Al2O3) and silicon dioxide (SiO2), which has been shown to be effective for decreasing the noise to enable more sensitive detection of DNA.15 TEM images of a Ga+ machined nanopore before and after ALD of alumina (Figure 2b), verified that performing ALD decreased nanopore diameter by a thickness specified by the number of deposition cycles (~11 nm of Al2O3). Figure 2c illustrates the I-V curves for a pore coated with Al2O3. The device exhibited linear I-V curves where conductance decreased and eventually saturated at low salt concentrations, as expected for a surface-charge dominated regime. The inset also contrasts the I-V curve of the device at 1 M KCl to a control device without a nanopore, demonstrating negligible leakage current in the latter device. Comparing the conductance as a function of electrolyte concentration of the Al2O3-coated nanopore with another device with a SiO2–coated nanopore (Figure 2d) demonstrates that the surface charge of the nanopore can be tuned via ALD. Fitting the data to a model for the conductance (Supplementary Section S1) yields estimated values of the effective nanopore diameters and surface charge (20 nm, −8 mC/m2 for SiO2; 19 nm, 40 mC/m2 for Al2O3), which is consistent with previous reports of SiO2 and Al2O3 nanopores. 16,17 Together, these results demonstrate the mechanical and electrical integrity of the membrane separating the two microchannels.
Microfluidics-Integrated Architecture Reduces Noise
Apart from the flexibility of the fabrication process, the device architecture that results from the membrane transfer approach has the potential to significantly improve the signal-to-noise ratio for single-molecule detection. Sensing nucleic acids and other biomolecules with nanopore systems, and with solid-state nanopores in particular, is generally limited by two coupled parameters; the measurement bandwidth (B) and total system noise (δIRMS)18 Increasing the sampling bandwidth to more accurately detect current changes through the nanopore is accompanied by a concomitant increase in system noise. Several groups have provided strong empirical evidence that the dominant source of noise in nanopore devices at high measurement bandwidths (above 10 kHz) is parasitic capacitance19,20, which stems from two leakage pathways: the electrolyte–Si–electrolyte pathway, and the electrolyte–SiNx membrane–electrolyte pathway. The device architecture described here almost completely eliminates the parasitic capacitance through both pathways. The first pathway is disrupted by replacement of the underlying Si support wafer, which has a relatively high dissipation factor, with a low dissipation factor material, PDMS, which has a dissipation factor similar to that of quartz.21 The second pathway is altered by reduction of the area of the membrane exposed to electrolyte solution to that defined by the width of the microfluidic channels (9-64 μm2), which decreases the total capacitance of the dielectric membrane. In contrast to previous schemes to reduce capacitance and system noise using elastomer gaskets20 (that limit the system to a single nanopore and cannot easily decrease membrane area below 1000 μm2) or multi-layer membranes 22 (that add complexity and cost to the fabrication process), the present configuration significantly reduces device capacitance with a single step that directly embeds a nanopore (or multiple nanopores) into a microfluidic system while eliminating the Si substrate and confining the exposed membrane area.
Development of System Noise Model
To explore the impact of the integrated device architecture on noise performance and to compare it with conventional nanopore systems, we modeled the independent sources of noise (Figure 3). Previously, a detailed mathematical treatment of relevant noise sources in nanopores was developed18, 23, and showed excellent agreement with experimental data. We used these previously developed noise models to explore the contributions of each noise source in conventional nanopore devices (Figure 3a) and to determine the potential limits of noise reduction in our device architecture (Figure 3b).
Figure 3.
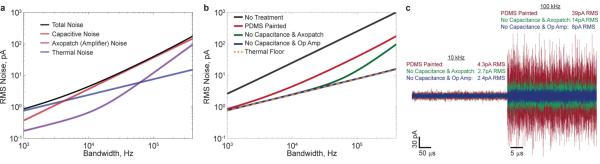
Theoretical calculations of different contributions to nanopore noise. (a) Estimates for individual sources of noise (nanopore resistance, membrane capacitance, and measurement or amplifier), and total RMS noise versus bandwidth obtained from a noise model for PDMS-painted membranes. (b) Estimates for RMS noise versus bandwidth for various device architectures (PDMS-painted, no capacitance and Axopatch, no capacitance and low-noise operational amplifier, and resistive thermal floor). (c) Simulated current traces for different device architectures, providing direct visualization of expected noise reduction from various treatments. The nanopore resistance is assumed to be 30 MΩ. A membrane capacitance of 7.4 pF and dissipation factor of 0.27 are assumed for the PDMS-painted case. The no treatment case reflects a membrane capacitance of 368 pF and dissipation factor of 0.27.
We first theoretically explore individual noise contributions and amalgamate each into a system noise model, which we verify by comparison with experiments in the next section. Each of the noise sources add in quadrature to yield the total RMS noise (δIRMS) as a function of bandwidth B18 (see Supplementary Section S2):
| (1) |
The first term, , is the resistive thermal noise contribution from the nanopore resistance, R. The second term, , is the contribution from the effective membrane and wafer capacitances, C, which also depends on the material properties through the dissipation factor, D. The last two terms are the contributions from measurement noise of the amplifier, , and from flicker noise, , respectively (the terms in brackets denote scaling dependences; see Supplementary Information section S2 for additional details). The noise performance of the amplifier used for these measurements was verified by comparing the RMS noise versus bandwidth for a high-quality resistor against the theoretical model (Supplementary Figures S2 and S3). Based on the assumptions of a nanopore resistance of 30 MΩ, the use of an Axopatch 200B amplifier, and state-of-the-art noise reduction methods (noise levels consistent with PDMS painting of the chip19 equivalently represented by a capacitance of 7.4 pF and a dissipation factor21 of 0.27), the contributions of each noise source are plotted versus bandwidth (Figure 3a). The contribution of flicker noise has been excluded in Figure 3a, primarily because flicker noise depends on the applied voltage and scales weakly with increasing bandwidth as the logarithm of the bandwidth and is not significant for high bandwidth measurements (above ~5 kHz in this case, see Figure S4). Furthermore, provided that the translocations are sufficiently fast, the low frequency noise provides little distortion of the translocation signal. It is clear from Figure 3a that above 10 kHz, capacitive noise is the dominant source of noise in typical solid-state nanopore devices.
Using the same noise model, we explore the total RMS noise as a function of bandwidth for nanopore devices with different amounts of parasitic capacitances expected in different scenarios (Figure 3b). The black and red curves in Figure 3b illustrate that reduction of the membrane area using PDMS gaskets decreases the total RMS noise by ~2x with respect to the no treatment case where no efforts are made to reduce parasitic capacitance (368 pF, 0.27 dissipation factor21). Complete removal of membrane capacitance further reduces noise, most notably at higher bandwidths (> 10 kHz) where the noise is now limited only by the Axopatch measurement instrument (green curve in Figure 3b). In this case, the noise properties of a nanopore system can be approximated to that of an ideal resistor (where the only relevant sources of noise are the resistive thermal and measurement noise). Finally, using a custom low-noise operational amplifier24 (blue curve in Figure 3b) the measurement noise contribution at 100 kHz is suppressed even further, and the noise can be approximated by only the resistive thermal noise. For typical nanopore systems that are currently in use, (where the current is measured with an Axopatch amplifier, as is also the case in the present study), eliminating the membrane capacitance is predicted to reduce RMS noise from 39 pA to 14 pA at 100 kHz, with the noise being dominated by the measurement noise (Figure 3c). With a custom low-noise amplifier24, the RMS noise at 100 kHz is expected to further decrease to < 10 pA. These results suggest that elimination of the membrane capacitance can effectively minimize system noise and enhance the signal-to-noise ratio at high bandwidths. Since high bandwidth sensing is limited by system noise, decoupling capacitive noise and high sampling bandwidths provides an efficient mechanism to sense nucleic acids with higher fidelity.
Systematic Reduction in Exposed Membrane Area Reduces Capacitive Noise
To explore the extent to which our device architecture eliminates capacitive noise and enables the nanopore system to behave as an ideal resistor, we experimentally measured the effect of the underlying Si substrate and exposed membrane area on the total capacitive noise. To ensure that the noise measured arose directly from the fluidic coupling through the substrate and membrane, all noise measurements were made in the absence of a fabricated nanopore and no applied bias (0 V). Given the large magnitude of the seal resistances (> 0.7 GΩ) and that the noise sources add in quadrature, the resistive noise contribution from the seal resistance (theoretical noise contribution of 0.76 pA) is expected to be negligible in comparison to the contributions from the amplifier and capacitance. Thus, any noise above the amplifier noise floor would arise from the device capacitance. We first constructed a control experiment by measuring system noise using a TEM grid with a 500 μm × 500 μm SiNx window (50 nm thick) and a large exposed membrane area (350 μm)2 in a custom flow cell, (Figure 4a, red curve). Using a 100 μm × 100 μm SiNx window (50 nm thick), and reducing the exposed membrane area to (70 μm)2 lowered the PSD and corresponding total RMS noise by an order of magnitude (Figure 4a, orange curve). The experiments in the flow cell are indicative of noise levels in conventional nanopore devices where the membrane is not integrated into the microfluidic architecture. Elimination of the silicon wafer backing and integration of membranes into the microfluidic architecture further lowered the PSD by another order of magnitude (Figure 4a, green curve).
Figure 4.
Measured noise characteristics of microfluidics-integrated and non-integrated membranes and nanopores. (a) Power-spectral density at no applied bias, sampled at 250 kHz and filtered at 100 kHz for ionic current measurements across: a (350 μm)2 exposed SiNx membrane area (red), a (70 μm)2 exposed SiNx membrane area, (orange), and a (6 μm)2 exposed membrane area between two perpendicular microchannels (green). (b) RMS noise over 10 s of data filtered at 100 kHz versus exposed membrane area. Horizontal line represents Axopatch 200B noise floor, obtained by measuring RMS noise in a 200 MΩ high-quality resistor at 0 V. (c) Comparison between predicted and experimental RMS noise as a function of bandwidth for the (70 μm)2 exposed membrane area (orange), and (6 μm)2 area in the microfluidics-integrated device (green). (d) Power-spectral density at no applied bias, sampled at 250 kHz and filtered at 100 kHz in 1 M KCl (dark red) and 10 mM KCl (dark green) with TBE in a 20 nm SiO2 coated nanopore in the microfluidics-integrated device, compared with 10 MΩ (light red) and 100 MΩ (light green) resistors. Nanopore resistances are also indicated.
We then systematically varied the exposed membrane area in a device with a 50 nm thick SiNx membrane (without nanopores or ALD) by modulating the microchannel width (Figure 4b), and found that at small exposed channel areas (< 6 μm)2, the noise from parasitic capacitance of the membrane was negligible compared to noise from the Axopatch measurement instrument. Given the amplifier noise of ~12 pA, we can estimate an upper bound for the capacitive noise contribution from a (15 μm)2 membrane area as 8 pA after a 100 kHz lowpass filter. The square root scaling of the RMS capacitive noise with membrane area implies a maximum capacitive noise contribution of ~2 pA for the 3-μm wide microchannel device at a 100 kHz bandwidth. This range of capacitive noise is lower than the resistive thermal noise for most relevant nanopore resistances (10 MΩ < R < 100 MΩ). Figure 4c depicts the variation of RMS noise with bandwidth of the non-integrated and the microfluidics-integrated devices with (70 μm)2 and (6 μm)2 membrane areas, respectively. A close agreement is observed between the noise predictions for the PDMS painted nanopore and the noise in the (70 μm)2 membrane area device, as well as between the theoretical predictions for the no capacitance case and the noise in the (6 μm)2 membrane area device. This agreement between the theoretical predictions and experimental results validates the utility of the theoretical noise model and further confirms that microfluidic integration reduces noise through elimination of the capacitive contribution.
Finally, we compared the power spectral densities of membrane devices with nanopores (20 nm effective diameter after ALD of SiO2) to high-quality resistors with low capacitance (< 1 pF) (Figure 4d). Introduction of the nanopore results in the addition of a resistance in parallel to the membrane capacitance, which adds thermal noise with a flat PSD (white noise) at zero voltage bias (Figure 4d). Under an applied bias, additional 1/f flicker noise is observed that dominates at lower frequencies but is small at higher frequencies (see Supplementary Section S2 and Figure S4). At high electrolyte concentrations (1 M KCl), and a nanopore resistance of ~10 MΩ, the microfluidics-integrated nanopore PSD indeed consists of a flat white noise spectrum (from the nanopore resistance) whose magnitude is dominated by the thermal noise commensurate with the resistance of the pore (dark and light red curves, Figure 4d). At lower electrolyte concentrations, the higher nanopore resistances result in low thermal noise. In this case, the noise levels are significantly above the expected thermal noise floor, with the dominant source of noise being the amplifier noise for both the nanopore device and the high-quality resistor (dark and light green curves, Figure 4d). Figure 4d demonstrates that the nanopore device behavior is similar to that of an ideal resistor, which implies that microfluidic integration has reduced the capacitive noise to negligible levels compared to other noise sources. The enhanced noise performance of the microfluidics-integrated nanopore device can be explained by an increase in the impedance (elimination of Si substrate and encapsulation in PDMS), and a concomitant decrease of the effective parasitic capacitance of the current pathways in parallel with the nanopore25 (reduction in membrane area) due to the integrated design architecture. As was noted above, the system noise in the microfluidic-integrated nanopore devices can be potentially further decreased with the use of low-noise amplifiers.24 The microfluidics-integrated device architecture could thus enable the potential of low-noise amplifiers to be fully exploited, which is not possible in the presence of significant levels of capacitive noise.
Single DNA Molecule Sensing
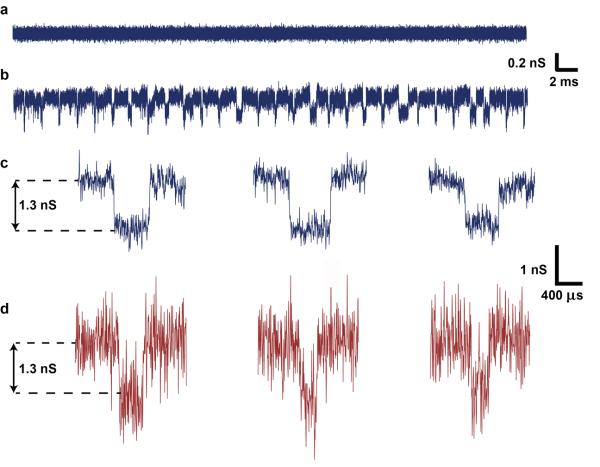
Finally, we demonstrate functionality of the microfluidics-integrated nanopores for sensing single dsDNA molecules, and show that the reduction in noise in the integrated device architecture results in enhanced signal-to-noise ratio compared with the conventional non-integrated architecture. DNA translocation experiments were performed in the custom flow cell (non-integrated) and integrated microfluidic devices with similar nanopore diameters, estimated at 7-9 nm after ALD of Al2O3. Prior to the addition of DNA, no transient conductance changes were observed (Figure 5a). A concentration of 1 μg/mL of 48502 basepair (bp) dsDNA molecules was then added into the devices. Upon application of a 100 mV bias across the nanopore, transient current decreases indicative of single molecule translocations26 were observed in both cases (Figure 5b). The translocation amplitudes (~1.3 nS) and translocation durations (~300 μs) were similar in both the flow cell and microfluidics-integrated devices, and were also consistent with previous reports 15, 17, 26. The fast translocations suggest weak nanopore-DNA interactions that cause reductions in translocation velocity, which is also consistent with the relatively large pore size. However, the microfluidics-integrated device significantly outperformed the non-integrated device in terms of the signal-to-noise ratio, which increased from 3 in the non-integrated device (Figure 5d) to 8 in the microfluidics-integrated nanopore device (Figure 5c), at 100 kHz bandwidth. The results demonstrate that the fabrication method developed here results in functional nanopores capable of detecting single DNA molecules with a significant reduction in the noise.
Figure 5.
Single DNA molecule detection in a microfluidics-integrated nanopore. (a) Open pore current for a 8 nm Al2O3-coated nanopore at a bias of 100 mV. (b) Concatenated current traces after the addition of λ–DNA (48502 bp), in 1 M KCl with TBE, translocating through the nanopore at a bias of 100 mV. (c) Typical translocation events observed through the nanopore. (d) Typical translocation events as measured in the custom flow cell (non-integrated) for a similar nanopore. Integration into the microfluidic device (c) produces significant improvement in the signal to noise ratio when compared to the flow cell (d).
CONCLUSION
In summary, we have developed microfluidics-integrated nanopore devices that can operate with low noise and sense single DNA molecules with high fidelity. The devices are fabricated using a simple, modular process that relies on transfer printing, whereby single nanopores in solid-state membranes are registered between microfluidic channels. Each step of the fabrication scheme is modular and can be independently adjusted to achieve specific functions. Thus, the assembly process is amenable to different nanopore fabrication schemes (TEM drilling, helium ion beam machining) and other materials such as graphene27,28 as well as incorporation of metallic contacts within the lumen of the nanopore29, yielding a powerful pathway to further engineer ionic and biomolecular transport. The device architecture reduces capacitive noise and may be an enabler for exploiting the capabilities of the recently developed low-noise amplifiers24 for nanopores. One of the rate-limiting aspects of current nanopore research has been bridging of microfluidic systems with nanopore sensors and as such, these findings have significant implications for nanopore usage in new formats such as active separation and concentration of specific single molecules for multiple assays. With the development of a simple method to integrate single or multiple solid-state nanopores into microfluidic networks, the present work is a significant step in the direction of rapid, portable biomolecule analysis.
Supplementary Material
Acknowledgements
This work made use of the MRSEC Shared Experimental Facilities at MIT, supported by the National Science Foundation under award number DMR-08-19762. Atomic Layer Deposition and SU-8 photolithography was performed at Harvard’s Center for Nanoscale Systems, which is supported by the National Science Foundation. This work was sponsored under NIH grant R21EB009180 and by the Department of the Air Force under Air Force Contract #FA8721-05-C-0002. Opinions, interpretations, recommendations and conclusions are those of the authors and are not necessarily endorsed by the United States Government.
Footnotes
Competing Financial Interests The authors declare no competing financial interests.
References
- (1).Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Predrag S, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. Nature Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Talaga D, Li J. J. Amer. Chem. Soc. 2009;131:9287–9297. doi: 10.1021/ja901088b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndic M. Nature Nanotech. 2010;5:807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- (4).Iqbal SM, Akin D, Bashir R. Nature Nanotech. 2007;2:243–248. doi: 10.1038/nnano.2007.78. [DOI] [PubMed] [Google Scholar]
- (5).Kowalczyk SW, Hall AR, Dekker C. Nano Lett. 2010;10:324–328. doi: 10.1021/nl903631m. [DOI] [PubMed] [Google Scholar]
- (6).Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Nature. 412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- (7).Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Nature Mater. 2003;2:537–540. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- (8).Cipriany BR, Murphy PJ, Hagarman JA, Cerf A, Latulippe DR, Levy SL, Benitez JJ, Tan CP, Topolancik J, Soloway PD, Craighead HG. Proc. Natl Acad. Sci. USA. 2012;109:8477–8482. doi: 10.1073/pnas.1117549109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kovarik ML, Jacobson SC. Anal. Chem. 2008;80:657–664. doi: 10.1021/ac701759f. [DOI] [PubMed] [Google Scholar]
- (10).Kuo TZ, Cannon DM, Chen Y, Tulock JJ, Shannon MA, Sweedler JV, Bohn PW. Anal. Chem. 2003;75:1861–1867. doi: 10.1021/ac025958m. [DOI] [PubMed] [Google Scholar]
- (11).Mosadegh B, Agarwal M, Torisawa Y, Takayama S. Lab On A Chip. 2010;10:1983–1986. doi: 10.1039/c003590d. [DOI] [PubMed] [Google Scholar]
- (12).Patel AA, Fucetola CP, Moon EE, Smith HI. J. Vac. Sci. Tech. B. 2011;29:06F402. [Google Scholar]
- (13).Patterson N, Adams DP, Hodges VC, Vasile MJ, Michael JR, Kotula PG. Nanotechnology. 2008;19:235304. doi: 10.1088/0957-4484/19/23/235304. [DOI] [PubMed] [Google Scholar]
- (14).Schiedt B, Auvray L, Bacri L, Oukhaled G, Madouri A, Bourhis E, Patriarche G, Pelta J, Jede R, Gierak J. J. Microelec. Engin. 2010;87:1300–1303. [Google Scholar]
- (15).Chen P, Mitsui T, Farmer DB, Golovchenko J, Gordon RG, Branton D. Nano Lett. 2004;4:1333–1337. doi: 10.1021/nl0494001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Storm AJ, Chen JH, Zandbergen HW, Dekker C. Phys. Rev. E. 2005;71:051903. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- (17).Venkatesan BM, Shah AB, Zuo JM, Bashir R. Adv. Func. Mater. 2010;20:1–10. doi: 10.1002/adfm.200902128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Uram JD, Ke K, Mayer M. ACS Nano. 2008;2:857–872. doi: 10.1021/nn700322m. [DOI] [PubMed] [Google Scholar]
- (19).Smeets RMM, Dekker NH, Dekker C. Nanotechnology. 2009;20:095501. doi: 10.1088/0957-4484/20/9/095501. [DOI] [PubMed] [Google Scholar]
- (20).Tabard-Cossa V, Trivedi D, Wiggin M, Jetha N, Marziali A. Nanotechnology. 2007;18:305505. [Google Scholar]
- (21).Naoki M, Kondo S. Polymer. 1983;24:1139–1144. [Google Scholar]
- (22).Dimitrov V, Mirsaidov U, Wang D, Sorsch T, Mansfield W, Miner J, Klemens F, Cirelli R, Yemenicioglu S, Timp G. Nanotechnology. 2010;21:065502. doi: 10.1088/0957-4484/21/6/065502. [DOI] [PubMed] [Google Scholar]
- (23).Smeets RMM, Keyser UF, Dekker NH, Dekker C. Proc. Natl Acad. Sci. USA. 2008;105:417–421. doi: 10.1073/pnas.0705349105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Rosenstein JK, Wanunu M, Merchant CA, Drndic M, Shepard KL. Nature Meth. 2012;9:487–492. doi: 10.1038/nmeth.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Waggoner PS, Kuan AT, Polonsky S, Peng H, Rossnagel SM. J. Vac. Sci. Tech. B. 2011;29:032206. [Google Scholar]
- (26).Venkatesan BM, Dorvel B, Yemenicioglu S, Watkins N, Petrov I, Bashir R. Adv. Mater. 2009;21:1–6. doi: 10.1002/adma.200803786. (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Merchant CA, Healy K, Wanunu M, Ray V, Peterman N, Bartel J, Fischbein MD, Venta K, Luo Z, Johnson CAT, Drndic M. Nano Lett. 2010;10:2915–2921. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- (28).Schneider GF, Kowalczyk SW, Calado VE, Pandraud G, Zandbergen H, Vandersypen LMK, Dekker C. Nano Lett. 2010;10:3163–3167. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- (29).Luan B, Peng H, Polonsky S, Rossnagel S, Stolovitsky G, Martyna G. Phys. Rev. Lett. 2010;104:238103. doi: 10.1103/PhysRevLett.104.238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.