Abstract
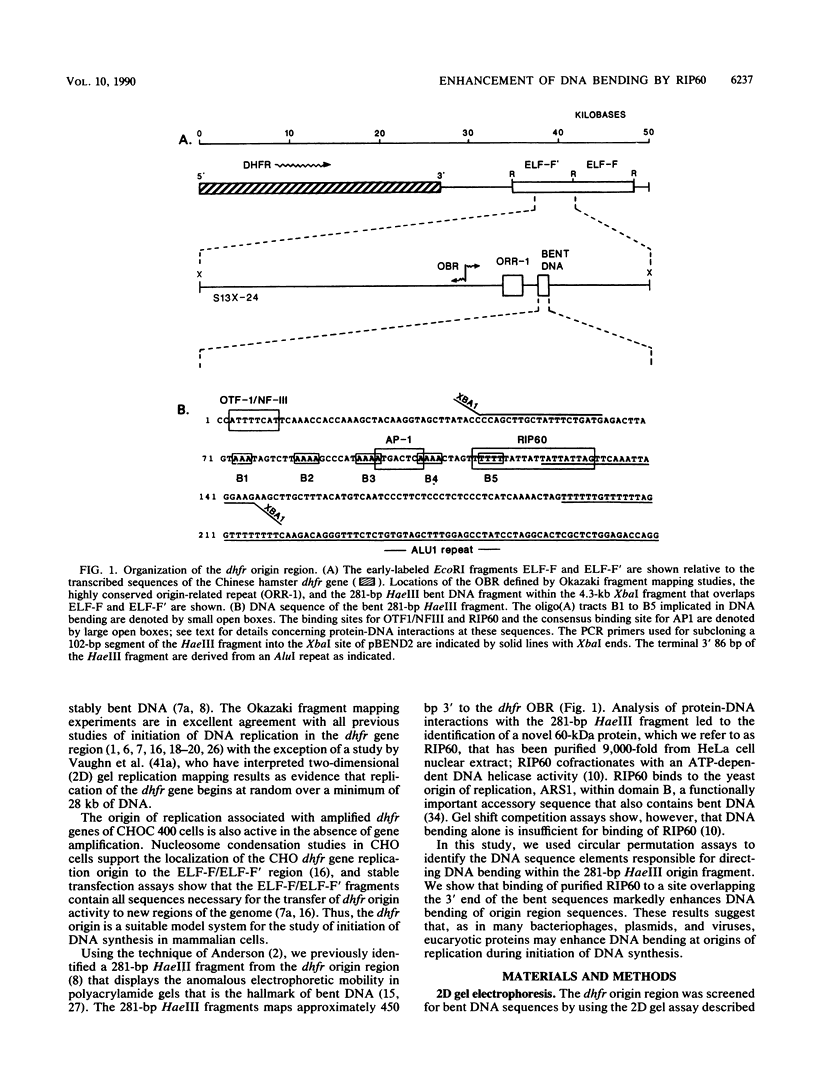
Replication of the Chinese hamster dihydrofolate (dhfr) gene initiates near a 281-bp HaeIII fragment of stably bent DNA that binds RIP60, a 60-kDa origin-specific DNA-binding protein that has been purified from HeLa cell nuclear extract (L. Dailey, M. S. Caddle, N. Heintz, and N. H. Heintz, Mol. Cell. Biol. 10:6225-6235, 1990). Circular permutation assays showed that stable DNA bending in the dhfr origin region fragment was due to the presence of five oligo (dA)3-4 tracts, designated bend elements B1 to B5, that are spaced 10 bp apart. DNA bending directed by elements B1 to B5, as assessed by anomolous migration of DNA fragments on polyacrylamide gels, was accentuated at 4 degrees C. Bend element B5, which is in inverse orientation relative to elements B1 to B4, overlaps an ATT-rich motif that comprises the RIP60 protein-binding site. Gel mobility shift assays with circularly permuted bent DNA fragments and purified RIP60 showed that RIP60 markedly enhanced DNA bending of the dhfr origin region sequences. These results suggest that, as in many plasmids, bacteriophages, and eucaryotic viruses, mammalian DNA-binding proteins may enhance DNA bending near origins of replication during initiation of DNA synthesis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anachkova B., Hamlin J. L. Replication in the amplified dihydrofolate reductase domain in CHO cells may initiate at two distinct sites, one of which is a repetitive sequence element. Mol Cell Biol. 1989 Feb;9(2):532–540. doi: 10.1128/mcb.9.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. N. Detection, sequence patterns and function of unusual DNA structures. Nucleic Acids Res. 1986 Nov 11;14(21):8513–8533. doi: 10.1093/nar/14.21.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel P., Imagawa M., Chiu R., Stein B., Imbra R. J., Rahmsdorf H. J., Jonat C., Herrlich P., Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987 Jun 19;49(6):729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C. Heteronomous DNA. Nucleic Acids Res. 1983 Jun 25;11(12):4141–4155. doi: 10.1093/nar/11.12.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur C. P., Knippers R. Protein-induced bending of the simian virus 40 origin of replication. J Mol Biol. 1988 Oct 20;203(4):1009–1019. doi: 10.1016/0022-2836(88)90125-8. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Selegue J. E., Heintz N. H. Isolation of the origin of replication associated with the amplified Chinese hamster dihydrofolate reductase domain. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7790–7794. doi: 10.1073/pnas.83.20.7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans W. C., Selegue J. E., Heintz N. H. Replication intermediates formed during initiation of DNA synthesis in methotrexate-resistant CHOC 400 cells are enriched for sequences derived from a specific, amplified restriction fragment. Biochemistry. 1986 Jan 28;25(2):441–449. doi: 10.1021/bi00350a025. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Vassilev L. T., Caddle M. S., Heintz N. H., DePamphilis M. L. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990 Sep 7;62(5):955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- Caddle M. S., Lussier R. H., Heintz N. H. Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J Mol Biol. 1990 Jan 5;211(1):19–33. doi: 10.1016/0022-2836(90)90008-A. [DOI] [PubMed] [Google Scholar]
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Dailey L., Caddle M. S., Heintz N., Heintz N. H. Purification of RIP60 and RIP100, mammalian proteins with origin-specific DNA-binding and ATP-dependent DNA helicase activities. Mol Cell Biol. 1990 Dec;10(12):6225–6235. doi: 10.1128/mcb.10.12.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
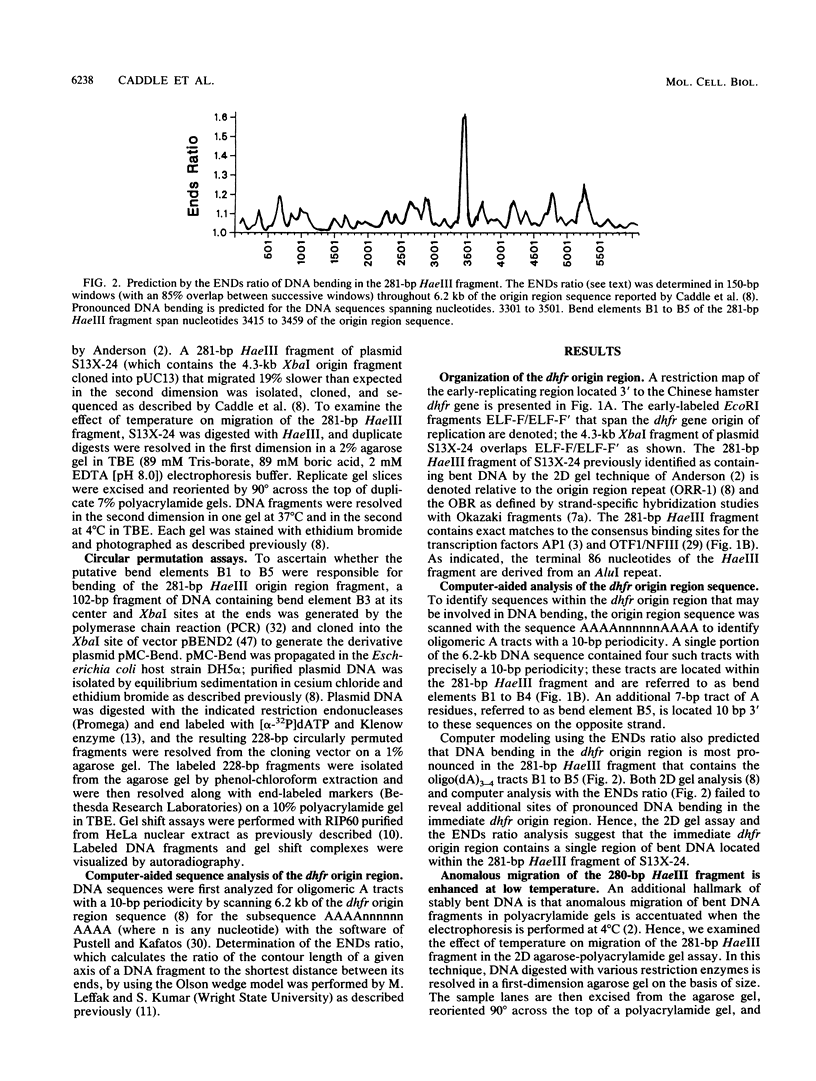
- Eckdahl T. T., Anderson J. N. Computer modelling of DNA structures involved in chromosome maintenance. Nucleic Acids Res. 1987 Oct 26;15(20):8531–8545. doi: 10.1093/nar/15.20.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckdahl T. T., Anderson J. N. Conserved DNA structures in origins of replication. Nucleic Acids Res. 1990 Mar 25;18(6):1609–1612. doi: 10.1093/nar/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Gerard R., Gluzman Y. Functional analysis of the role of the A + T-rich region and upstream flanking sequences in simian virus 40 DNA replication. Mol Cell Biol. 1986 Dec;6(12):4570–4577. doi: 10.1128/mcb.6.12.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough G. W., Lilley D. M. DNA bending induced by cruciform formation. Nature. 1985 Jan 10;313(5998):154–156. doi: 10.1038/313154a0. [DOI] [PubMed] [Google Scholar]
- Handeli S., Klar A., Meuth M., Cedar H. Mapping replication units in animal cells. Cell. 1989 Jun 16;57(6):909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- Hay R. T., DePamphilis M. L. Initiation of SV40 DNA replication in vivo: location and structure of 5' ends of DNA synthesized in the ori region. Cell. 1982 Apr;28(4):767–779. doi: 10.1016/0092-8674(82)90056-3. [DOI] [PubMed] [Google Scholar]
- Heintz N. H., Hamlin J. L. An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4083–4087. doi: 10.1073/pnas.79.13.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. H., Milbrandt J. D., Greisen K. S., Hamlin J. L. Cloning of the initiation region of a mammalian chromosomal replicon. 1983 Mar 31-Apr 6Nature. 302(5907):439–441. doi: 10.1038/302439a0. [DOI] [PubMed] [Google Scholar]
- Heintz N. H., Stillman B. W. Nuclear DNA synthesis in vitro is mediated via stable replication forks assembled in a temporally specific fashion in vivo. Mol Cell Biol. 1988 May;8(5):1923–1931. doi: 10.1128/mcb.8.5.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson E. A., Fritze C. E., Folk W. R., DePamphilis M. L. The origin of bidirectional DNA replication in polyoma virus. EMBO J. 1987 Jul;6(7):2011–2018. doi: 10.1002/j.1460-2075.1987.tb02465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepsel R. R., Khan S. A. Static and initiator protein-enhanced bending of DNA at a replication origin. Science. 1986 Sep 19;233(4770):1316–1318. doi: 10.1126/science.3749879. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. The role of the leucine zipper in the fos-jun interaction. Nature. 1988 Dec 15;336(6200):646–651. doi: 10.1038/336646a0. [DOI] [PubMed] [Google Scholar]
- Laundon C. H., Griffith J. D. Curved helix segments can uniquely orient the topology of supertwisted DNA. Cell. 1988 Feb 26;52(4):545–549. doi: 10.1016/0092-8674(88)90467-9. [DOI] [PubMed] [Google Scholar]
- Leu T. H., Hamlin J. L. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol Cell Biol. 1989 Feb;9(2):523–531. doi: 10.1128/mcb.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Patel I., Bastia D. Conformational changes in a replication origin induced by an initiator protein. Cell. 1985 Nov;43(1):189–197. doi: 10.1016/0092-8674(85)90023-6. [DOI] [PubMed] [Google Scholar]
- O'Neill E. A., Fletcher C., Burrow C. R., Heintz N., Roeder R. G., Kelly T. J. Transcription factor OTF-1 is functionally identical to the DNA replication factor NF-III. Science. 1988 Sep 2;241(4870):1210–1213. doi: 10.1126/science.3413485. [DOI] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A convenient and adaptable package of computer programs for DNA and protein sequence management, analysis and homology determination. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):643–655. doi: 10.1093/nar/12.1part2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramstein J., Lavery R. Energetic coupling between DNA bending and base pair opening. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7231–7235. doi: 10.1073/pnas.85.19.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Schroth G. P., Cook G. R., Bradbury E. M., Gottesfeld J. M. Transcription factor IIIA induced bending of the Xenopus somatic 5S gene promoter. Nature. 1989 Aug 10;340(6233):487–488. doi: 10.1038/340487a0. [DOI] [PubMed] [Google Scholar]
- Snyder M., Buchman A. R., Davis R. W. Bent DNA at a yeast autonomously replicating sequence. Nature. 1986 Nov 6;324(6092):87–89. doi: 10.1038/324087a0. [DOI] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Struhl K. The DNA-binding domains of the jun oncoprotein and the yeast GCN4 transcriptional activator protein are functionally homologous. Cell. 1987 Sep 11;50(6):841–846. doi: 10.1016/0092-8674(87)90511-3. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Why bend DNA? Cell. 1990 Jan 26;60(2):177–180. doi: 10.1016/0092-8674(90)90729-x. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Curved DNA. CRC Crit Rev Biochem. 1985;19(2):89–106. doi: 10.3109/10409238509082540. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N., Sussman J. L. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T., Matsubara K. Multiple initiation sites of DNA replication flanking the origin region of lambda dv genome. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7402–7406. doi: 10.1073/pnas.81.23.7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umek R. M., Linskens M. H., Kowalski D., Huberman J. A. New beginnings in studies of eukaryotic DNA replication origins. Biochim Biophys Acta. 1989 Jan 23;1007(1):1–14. doi: 10.1016/0167-4781(89)90123-1. [DOI] [PubMed] [Google Scholar]
- Vaughn J. P., Dijkwel P. A., Hamlin J. L. Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell. 1990 Jun 15;61(6):1075–1087. doi: 10.1016/0092-8674(90)90071-l. [DOI] [PubMed] [Google Scholar]
- Williams J. S., Eckdahl T. T., Anderson J. N. Bent DNA functions as a replication enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2763–2769. doi: 10.1128/mcb.8.7.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Direct evidence for DNA bending at the lambda replication origin. Science. 1987 Apr 24;236(4800):416–422. doi: 10.1126/science.2951850. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature. 1985 Oct 3;317(6036):451–453. doi: 10.1038/317451a0. [DOI] [PubMed] [Google Scholar]
- Zwieb C., Kim J., Adhya S. DNA bending by negative regulatory proteins: Gal and Lac repressors. Genes Dev. 1989 May;3(5):606–611. doi: 10.1101/gad.3.5.606. [DOI] [PubMed] [Google Scholar]