Abstract
Overexpression of the NEDD9/HEF1/Cas-L scaffolding protein is frequent, and drives invasion and metastasis in breast, head and neck, colorectal, melanoma, lung, and other types of cancer. We have examined the consequences of genetic ablation of Nedd9 in the MMTV-HER2/ERBB2/neu mouse mammary tumor model. Unexpectedly, we found that only a limited effect on metastasis in MMTV-neu;Nedd9−/− mice compared to MMTV-neu;Nedd9+/+ mice, but instead a dramatic reduction in tumor incidence (18% versus 80%), and a significantly increased latency until tumor appearance. Orthotopic reinjection and tail vein injection of cells arising from tumors, coupled with in vivo analysis, indicated tumors arising in MMTV-neu;Nedd9−/− mice had undergone mutational selection that overcame the initial requirement for Nedd9. To better understand the defects in early tumor growth, we compared mammary progenitor cell pools from MMTV-neu;Nedd9−/− versus MMTV-neu;Nedd9+/+ mice. The MMTV-neu;Nedd9−/− genotype selectively reduced both the number and colony-forming potential of mammary luminal epithelial progenitor cells, while not affecting basal epithelial progenitors. MMTV-neu;Nedd9−/−mammospheres had striking defects in morphology and cell polarity. All of these defects were seen predominantly in the context of the HER2/neu oncogene, and were not associated with randomization of the plane of mitotic division, but rather with depressed expression the cell attachment protein FAK, accompanied by increased sensitivity to small molecule inhibitors of FAK and SRC. Surprisingly, in spite of these significant differences, only minimal changes were observed in the gene expression profile of Nedd9−/− mice, indicating critical Nedd9-dependent differences in cell growth properties were mediated via post-transcriptional regulation of cell signaling. Coupled with emerging data indicating a role for NEDD9 in progenitor cell populations during the morphogenesis of other tissues, these results indicate a functional requirement for NEDD9 in the growth of mammary cancer progenitor cells.
Keywords: breast cancer, HER2, mammary precursor cells, drug resistance
Introduction
Breast cancer is the most common malignancy in women, accounting for 1 in 4 total cancer diagnoses, and over 250,000 cases in 2009 in the United States (1). As with many cancers, mortality is predominantly associated with an initial diagnosis of invasive or metastatic disease, and initial or acquired drug resistance. A high percentage of breast cancers have been linked to aberrant expression and/or activity of the estrogen receptor (ER), the progesterone receptor (PR) or the human epidermal growth factor receptor 2 (HER2/ErbB2) (2, 3). HER2 overexpression occurs in 10–34% of patients, and correlates with aggressive disease (4–6). Given the importance of HER2-directed therapies in the clinic (7), it is of considerable interest to better understand the biology of HER2-dependent tumors.
The NEDD9 (also known as HEF1 and CAS-L, (8, 9)) scaffolding protein assembles protein complexes that regulate cell adhesion, migration, division, and survival (reviewed in (10)). Increased expression of NEDD9 has been associated with invasion and metastasis for multiple cancer cell types, including among others glioblastoma, melanoma, colon, and head and neck squamous cell carcinoma (10, 11). One recent report suggests that NEDD9 is similarly upregulated in higher stages of human breast cancer, particularly in a subset of HER2+ and triple negative tumors (12). Mechanistically, we have shown that overexpressed NEDD9 positively regulates migration and invasion of MCF7 cells (13) and other mammary tumor cells (14, 15), promotes expression of matrix metalloproteases (13), and decreases the expression of the epithelial marker E-cadherin. E-cadherin loss typifies the epithelial-mesenchymal transition (EMT) that is a hallmark of tumor invasion, and has also been associated with NEDD9 function (12, 16, 17). Taken together, these data suggest that upregulation of NEDD9 promotes breast tumor aggressiveness.
In contrast to the well-documented role of overexpressed NEDD9, the question of whether there is an actual requirement for NEDD9 during tumor metastasis has not been well addressed. The MMTV-neu (rat HER2/ErbB2) mouse model produces mammary tumors that in many respects recapitulate features of human HER2+ breast cancer (5, 18). Further supporting specific physiological relevance of this model to studies of NEDD9, recent reports indicate that overexpression or ligand stimulation of HER2 induces NEDD9 expression and phosphorylation (associated with protein activity) (19), while earlier work showed NEDD9 overexpression induces HER2 expression, indicating a feedback loop (13). In this study, we crossed MMTV-neu mice to a Nedd9−/− strain of mice (20), and analyzed the development of mammary tumors. Unexpectedly, although loss of Nedd9 had relatively little effect on tumor metastasis, we found that MMTV-neu;Nedd9−/− mice were largely resistant to neu-induced tumor formation, reflecting an important and previously undetected role for NEDD9 at early stages of tumor growth. Extending this study to a direct analysis of the mammary progenitor cell pools from which HER2-dependent tumors originate, we for the first time describe a requirement for NEDD9 in controlling the growth of pre-tumorous luminal progenitor cells.
Results
Nedd9−/− mice are strikingly resistant to HER2/neu-induction of mammary tumors
Comparison of the incidence and time to appearance of mammary tumor formation in Nedd9−/−or Nedd9+/+ mice in the presence or absence of the MMTV-neu transgene revealed a highly significant difference (P = 2.17e-09) (Figure 1A). In 800 days of observation, approximately 80% of MMTV-neu;Nedd9+/+ mice developed mammary tumors with an average latency of 339 days. In contrast, only 18% of MMTV-neu;Nedd9−/− mice developed tumors, with an average latency of 416 days for this small group of mice. However, once formed, the tumors in MMTV-neu;Nedd9−/− mice grew at least as rapidly as those in MMTV-neu;Nedd9+/+ mice (Figure 1B). Histopathological analysis revealed no gross qualitative differences between comparably sized tumors isolated from MMTV-neu;Nedd9+/+ versus MMTV-neu;Nedd9−/− mice (Figure 1C). Not all mice with primary tumors had associated lung metastases at time of euthanasia (13/37 for MMTV-neu;Nedd9+/+, and 4/8 for MMTV-neu;Nedd9−/−). However, among mice with detectable metastases, the number observed was statistically equivalent between genotypes (P=0.5101) (Figure 1D). These results emphasized a critical requirement for Nedd9 prior to the formation of detectable neu-induced tumors, but either a less important role in later stages of tumor growth, or alternatively, the acquisition by tumors of compensating genetic or epigenetic changes that reduce the requirement for Nedd9.
Figure 1. A Comparison of in vivo growth of MMTV-neu;Nedd9+/+ versus MMTV-neu;Nedd9−/− primary tumors and metastases.
A. Kaplan-Meier curve indicates a significant increase in latency and decrease in incidence for tumor formation in MMTV-neu;Nedd9−/− (n = 8 from 46 total) versus MMTV-neu;Nedd9+/+ (n = 39 from 44 total) mice (P = 2.17e-09). B. Kaplan Meier curve indicates days intervening from initial detection of tumor to sacrifice for MMTV-neu;Nedd9−/− versus MMTV-neu;Nedd9+/+ mice (p = 0.01). C. Representative H&E-stained mammary tumors from MMTV-neu;Nedd9+/+ and MMTV-neu;Nedd9−/− tumors, at magnification 20X. D. The number of metastases per lung parenchyma cross sectional area (cm2) are not significantly different between MMTV-neu;Nedd9+/+ and MMTV-neu;Nedd9−/− mice with large primary tumors (p=0.51; mean and SEM are indicated as calculated using Wilcoxon rank sum tests).
Primary cell lines derived from tumors isolated from MMTV-neu;Nedd9+/+ and MMTV-neu;Nedd9−/− mice have similar growth profiles in vitro and in vivo
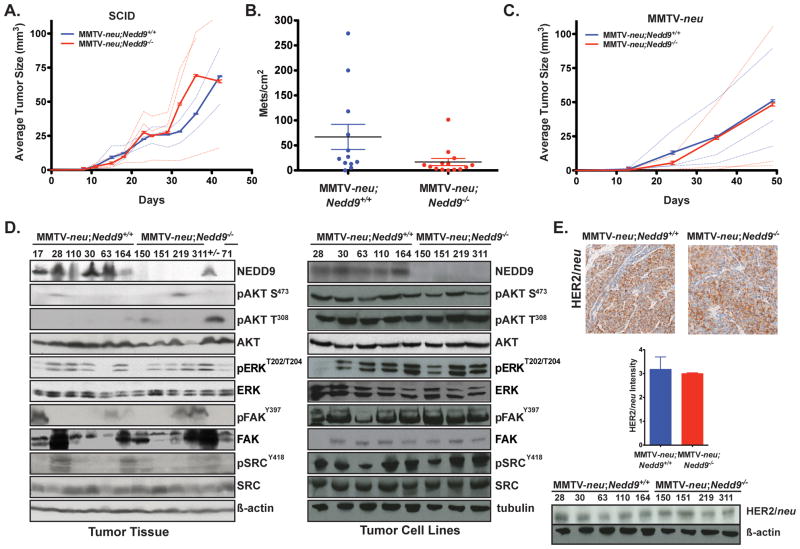
We next generated multiple cell lines from primary MMTV-neu;Nedd9+/+ and MMTV-neu;Nedd9−/− tumors for further analysis. Extensive typing of 3 cell lines of each genotype in vitro indicated comparable rates of anchorage-dependent proliferation (p = 0.49), growth on fibroblast-derived 3D matrix to simulate an in vivo microenvironment (21) (p = 0.57), and rate of attachment (P = 0.77), and a marginal difference in soft agar growth (p = 0.05) between genotypes (Supplemental Figure S1). To determine whether reduced ability to form tumors in vivo persisted in isolated tumor cells, we next compared the behavior of MMTV-neu;Nedd9+/+ and MMTV-neu;Nedd9−/− cell lines in mammary orthotopic xenograft analysis, injecting 5 SCID mice for each of the 6 cell lines analyzed in vitro. In this case, Nedd9 genotype did not influence the rate of xenograft growth (p = 0.1652) (Figure 2A). However, MMTV-neu;Nedd9−/− cell lines were somewhat less effective in colonizing the lung than MMTV-neu;Nedd9+/+ lines following tail vein injection (p=0.042) (Figure 2B). Because NEDD9 supports chemotaxis of B-lymphocytes (20), the difference between the growth of primary tumor cells and growth of xenografts might arise from the lack of a normal immune system in SCID mice. Therefore, we also tested the growth of the same MMTV-neu tumor cell lines in orthotopic xenograft analysis in syngeneic MMTV-neu mice (20). There was again not a significant difference in growth of MMTV-neu;Nedd9+/+ and MMTV-neu;Nedd9−/− cell lines in tumor formation (p=0.2120) (Figure 2C). These results do not exclude a role for Nedd9 in the stroma of mammary tissue: however, consistently, studies have found that Nedd9 expression is extremely low in cultured fibroblasts in contrast to epithelial or lymphoid cells, and immunohistochemistry of breast tumors has shown negative staining in tumor-adjacent stroma (e.g. (12)), arguing against such a role.
Figure 2. Comparison of in vivo growth of MMTV-neu;Nedd9+/+ versus MMTV-neu;Nedd9−/− cell lines in orthotopic xenograft and tail vein injection, and analysis of genome stability.
A. MMTV-neu;Nedd9−/− (red) xenografts versus MMTV-neu;Nedd9+/+ (blue) xenografts in SCID mice (p=0.1652). Data shown reflects each cell line according to genotype; solid lines represent average of 3 lines of each genotype. B. Quantification of lung metastases/cm2, averaged from 5 mice/cell line (30 mice), 28 days after tail vein injection. MMTV-neu;Nedd9−/− tumor cells were less metastatic to lung than MMTV-neu;Nedd9+/+ cells (p=0.042). C. MMTV-neu;Nedd9−/− (red) xenografts versus MMTV-neu;Nedd9+/+ (blue) xenografts in MMTV-neu mice (p=0.2120). Data shown reflects each cell line according to genotype; solid lines represent average of 3 lines of each genotype. D. Western analysis of protein lysates of primary tumor tissue from MMTV-neu;Nedd9+/+ versus MMTV-neu;Nedd9−/− primary tumors (left) indicates no consistent differences in expression or phosphorylation (reflecting activation) for proteins shown. Representative Westerns blots from 3 experiments. +/−, mouse initially genotyped as Nedd9−/−, but subsequently re-genotyped as Nedd9+/− and excluded from all analyses. Western analysis with mammary cell lines derived from tumors indicated (right). E. Top, immunohistochemical staining for HER2/neu from representative H&E counterstained primary tumors. Bottom left, quantification on a scale of 1–4 for all tumors indicates no significant difference (p=0.33). Bottom right, western blot analysis of the primary mammary cell lines indicates similar HER2/neu levels, independent of Nedd9 genotype.
Based on these results, we examined the expression and/or activation of a group of HER2-effectors and NEDD9 signaling partners in MMTV-neu;Nedd9+/+ and MMTV-neu;Nedd9−/−primary tumors and cell lines (Figure 2D). No consistent pattern in activation of AKT, ERK, SRC, or FAK was observed corresponding to genotype in the tumors; rather, sporadic activation of these proteins was observed (Figure 2D, left). In contrast, all pathways were similarly active in cell lines derived from each of the tumors (Figure 2D, right panel). We also confirmed that Nedd9 genotype did not affect expression of the HER2/neu driver oncogene (Figure 2E). Taken together, these data suggested secondary events had occurred to permit proliferation of tumors, compensating for the initial growth defect associated with lack of Nedd9. Comparative genomic hybridization (CGH) analysis of MMTV-neu derived cell lines excluded the occurrence of chromosomal rearrangements, amplification, or deletions (Supplementary Figure S2A, B), indicating such any compensating events likely were more subtle DNA-based changes, or epigenetic.
Reduced mammary luminal progenitor cell populations in Nedd9−/− and MMTV-neu;Nedd9−/− mice
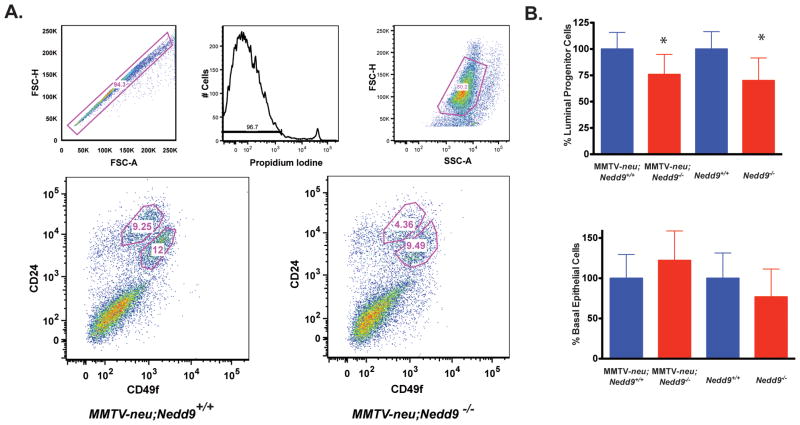
Nedd9 most greatly influenced the initiation stages of MMTV-neu-induced tumor formation. To better investigate the reason for this early defect, we evaluated the progenitor cell populations of the mammary glands from which tumors formed. HER2-positive human breast tumors and MMTV-neu-induced tumors originate from mammary luminal epithelial progenitor cells (4, 18, 22, 23), with the neu protein first detectable in mammary tissue at 4 months of age (24), 2–3 months prior to the average time for palpable tumor formation in the MMTV-neu;Nedd9+/+ mice ((24)). We analyzed cells isolated from the mammary glands of 4 month old mice containing or lacking the MMTV-neu transgene. After enriching for epithelial cells, we used FACS to sort luminal progenitor cells (CD24highCD49fmed/low) and basal progenitor (CD24medCD49fhigh) (25), analyzing the relative proportion of each progenitor cell population to the overall mammary epithelial pool (Figure 3). The CD24highCD49fmed/low luminal progenitor population was proportionally reduced by 25% in MMTV-neu;Nedd9−/− compared to MMTV-neu;Nedd9+/+ mammary epithelial cells (P=0.018) and by 30% in Nedd9−/− versus Nedd9+/+ mammary epithelial cells (P=0.0147) (Figure 3A, 3B top). By contrast, Nedd9 genotype did not affect the frequency of CD24medCD49fhigh basal progenitor cells, regardless of neu genotype (p=0.744, neu+, p=0.648, neu−) (Figure 3A, 3B bottom). To determine whether these differences persist in the cell lines generated from primary tumors, we performed similar analysis of CD24 and CD49f expression for three lines of each genotype (Supplementary Figure S3A). In each case, the only population detected was a novel CD24highCD49fhigh cell class, paralleling other results reported with MMTV-neu cell lines, and suggested to reflect differentiation events induced by sustained cell culture (26, 27). We therefore focused on more in depth analysis of progenitor pools.
Figure 3. Comparative analysis of mammary progenitor cells in MMTV-neu;Nedd9−/−, MMTV-neu;Nedd9+/+, Nedd9+/+, and Nedd9−/− mice.
A. Top, gating plots of FACS sorted mammary epithelial cells. Bottom, representative FACS plot showing relative proportion of CD24high;CD49flow cells harvested from at least 7 independent experiments. B. Top, quantification of percentage of luminal cell progenitor population within bulk mammary epithelial cells was reduced in Nedd9−/− mice positive (p=0.018) and negative (p=0.015) for the MMTV-neu transgene. Bottom, similar analysis performed for the CD24medCD49fhigh basal cell progenitor pool indicates no significant difference based on Nedd9 genotype (p= 0.744 and 0.648, for neu positive and negative). * represents statistical significance by two-tailed t-test < 0.05.
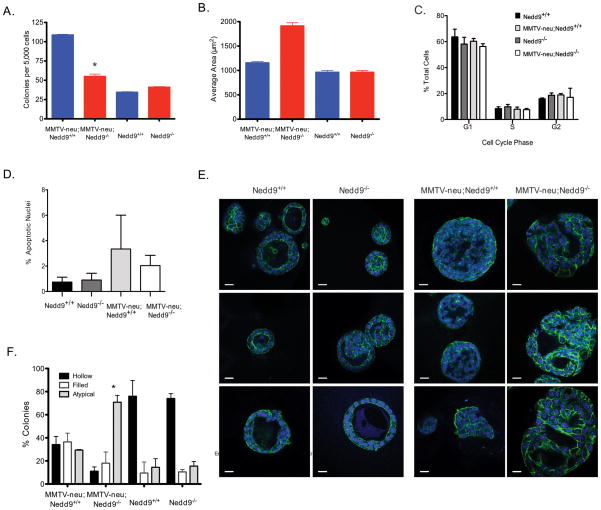
Although the reduced proportional population of luminal (CD24highCD49fmed/low) progenitor cells associated with the MMTV-neu;Nedd9−/− genotype is consistent with the much lower incidence of tumor formation in these mice (Figure 1A), it is not sufficient to explain the magnitude of the reduction in tumor growth unless additional functional impairments are also present. We analyzed the ability of the luminal progenitor cells from all genotypes to form three-dimensional mammospheres in Matrigel (Figure 4). Cells isolated from MMTV-neu;Nedd9−/− mice were less likely to form mammospheres compared to the MMTV-neu;Nedd9+/+ cells (<52%, P<0.0001) (Figure 4A). However, mammospheres that formed from MMTV-neu;Nedd9−/− luminal cells were larger in area (extent of spread in the horizontal axis) than the corresponding MMTV-neu;Nedd9+/+ mammospheres (Figure 4B, p = 0.0022). Nedd9 genotype did not significantly affect proliferation (Figure 4C) nor rate of apoptosis (Figure 4D) of cells in mammospheres, regardless of the status of the neu transgene. We therefore examined mammosphere organization, as an alternative explanation for the increased spread.
Figure 4. Mammosphere growth and morphology in cells with MMTV-neu;Nedd9−/−, MMTV-neu;Nedd9+/+, Nedd9+/+, and Nedd9−/− genotypes.
A. Average number of mammosphere colonies per 5,000 luminal progenitor cells seeded. Error bars indicate SEM. (p<0.0001). B. Quantification of average area (μm2) of each mammosphere colony indicates increase in overall area of mammospheres formed by MMTV-neu;Nedd9−/− versus MMTV-neu;Nedd9+/+ luminal progenitor cells (average 1623 μm2 vs. 922 μm2, p= 0.0022). C. Cell cycle compartmentalization of cells with genotypes indicated, determined by Guava analysis of cells recovered after growth in Matrigel. No significant differences were predicted by genotype. D. Frequency of condensed, pycnotic nuclei, reflecting apoptosis, in cells grown in Matrigel. No significant differences were associated with Nedd9 genotype; presence of MMTV-neu slightly increased rate of apoptosis. E. Immunofluorescence of luminal progenitor cell-derived colonies after 7–10 days in Matrigel. At least 3 independent experiments were performed. Green, phalloidin-488 indicates actin; blue, DAPI. Magnification, 40× oil; scale bar, 50 μm. F. Quantification of hollow, filled, or atypical colonies, from representative images shown in E. Error bars indicate SEM.
In analyzing colony morphology, there was little difference in the percentage of mammospheres from either genotype to have a hollow or solid lumen (p=0.3, filled; p=0.09, hollow). However, the MMTV-neu;Nedd9−/− mammospheres were markedly more likely to be characterized by multiple irregularly shaped joined lumens (“atypical”, 70.8% vs 29.4% in MMTV-neu;Nedd9+/+; p=0.02) (Figure 4E, 4F). Intriguingly, there were no significant differences between colony formation, average area, or morphology based on Nedd9 status in the absence of the MMTV-neu transgene (p = 0.63, 0.88, and 0.54, respectively).
Mammospheres derived from MMTV-neu;Nedd9−/− mammary luminal epithelial cells have defective cell adhesion signaling
In migrating cells, Nedd9 interacts with FAK and SRC to control focal adhesion dynamics and organize the actin cytoskeleton (10). Recent studies of FAK in mammary progenitor cells have found that absence of FAK causes morphological disordering of the mammary gland, and reduces tumor growth (28, 29). We analyzed expression and localization of FAK, SRC, and polymerized F-actin in mammospheres. Quantification of staining intensity found in mammospheres of distinct genotypes indicated a consistent reduction in levels of FAK and of F-actin in MMTV-neu;Nedd9−/− versus MMTV-neu;Nedd9+/+ mammospheres (Figures 5A, B). As an alternative means of probing robustness of FAK and SRC signaling, we treated mammospheres with inhibitors of FAK (Figure 5C) and SRC (Figure 5D), to determine if Nedd9 genotype influenced the ability of these compounds to limit cell growth. These studies confirmed that lower doses of the FAK inhibitor PF-573228 and the SRC inhibitor dasatinib impaired growth of colonies from Nedd9−/− progenitor cells at lower concentrations than at from Nedd9+/+ cells. This distinction was minimized or absent in tumor cell lines derived from mammary tumors, suggesting it had been selected against during tumor outgrowth (Supplemental Figure S3B, S3C).
Figure 5. Defective FAK and SRC signaling in MMTV-neu;Nedd9−/− versus MMTV-neu;Nedd9+/+ mammospheres.
A. Colonies visualized with antibodies to SRC or FAK, or with phalloidin, are shown as (a) image stack to represent entire mammosphere, (b) confocal section, and (c) pseudocolored image, where yellow represents the highest signal intensity and purple represents the lowest. Scale bar, 100 μm. B. Metamorph-based quantification of signal intensity in mammospheres in mammospheres, averaged from 8–11colonies per genotype. P values, *1. 0.0145, *2. 0.0659, *3. 0.0056, relative to Nedd9+/+ control. C. Quantification of average area (μm2) of each mammosphere colony formed from luminal progenitor cells, following plating and maintenance in Matrigel with indicated doses of vehicle or PF-573228 for 10 days. Data are expressed in arbitrary scale. Significant P values (for drug-treated in comparison to vehicle treated for each genotype: 1) 0.770, 2) 0.9650, 3) 1.826E-17, 4) 0.0002, 5) 0.0155, 6) 4.0336E-17, 7) 0.6561, 8) 0.1916, 9) 2.225E-14, 10) 0.6718, 11) 0.00084, and 12) 1.677E-12. D. Analysis as in C, but instead following treatment with the SRC inhibitor dasatinib. Significant P-values include 1) 0.3395, and 2) 6.668E-16. E. Left, representative images of mitotic spindles visualized with antibodies to a-tubulin in mammospheres grown for 6 days in Matrigel; insets represent expanded images. Right, average orientation of the spindle to the plane of the cell surface, reported as in (57). Bold lines represent average and dashed lines represent +/− standard deviation. No significant difference was observed between the MMTV-neu;Nedd9−/−versus MMTV-neu;Nedd9+/+ genotypes.
All of these changes were consistent with defects in cell adhesion signaling affecting mammosphere organization, and by extrapolation, absence of NEDD9 limiting tumor growth through depression of FAK signaling. An alternative mechanism for the generation of disorganized mammospheres would be if NEDD9 affected the plane of mitotic division in reference to the epithelial sheet (from parallel to perpendicular or disorganized), and we have previously demonstrated NEDD9 can influence mitotic progression (30, 31). However, quantification of mitotic planes of mammosphere cells indicated no effect of Nedd9 on this property (Figure 5D), suggesting this mechanism did not contribute to the observed defects.
Given the profound changes in mammosphere growth associated with a Nedd9−/−genotype, we performed gene expression analysis of the luminal progenitor cell populations of the four genotypic cohorts to ask if these differences were reflected in changes in mRNA profile (Supplemental Figure S4). Comparison of the Nedd9+/+ versus the Nedd9−/− genotypes (using a paired t-test to isolate neu status) showed 64 probes with p<0.05 after multiple testing correction and with a log fold change >1 (i.e., a doubling or halving of expression), with the top four being probes for Nedd9 (mean log fold change of 3.2). These included a number of genes associated with genomic stability, including Smc4, Plk4, and CenpE; however, although direct qRT-PCR analysis confirmed these as modestly upregulated, results did not rise to statistical significance (Supplemental Figure S5A). However, Gene Set Analysis (GSA) of the entire group of differentially expressed genes, based on gene annotations in the KEGG, Biocarta, and Reactome databases, identified significant enrichment of the set for genes annotated for mitosis, replicative stress, and centrosome maturation (Supplemental Figure S5B). Inspection of individual genes that were specifically affected by Nedd9−/− genotype only in the context of the MMTV-neu transgene suggested modest downregulation of Idh1, and upregulation of Zeb2, Park2, and Ddr2, genes linked to control of stem cell identity, tumor invasiveness, and drug responsiveness. However, these changes were again not statistically significant following multiple testing correction.
Discussion
The results of this study support three main conclusions. First, they reveal a very substantial requirement for Nedd9 during early stages of the growth of HER2/neu-dependent tumors. While no effect on metastatic potential is seen with primary tumors, this may reflect the small number of MMTV-HER2/neu;Nedd9−/− mice developing primary tumors available for analysis: in contrast, a significant reduction in lung colonization following tail vein injection of MMTV-HER2/neu;Nedd9−/− tumor cells is observed. Second, these results are the first to demonstrate a role for Nedd9 in supporting the abundance and mammosphere-forming potential of mammary luminal progenitor cells. These results support the idea that Nedd9 action is intrinsic to mammary epithelial cells, rather than based on activity in the mammary stromal compartment. Third, they indicated that the defects in mammosphere growth likely involve perturbation of crucial cellular attachment signaling pathways involving FAK and SRC, and surprisingly, do not involve significant reprogramming of the gene expression profile of cells lacking Nedd9.
There are no gross differences in the mammary gland development of Nedd9−/− mice, in the absence of a driver oncogene (15). Here, our data indicated that while the absence of Nedd9 consistently reduced the size of luminal precursor pools, with or without such a driver, changes in these cells relevant to mammosphere growth rate, morphology, and signaling were contingent on the simultaneous presence of HER2/neu. Independently, studies in human tumor cells have shown that NEDD9 expression is particularly important in supporting activity of the ERBB pathway, with depletion of NEDD9 increasing the sensitivity of tumor cells to ERBB1/EGFR-targeting drugs (32, 33). It is also of interest that a recent study profiling transcript levels in a human mammary luminal epithelial model of ERBB2 overexpression demonstrated that NEDD9 was strongly upregulated, together with multiple genes involved in growth factor signaling, proliferation, and cytoskeletal control (34). Although some MMTV-neu;Nedd9−/− tumors arose in our study, indicating they have been able to partially compensate for absence of Nedd9, their activation of relevant signaling pathways is clearly erratic (Figure 2D), and suggests that a diverse set of compensatory mutations have occurred.
In addition to indicating differences in cell proliferation, analysis of mammospheres revealed striking differences in cell organization in MMTV-neu derived cells lacking Nedd9, accompanied by decreased FAK, and increased evidence of actin stress fibers (Figure 5). Nedd9 directly binds and cooperates with FAK to regulate SRC activation and cytoskeletal dynamics (35–37). A number of groups have studied the consequences of FAK knockout for tumor initiation, progression, and mammary morphogenesis (28, 29, 38–40). A common finding was the impact of FAK knockout on mammary progenitor pools, with one study noting that loss of FAK blocked tumor formation driven by transgene-induced activation of the EGFR effectors PI3K and Ras, without affecting normal mammary epithelial cells in the absence of a transgene (40), similar to the results with Nedd9 knockout. Another study emphasized that loss of FAK in cultured mammary epithelial cells led to aberrant branching morphogenesis, with a striking increase in actin stress fibers arising from hyperactivation of Rho kinase (29), similar to our findings. The fact that Nedd9-null cells are sensitized to drug inhibition of FAK or SRC implies a synthetic lethal relationship, similar to that we have demonstrated in earlier genetic experiments in Drosophila, where absence of Nedd9 or FAK or Src could be tolerated in isolation, but not in combination (37), and again compatible with the idea of a fundamentally linked function. Finally, studies in cultured human breast cell lines have shown that integrity of NEDD9-FAK-SRC signaling at focal adhesions is important for both for cell shape control and movement, but also for integrin-dependent survival functions, due to inhibition of anoikis (41, 42), with these processes fundamentally linked: our data imply this linkage is maintained in mammary tumor progenitor cells. Taken in sum, these data suggest the subset of human breast cancers in which NEDD9 is overexpressed (12) are likely to be refractory to inhibition of FAK and SRC, as well as inhibitors of ERBB family members, which has clear implications for clinical treatment.
Although NEDD9 has not previously been studied in mammary progenitor cells, a functional role of Nedd9 in support of progenitor identity is also compatible with other studies of NEDD9 expression that suggest a role in non-differentiated cell lineages. For example, NEDD9 is highly expressed and tightly regulated by integrin-dependent signals in neural crest progenitor cells that differentiate into glia, peripheral neurons, and pigment cells, and in immature blood cells (43–45), and based on this study is likely to support this differentiation process. However, it is possible that significant differences in NEDD9 function exist between cells and tumors of different lineages. Our results differ strikingly from two independent studies of the contribution of Nedd9 to tumor development. In the context of overexpressed Bcr/Abl as oncogenic lesion, absence of Nedd9 accelerated the progression of myeloid leukemia (46); in the context of the overexpressed polyoma virus middle T antigen, absence of Nedd9 initially moderately delayed tumor growth, but then selected hyper-aggressive tumors (15, 33). We hypothesize that one possible explanation for the differences observed between the MMTV-PyVT and MMTV-neu models lies in the comparative oncogenic potential of the PyVT and neu oncogenes. PyVT is pleiotropic, targeting of Shc, c-Src, PP2A, 14-3-3, and PI3K (47). In contrast, ERBB2/HER2/neu stimulates canonical receptor tyrosine kinase (RTK) effector pathways, which activate PI3K, Ras, and PLC-γ, but are less potent in tumor formation in mice (24, 48, 49); in this context, most Nedd9 deficient progenitor cells cannot grow enough to produce tumors, or select highly aggressive clonal descendents. The explanation for the even more striking difference with the BCR/Abl model, in which Nedd9 is tumor suppressive rather than oncogenic, is at this time obscure. Although NEDD9 clearly has important function in myeloid cells (20, 50–52), no systematic analysis of relevant differences in NEDD9-dependent signaling between these and mammary cells has been undertaken. In sum, these data provide a strong justification for future analysis of the role of NEDD9 in the defective signaling of transformed mammary epithelial progenitor populations that initiate human breast cancer.
Materials and Methods
Mouse strains
All experiments involving mice were pre-approved by the FCCC Institutional Animal Care and Use Committee (IACUC). C57Bl/6 Nedd9−/+ mice (20), were backcrossed to the FVB genetic background for over 4 generations (as suggested by (53)), then mated to FVB/N-Tg(MMTVNEU)202ML/J mice (Jackson Laboratories, Bar Harbor, ME) to generate siblings for analysis. All pups were weaned at age 21–23 days, genotyped and segregated. All mice used for tumor analysis were virgin females.
Tumor growth assessment
Mice were examined weekly for tumor onset by palpation of all 10 mammary glands as described previously (15). We used log-rank tests to test for differences between Nedd9 groups in age at euthanasia and age at which tumors developed. We tested for differences in tumor volume over time using linear regression model with interactions estimated by Generalized Estimating Equations (GEE) assuming an exchangeable working correlation matrix (54), with robust standard errors) with age and Nedd9 group entered as covariates. We used Kaplan Meier curves for figures 1A and 1B. We used Wilcoxon rank-sum tests for comparisons of primary and tail-vein metastases. For analysis of xenograft growth in SCID or MMTV-neu mice, we tested for differences in tumor volume using one-way analysis of variation (ANOVA) and a Bonferonni post hoc test.
Analysis of tumor signaling and derivation of cell lines
Mice were euthanized in accordance with IACUC guidelines using isofluorane. The largest tumor and lungs were excised from each tumor-bearing mouse, divided in half, and processed either for pathology or Western analysis as described previously (15). Mammary tumor sections were immunostained with antibodies to ErbB2 (DAKO, Carpinteria, CA). Images were acquired at 10× and 20× using a Nikon Eclipse E600 microscope. Lung metastases were also analyzed as described (15). All analyses were performed by a board-certified pathologist (AK-S) blinded to sample identity. Tumor sections were homogenized immediately for protein and 50 μg of lysate was used for Western analysis as described (15). Mammary tumors from mice were dissected and processed for cell lines as previously described (15). Primary tumor cell lines derived from mammary tumors were grown in 0.1% gelatin-coated T75 flasks in low Ca2+ medium with 5% horse serum as described in (33).
Orthotopic and tail vein injections
For 3 independently derived cell lines of each genotype, 106 cells in 200 μl PBS were injected (bilateral inguinal) into the fourth mammary fat pad of 5 young SCID or MMTV-neu mice using a Hamilton syringe. Mice were examined twice weekly for mammary tumor onset. Tumor measurement, euthanasia, and subsequent processing were performed as described above for primary tumor analysis (33). In a lung-seeding assay, for 3 cell lines per genotype, 106 cells in PBS were injected into the lateral tail vein of 5 SCID mice. Mice were monitored for 4 weeks for any signs of distress, and then euthanized. The lungs and livers were extracted and processed for pathology and analyzed as in (33).
Analysis of mammary progenitor cells and mammosphere growth
Non-tumor bearing mice were euthanized at ~ 4 months old. Thoracic and inguinal mammary glands were removed epithelial cells were isolated using published methods (55) and commercially available reagents (Stem Cell Technologies, Vancouver, British Columbia) and FACS data was analyzed using FlowJo Software (TreeStar, Ashland OR): for full details, please refer to Supplementary Material. Primary mammary epithelial cells sorted by FACS were seeded in Epicult-B medium containing 5% FBS, supplements, EGF and FGF-8b (Stem Cell Technologies, Vancouver, BC) at a density of approximately 53 cells/well in a 8-well chamber slide coated with 35 μl Matrigel (BD Biosciences, San Jose, CA).
After 6 (for alpha-tubulin-stained cells for mitotic analysis) or 10 days (all other mammospheres), cells were fixed and stained as in (56) using indicated antibodies, and imaged using an Inverted Nikon TE2000 with Nikon C1 confocal scanhead at 40× or 60× (alpha-tubulin only) magnification (Nikon, Melville, NY). For each antibody, acquisition settings including laser power, gain, and pinhole settings were all kept consistent between experiments. To quantify F-actin, FAK and SRC staining of 3D reconstructed mammosphere z-stacks Metamorph’s Integrated Morphometry Analysis (total intensity and total area function) was used, with outputs consisting of ratio of total intensity to total area providing integrated intensity readouts. In other to identify positive staining areas, grayscale monochromatic images were assigned individual thresholds (mask) that were individually set for each marker. Images representative of individual 2D slices were obtained from the original stacks used for 3D reconstruction and served as controls. 3D reconstructed and 2D images were also pseudo-colored to highlight differences in staining intensities. The colors ranged between cold colors representing low intensity (i.e., black = 0) and warm colors for high intensities (i.e., 255= white). Following pseudo-coloring, quantification was performed by analysis of multiple colonies for each genotype. Finally, all images were processed in Photoshop (Adobe, San Jose, CA) using identical image input levels followed by identical smart sharpen filter radius and amount changes for each marker set. A two-tailed t-test was used to determine significance.
To measure angle of mitotic spindles, colonies were stained with α-tubulin and e-cadherin (Cell Signaling) for immunofluorescence. Pictures were taken at 1 μm sections and specific z-stacks with identifiable mitotic spindles at the basal edge of the mammospheres were analyzed in Photoshop by drawing a line through the spindle poles and measuring its angle to the basal surface. Angles were noted between 0° and 90° and averaged by genotype. A two-tailed t-test was used to determine significance.
For cell cycle assays, mammosphere colonies were seeded as mentioned previously. On day 10, mammosphere colonies were collected as described in dispase protocol (protocol from manufacturer, BD Biosciences, San Jose, CA). Cell cycle compartmentalization was measured using a Guava Personal Cell Analysis-96 system (Guava Technologies/ Millipore, Billerica, MA). To determine amounts of apoptotic cells in mammospheres, DAPI-stained images taken approximately 8 μm apart were analyzed from z-stacks of 1 μm sections. Total nuclei and pycnotic nuclei were counted and the percentage was analyzed in regard to Nedd9 genotype.
Drug inhibition studies
For drug treatment experiments, tumor cell lines were seeded into 96-well plate. After 24 hours, vehicle (0.2% DMSO) or drugs (the SRC inhibitor dasatinib, obtained from the Fox Chase Cancer Center pharmacy, and the FAK inhibitor PF-573228 (Sigma-Aldrich, St. Louis, MO) were added to the medium. After 72 hours, cell viability was assessed by CellTiter-Blue® (Promega) assay. In parallel, to confirm drug inhibition of targets, cells were seeded into 12-well plate, treated with drug at 24 hours after plating, and collected after 72 hours for Western analysis. Antibodies for Western blotting targeted phospho-SRC(Y418), SRC, phospho-FAK(Y397), and FAK (Cell Signaling, Danvers, MA). Proteins were visualized using the West-Pico system (Pierce, Rockford, IL). For drug treatment experiments with progenitor cells, 24 hours after seeding into Matrigel, progenitor cells were treated with vehicle or drugs, as for tumor cell lines. Media was changed every 4 day. Colonies were fixed at day 10 and stained with DAPI. Area, shape, 3D reconstruction of 1 μm z-stacks and color/wave length separation of individual colonies were measured using Metamorph software.
Supplementary Material
Acknowledgments
We are grateful to Emmanuelle Nicolas (Genotyping and Real Time-PCR Facility of FCCC), Jianming Pei (Cytogenetics and Chromosome Microarray Analysis Facility of FCCC) and Yue-Sheng Liu (Expression Microarray Facility of FCCC), Anthony Lerro and Jackie Valvardi (Laboratory Animal Facility of FCCC), James Oesterling (Cell Sorting Facility of FCCC), Jin Fang (Cell Culture Facility of FCCC), Catherine Renner (Histopathology Facility), Marina Boushra (from Cornell University), Christiaan Honig (from Germantown Academy) and Nicolas G. Day (from St. John’s College) for technical help. We particularly acknowledge Edna Cukierman for expert advice with analysis of microscopic images. R01s CA63366 and CA113342, and Pennsylvania Tobacco Settlement funding (to EAG); and NIH core grant CA-06927 and the Pew Charitable Fund (to Fox Chase Cancer Center) supported this work. JLL was supported by NIH T32 CA-009035 and F32 CA-150553; EI by the American Associates, Ben-Gurion University of the Negev; EP by the Fox Chase Cancer Center Undergraduate Student Summer Fellowship Program. MFO was supported by R21 LM009382.
Footnotes
Conflict of Interest.
The authors declare no conflict of interest.
References
- 1.Pontier SM, Muller WJ. Integrins in mammary-stem-cell biology and breast-cancer progression - a role in cancer stem cells? J Cell Sci. 2009 Jan 15;122(2):207–14. doi: 10.1242/jcs.040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [101038/35021093] [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou C, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. PNAS. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987 Jan 9;235(4785):177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 5.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989 May 12;244(4905):707–12. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 6.Moasser MM. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene. 2007;26(45):6469–87. doi: 10.1038/sj.onc.1210477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baselga J. Treatment of HER2-overexpressing breast cancer. Annals of Oncology 2010. 2010 Oct 1;21(suppl 7):vii36–vii40. doi: 10.1093/annonc/mdq421. [DOI] [PubMed] [Google Scholar]
- 8.Law SF, Estojak J, Wang B, Mysliwiec T, Kruh G, Golemis EA. Human enhancer of filamentation 1, a novel p130cas-like docking protein, associates with focal adhesion kinase and induces pseudohyphal growth in Saccharomyces cerevisiae. Mol Cell Biol. 1996 Jul;16(7):3327–37. doi: 10.1128/mcb.16.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minegishi M, Tachibana K, Sato T, Iwata S, Nojima Y, Morimoto C. Structure and function of Cas-L, a 105-kD Crk-associated substrate-related protein that is involved in beta 1 integrin-mediated signaling in lymphocytes. J Exp Med. 1996 Oct 1;184(4):1365–75. doi: 10.1084/jem.184.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tikhmyanova N, Little JL, Golemis EA. CAS proteins in normal and pathological cell growth control. Cell Mol Life Sci. 2010 Apr;67(7):1025–48. doi: 10.1007/s00018-009-0213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Neill GM, Seo S, Serebriiskii IG, Lessin SR, Golemis EA. A new central scaffold for metastasis: parsing HEF1/Cas-L/NEDD9. Cancer Res. 2007 Oct 1;67(19):8975–9. doi: 10.1158/0008-5472.CAN-07-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong C, Wang C, Wang L, Ma M, Niu C, Sun X, et al. NEDD9 Is a Positive Regulator of Epithelial-Mesenchymal Transition and Promotes Invasion in Aggressive Breast Cancer. PLoS One. 2011;6(7):e22666. doi: 10.1371/journal.pone.0022666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fashena SJ, Einarson MB, O’Neill GM, Patriotis C, Golemis EA. Dissection of HEF1-dependent functions in motility and transcriptional regulation. J Cell Sci. 2002 Jan 1;115(Pt 1):99–111. doi: 10.1242/jcs.115.1.99. [DOI] [PubMed] [Google Scholar]
- 14.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992 Mar;12(3):954–61. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izumchenko E, Singh MK, Plotnikova OV, Tikhmyanova N, Little JL, Serebriiskii IG, et al. NEDD9 promotes oncogenic signaling in mammary tumor development. Cancer Res. 2009 Sep 15;69(18):7198–206. doi: 10.1158/0008-5472.CAN-09-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tikhmyanova N, Golemis EA. NEDD9 and BCAR1 negatively regulate E-cadherin membrane localization, and promote E-cadherin degradation. PLoS One. 2011;6(7):e22102. doi: 10.1371/journal.pone.0022102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008 Oct 31;135(3):510–23. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 18.Lim E, Wu D, Pal B, Bouras T, Asselin-Labat ML, Vaillant F, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12(2):R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagashima T, Oyama M, Kozuka-Hata H, Yumoto N, Sakaki Y, Hatakeyama M. Phosphoproteome and transcriptome analyses of ErbB ligand-stimulated MCF-7 cells. Cancer Genomics Proteomics. 2008 May-Aug;5(3–4):161–8. [PubMed] [Google Scholar]
- 20.Seo S, Asai T, Saito T, Suzuki T, Morishita Y, Nakamoto T, et al. Crk-associated substrate lymphocyte type is required for lymphocyte trafficking and marginal zone B cell maintenance. J Immunol. 2005 Sep 15;175(6):3492–501. doi: 10.4049/jimmunol.175.6.3492. [DOI] [PubMed] [Google Scholar]
- 21.Castello-Cros R, Khan DR, Simons J, Valianou M, Cukierman E. Staged stromal extracellular 3D matrices differentially regulate breast cancer cell responses through PI3K and beta1-integrins. BMC Cancer. 2009;9:94. doi: 10.1186/1471-2407-9-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaillant F, Asselin-Labat ML, Shackleton M, Lindeman GJ, Visvader JE. The emerging picture of the mouse mammary stem cell. Stem Cell Rev. 2007 Jun;3(2):114–23. doi: 10.1007/s12015-007-0018-2. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci U S A. 2003 Dec 23;100(26):15853–8. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10578–82. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006 Feb 23;439(7079):993–7. doi: 10.1038/nature04496. [Research Support, Non-U.S. Gov’t] [DOI] [PubMed] [Google Scholar]
- 26.Lo PK, Kanojia D, Liu X, Singh UP, Berger FG, Wang Q, et al. CD49f and CD61 identify Her2/neu-induced mammary tumor-initiating cells that are potentially derived from luminal progenitors and maintained by the integrin-TGFbeta signaling. Oncogene. 2012 May 24;31(21):2614–26. doi: 10.1038/onc.2011.439. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller PJ, Lin A, Arendt LM, Klebba I, Jones AD, Rudnick JA, et al. Mapping the cellular and molecular heterogeneity of normal and malignant breast tissues and cultured cell lines. Breast Cancer Res. 2010;12(5):R87. doi: 10.1186/bcr2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lahlou H, Sanguin-Gendreau V, Zuo D, Cardiff RD, McLean GW, Frame MC, et al. Mammary epithelial-specific disruption of the focal adhesion kinase blocks mammary tumor progression. Proc Natl Acad Sci U S A. 2007 Dec 18;104(51):20302–7. doi: 10.1073/pnas.0710091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Miltenburg MH, Lalai R, de Bont H, van Waaij E, Beggs H, Danen EH, et al. Complete focal adhesion kinase deficiency in the mammary gland causes ductal dilation and aberrant branching morphogenesis through defects in Rho kinase-dependent cell contractility. Faseb J. 2009 Oct;23(10):3482–93. doi: 10.1096/fj.08-123398. [Research Support, Non-U.S. Gov’t] [DOI] [PubMed] [Google Scholar]
- 30.Dadke D, Jarnik M, Pugacheva EN, Singh MK, Golemis EA. Deregulation of HEF1 impairs M-phase progression by disrupting the RhoA activation cycle. Mol Biol Cell. 2006 Mar;17(3):1204–17. doi: 10.1091/mbc.E05-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pugacheva EN, Golemis EA. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol. 2005 Oct;7(10):937–46. doi: 10.1038/ncb1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Astsaturov I, Ratushny V, Sukhanova A, Einarson MB, Bagnyukova T, Zhou Y, et al. Synthetic Lethal Screen of an EGFR-Centered Network to Improve Targeted Therapies. Science Signaling 2010. 2010 Sep 21;3(140) doi: 10.1126/scisignal.2001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh MK, Izumchenko E, Klein-Szanto AJ, Egleston BL, Wolfson M, Golemis EA. Enhanced genetic instability and dasatinib sensitivity in mammary tumor cells lacking NEDD9. Cancer Res. 2010 Nov 1;70(21):8907–16. doi: 10.1158/0008-5472.CAN-10-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worthington J, Bertani M, Chan H-L, Gerrits B, Timms J. Transcriptional profiling of ErbB signalling in mammary luminal epithelial cells - interplay of ErbB and IGF1 signalling through IGFBP3 regulation. BMC Cancer. 2010;10(1):490. doi: 10.1186/1471-2407-10-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Seventer GA, Salman HJ, Law SF, O’Neill GM, Mullen MM, Franz AA, et al. Focal adhesion kinase regulates beta1 integrin dependent migration through an HEF1 effector pathway. Eur J Imm. 2001;31:1417–27. doi: 10.1002/1521-4141(200105)31:5<1417::AID-IMMU1417>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 36.Law SF, Estojak J, Wang B, Mysliwiec T, Kruh GD, Golemis EA. Human Enhancer of Filamentation 1 (HEF1), a novel p130Cas-like docking protein, associates with FAK, and induces pseudohyphal growth in yeast. Mol Cell Biol. 1996;16:3327–37. doi: 10.1128/mcb.16.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tikhmyanova N, Tulin AV, Roegiers F, Golemis EA. Dcas supports cell polarization and cell-cell adhesion complexes in development. PLoS One. 2010;5(8) doi: 10.1371/journal.pone.0012369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo M, Fan H, Nagy T, Wei H, Wang C, Liu S, et al. Mammary epithelial-specific ablation of the focal adhesion kinase suppresses mammary tumorigenesis by affecting mammary cancer stem/progenitor cells. Cancer Res. 2009 Jan 15;69(2):466–74. doi: 10.1158/0008-5472.CAN-08-3078. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Provenzano PP, Inman DR, Eliceiri KW, Beggs HE, Keely PJ. Mammary epithelial-specific disruption of focal adhesion kinase retards tumor formation and metastasis in a transgenic mouse model of human breast cancer. Am J Pathol. 2008 Nov;173(5):1551–65. doi: 10.2353/ajpath.2008.080308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pylayeva Y, Gillen KM, Gerald W, Beggs HE, Reichardt LF, Giancotti FG. Ras- and PI3K-dependent breast tumorigenesis in mice and humans requires focal adhesion kinase signaling. J Clin Invest. 2009 Feb;119(2):252–66. doi: 10.1172/JCI37160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Law SF, O’Neill GM, Fashena SJ, Einarson MB, Golemis EA. The docking protein HEF1 is an apoptotic mediator at focal adhesion sites. Mol Cell Biol. 2000;20:5184–95. doi: 10.1128/mcb.20.14.5184-5195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Neill GM, Golemis EA. Proteolysis of the docking protein HEF1 and implications for focal adhesion dynamics. Mol Cell Biol. 2001;21:5094–108. doi: 10.1128/MCB.21.15.5094-5108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aquino JB, Lallemend F, Marmigere F, Adameyko II, Golemis EA, Ernfors P. The retinoic acid inducible Cas-family signaling protein Nedd9 regulates neural crest cell migration by modulating adhesion and actin dynamics. Neuroscience. 2009 Sep 15;162(4):1106–19. doi: 10.1016/j.neuroscience.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogel T, Ahrens S, Buttner N, Krieglstein K. Transforming growth factor beta promotes neuronal cell fate of mouse cortical and hippocampal progenitors in vitro and in vivo: identification of Nedd9 as an essential signaling component. Cereb Cortex. 2010 Mar;20(3):661–71. doi: 10.1093/cercor/bhp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tondeur S, Pangault C, Le Carrour T, Lannay Y, Benmahdi R, Cubizolle A, et al. Expression map of the human exome in CD34+ cells and blood cells: increased alternative splicing in cell motility and immune response genes. PLoS One. 2010;5(2):e8990. doi: 10.1371/journal.pone.0008990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seo S, Nakamoto T, Takeshita M, Lu J, Sato T, Suzuki T, et al. Crk-associated substrate lymphocyte type regulates myeloid cell motility and suppresses the progression of leukemia induced by p210Bcr/Abl. Cancer Sci. 2011 Dec;102(12):2109–17. doi: 10.1111/j.1349-7006.2011.02066.x. [Research Support, Non-U.S. Gov’t] [DOI] [PubMed] [Google Scholar]
- 47.Dilworth SM. Polyoma virus middle T antigen and its role in identifying cancer-related molecules. Nat Rev Cancer. 2002;2(12):951–6. doi: 10.1038/nrc946. [101038/nrc946] [DOI] [PubMed] [Google Scholar]
- 48.Dankort DL, Muller WJ. Signal transduction in mammary tumorigenesis: a transgenic perspective. Oncogene. 2000 Feb 21;19(8):1038–44. doi: 10.1038/sj.onc.1203272. [DOI] [PubMed] [Google Scholar]
- 49.Li B, Rosen JM, McMenamin-Balano J, Muller WJ, Perkins AS. neu/ERBB2 cooperates with p53–172H during mammary tumorigenesis in transgenic mice. Mol Cell Biol. 1997 Jun;17(6):3155–63. doi: 10.1128/mcb.17.6.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Astier A, Manie S, Avraham H, Hirai H, Law SF, Zhang Y-Z, et al. The related adhesion focal tyrosine kinase differentially phosphorylates p130Cas and the Cas-like protein, p105HEF1. J Biol Chem. 1997;272:19719–30. doi: 10.1074/jbc.272.32.19719. [DOI] [PubMed] [Google Scholar]
- 51.Astier A, Manie SN, Law SF, Canty T, Hagheyeghi N, Druker BJ, et al. Association of the Cas-like molecule HEF1 with CrkL following integrin and antigen receptor signaling in B cells. Possible relevance to neoplastic lymphohematopoietic cells. Leuk Lymph. 1997;28:65–72. doi: 10.3109/10428199709058332. [DOI] [PubMed] [Google Scholar]
- 52.Manie SN, Beck ARP, Astier A, Law SF, Canty T, Hirai H, et al. Involvement of p130Cas and p105HEF1, a novel Cas-like docking protein, in a cytoskeleton-dependent signaling pathway initiated by ligation of integrin or antigen receptor on human B cells. J Biol Chem. 1997;272:4230–6. doi: 10.1074/jbc.272.7.4230. [DOI] [PubMed] [Google Scholar]
- 53.Lifsted T, Le Voyer T, Williams M, Muller W, Klein-Szanto A, Buetow KH, et al. Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int J Cancer. 1998 Aug 12;77(4):640–4. doi: 10.1002/(sici)1097-0215(19980812)77:4<640::aid-ijc26>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 54.Shults J, Ratcliffe SJ, Leonard M. Improved generalized estimating equastion analysis via xtqls for quasi-least squares in STATA. The STATA Journal. 2007;7:147–66. [Google Scholar]
- 55.Stingl J, Emerman JT, Eaves CJ. Enzymatic dissociation and culture of normal human mammary tissue to detect progenitor activity. Methods Mol Biol. 2005;290:249–63. doi: 10.1385/1-59259-838-2:249. [DOI] [PubMed] [Google Scholar]
- 56.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003 Jul;30(3):256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 57.Hao Y, Du Q, Chen X, Zheng Z, Balsbaugh JL, Maitra S, et al. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical Pins. Curr Biol. 2010 Oct 26;20(20):1809–18. doi: 10.1016/j.cub.2010.09.032. [Research Support, N.I.H., Extramural] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.