Abstract
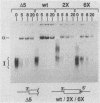
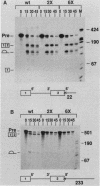
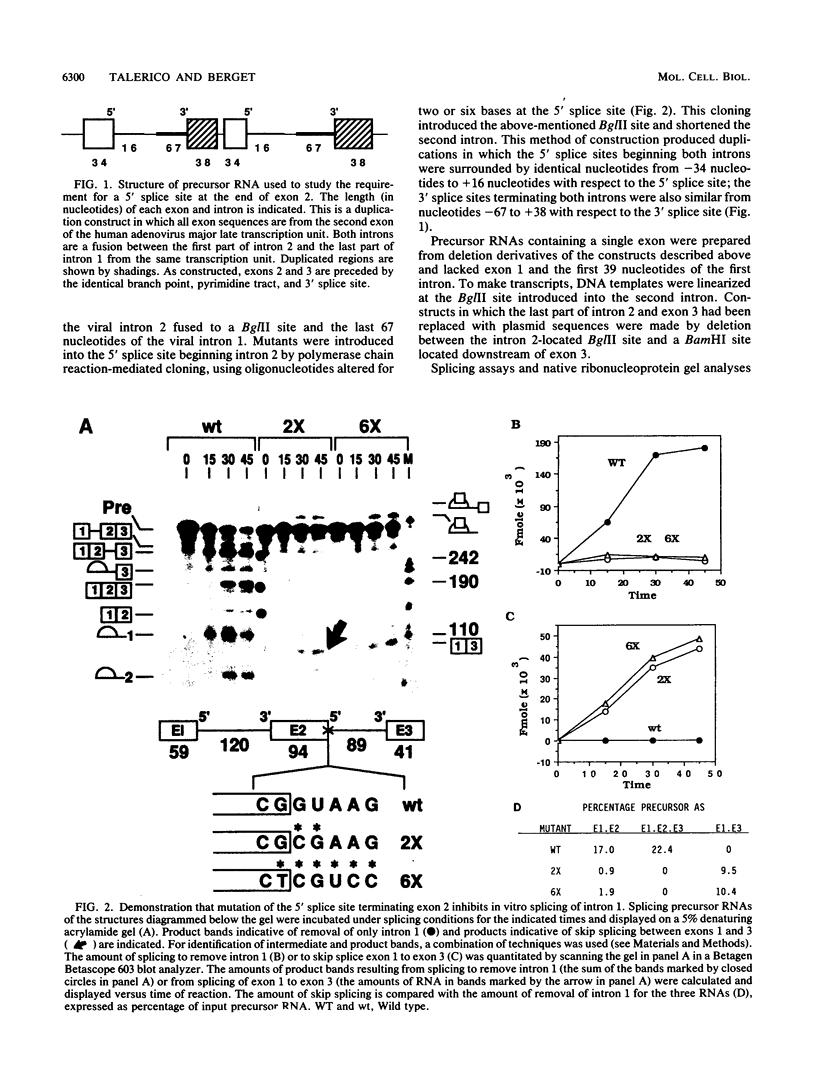
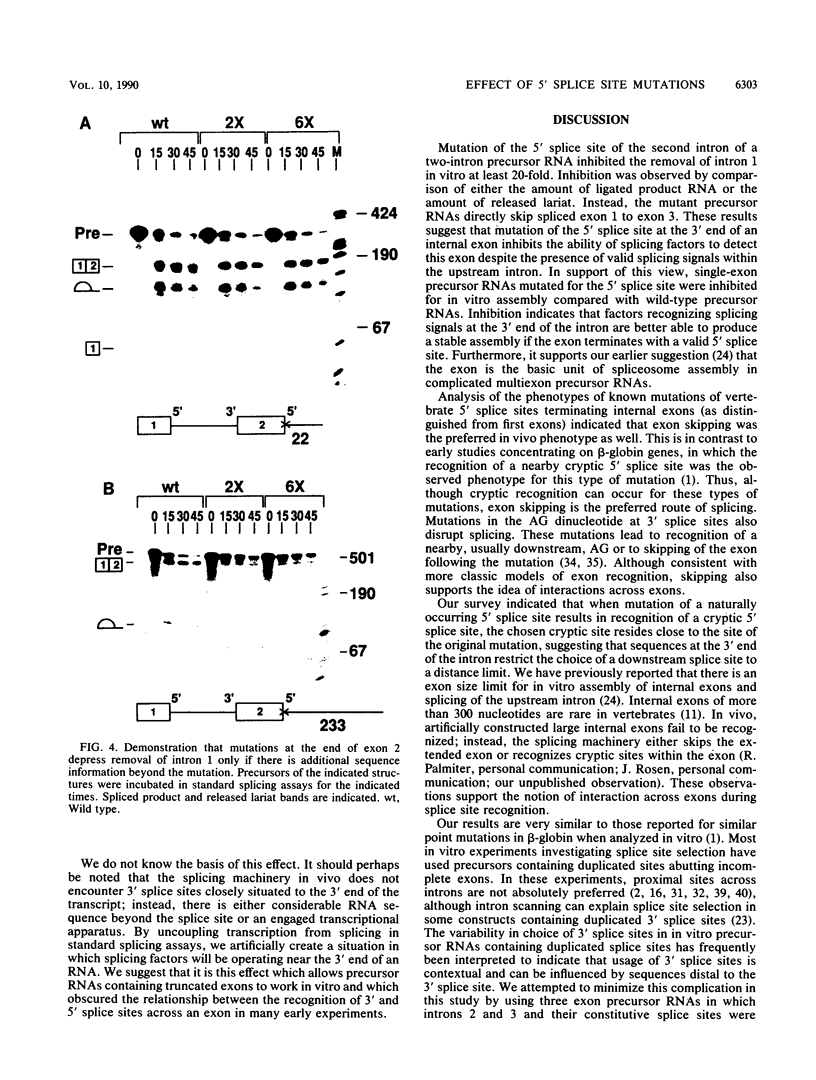
Three exon constructs containing identical intron and exon sequences were mutated at the 5' splice site beginning intron 2 and assayed for the effect of the mutation on splicing of the upstream intron in vitro. Alteration of two or six bases within the 5' splice site reduced removal of intron 1 at least 20-fold, as determined by quantitation of either spliced product or released lariat RNA. The prominent product was skip splicing of exon 1 to exon 3. Examination of complex formation indicated that mutation of the 5' splice site terminating exon 2 depressed the ability of precursor RNAs containing just the affected exon to direct assembly in vitro. These results suggest that mutation at the end of an internal exon inhibits the ability of the exon to be recognized by splicing factors. A comparison of the known vertebrate 5' splice site mutations in which the mutation resides at the end of an internal exon indicated that exon skipping is the preferred phenotype for this type of mutation, in agreement with the in vitro observation reported here. Inhibition of splicing by mutation at the distal and of the exon supports the suggestion that exons, rather than splice sites, are the recognition units for assembly of the spliceosome.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Aebi M., Hornig H., Weissmann C. 5' cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5' splice region, not by the conserved 5' GU. Cell. 1987 Jul 17;50(2):237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- Bakhshi A., Guglielmi P., Siebenlist U., Ravetch J. V., Jensen J. P., Korsmeyer S. J. A DNA insertion/deletion necessitates an aberrant RNA splice accounting for a mu heavy chain disease protein. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2689–2693. doi: 10.1073/pnas.83.8.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt C. R., Morrison S. L., Birshtein B. K., Milcarek C. Loss of a consensus splice signal in a mutant immunoglobulin gene eliminates the CH1 domain exon from the mRNA. Mol Cell Biol. 1984 Jul;4(7):1270–1277. doi: 10.1128/mcb.4.7.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Liao Y. C., Smith D. H., Barrera-Saldaña H. A., Gelinas R. E., Seeburg P. H. The human growth hormone locus: nucleotide sequence, biology, and evolution. Genomics. 1989 May;4(4):479–497. doi: 10.1016/0888-7543(89)90271-1. [DOI] [PubMed] [Google Scholar]
- Dobkin C., Bank A. Reversibility of IVS 2 missplicing in a mutant human beta-globin gene. J Biol Chem. 1985 Dec 25;260(30):16332–16337. [PubMed] [Google Scholar]
- Gallego M. E., Nadal-Ginard B. Myosin light-chain 1/3 gene alternative splicing: cis regulation is based upon a hierarchical compatibility between splice sites. Mol Cell Biol. 1990 May;10(5):2133–2144. doi: 10.1128/mcb.10.5.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R. A., Nguyen P. N., Edwards A., Civitello A. B., Caskey C. T. Multiplex DNA deletion detection and exon sequencing of the hypoxanthine phosphoribosyltransferase gene in Lesch-Nyhan families. Genomics. 1990 Jun;7(2):235–244. doi: 10.1016/0888-7543(90)90545-6. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hawkins J. D. A survey on intron and exon lengths. Nucleic Acids Res. 1988 Nov 11;16(21):9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka Y., Palella T. D., O'Toole T. E., Tarlé S. A., Kelley W. N. Human adenine phosphoribosyltransferase. Identification of allelic mutations at the nucleotide level as a cause of complete deficiency of the enzyme. J Clin Invest. 1987 Nov;80(5):1409–1415. doi: 10.1172/JCI113219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges P. E., Rosenberg L. E. The spfash mouse: a missense mutation in the ornithine transcarbamylase gene also causes aberrant mRNA splicing. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4142–4146. doi: 10.1073/pnas.86.11.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Kudo S., Fukuda M. Structural organization of glycophorin A and B genes: glycophorin B gene evolved by homologous recombination at Alu repeat sequences. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4619–4623. doi: 10.1073/pnas.86.12.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühne T., Wieringa B., Reiser J., Weissmann C. Evidence against a scanning model of RNA splicing. EMBO J. 1983;2(5):727–733. doi: 10.1002/j.1460-2075.1983.tb01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A. I., Konarska M. M., Sharp P. A. A mutational analysis of spliceosome assembly: evidence for splice site collaboration during spliceosome formation. Genes Dev. 1987 Aug;1(6):532–543. doi: 10.1101/gad.1.6.532. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Marvit J., DiLella A. G., Brayton K., Ledley F. D., Robson K. J., Woo S. L. GT to AT transition at a splice donor site causes skipping of the preceding exon in phenylketonuria. Nucleic Acids Res. 1987 Jul 24;15(14):5613–5628. doi: 10.1093/nar/15.14.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Urlaub G., Chasin L. Spontaneous splicing mutations at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1986 Jun;6(6):1926–1935. doi: 10.1128/mcb.6.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M., Pettersson I., Hinterberger M., Karmas A., Steitz J. A. The U1 small nuclear RNA-protein complex selectively binds a 5' splice site in vitro. Cell. 1983 Jun;33(2):509–518. doi: 10.1016/0092-8674(83)90432-4. [DOI] [PubMed] [Google Scholar]
- Ohno K., Suzuki K. Multiple abnormal beta-hexosaminidase alpha chain mRNAs in a compound-heterozygous Ashkenazi Jewish patient with Tay-Sachs disease. J Biol Chem. 1988 Dec 5;263(34):18563–18567. [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988 Nov 18;242(4881):1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- Seidman J. G., Leder P. A mutant immunoglobulin light chain is formed by aberrant DNA- and RNA-splicing events. Nature. 1980 Aug 21;286(5775):779–783. doi: 10.1038/286779a0. [DOI] [PubMed] [Google Scholar]
- Seraphin B., Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989 Oct 20;59(2):349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Sikder S. K., Kabat E. A., Morrison S. L. Alternative splicing patterns in an aberrantly rearranged immunoglobulin kappa-light-chain gene. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4045–4049. doi: 10.1073/pnas.82.12.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatei K., Takemura K., Tanaka H., Masaki T., Ohshima Y. Recognition of 5' and 3' splice site sequences in pre-mRNA studied with a filter binding technique. J Biol Chem. 1987 Aug 25;262(24):11667–11674. [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Structural and functional defects in beta-thalassemia. Prog Clin Biol Res. 1983;134:99–121. [PubMed] [Google Scholar]
- Treisman R., Proudfoot N. J., Shander M., Maniatis T. A single-base change at a splice site in a beta 0-thalassemic gene causes abnormal RNA splicing. Cell. 1982 Jul;29(3):903–911. doi: 10.1016/0092-8674(82)90452-4. [DOI] [PubMed] [Google Scholar]
- Tromp G., Prockop D. J. Single base mutation in the pro alpha 2(I) collagen gene that causes efficient splicing of RNA from exon 27 to exon 29 and synthesis of a shortened but in-frame pro alpha 2(I) chain. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5254–5258. doi: 10.1073/pnas.85.14.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulfendahl P. J., Kreivi J. P., Akusjärvi G. Role of the branch site/3'-splice site region in adenovirus-2 E1A pre-mRNA alternative splicing: evidence for 5'- and 3'-splice site co-operation. Nucleic Acids Res. 1989 Feb 11;17(3):925–938. doi: 10.1093/nar/17.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil D., Bernard M., Combates N., Wirtz M. K., Hollister D. W., Steinmann B., Ramirez F. Identification of a mutation that causes exon skipping during collagen pre-mRNA splicing in an Ehlers-Danlos syndrome variant. J Biol Chem. 1988 Jun 25;263(18):8561–8564. [PubMed] [Google Scholar]
- Weil D., D'Alessio M., Ramirez F., Steinmann B., Wirtz M. K., Glanville R. W., Hollister D. W. Temperature-dependent expression of a collagen splicing defect in the fibroblasts of a patient with Ehlers-Danlos syndrome type VII. J Biol Chem. 1989 Oct 5;264(28):16804–16809. [PubMed] [Google Scholar]
- Weil D., D'Alessio M., Ramirez F., de Wet W., Cole W. G., Chan D., Bateman J. F. A base substitution in the exon of a collagen gene causes alternative splicing and generates a structurally abnormal polypeptide in a patient with Ehlers-Danlos syndrome type VII. EMBO J. 1989 Jun;8(6):1705–1710. doi: 10.1002/j.1460-2075.1989.tb03562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Zapp M. L., Berget S. M. Evidence for nuclear factors involved in recognition of 5' splice sites. Nucleic Acids Res. 1989 Apr 11;17(7):2655–2674. doi: 10.1093/nar/17.7.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zillmann M., Rose S. D., Berget S. M. U1 small nuclear ribonucleoproteins are required early during spliceosome assembly. Mol Cell Biol. 1987 Aug;7(8):2877–2883. doi: 10.1128/mcb.7.8.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillmann M., Zapp M. L., Berget S. M. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol Cell Biol. 1988 Feb;8(2):814–821. doi: 10.1128/mcb.8.2.814. [DOI] [PMC free article] [PubMed] [Google Scholar]