Abstract
The purposes of this study were to evaluate for differences in phenotypic and genotypic characteristics in women who did and did not develop lymphedema (LE) following breast cancer treatment. Breast cancer patients completed a number of self-report questionnaires. LE was evaluated using bioimpedance spectroscopy. Genotyping was done using a custom genotyping array. No differences were found between patients with (n = 155) and without LE (n = 387) for the majority of the demographic and clinical characteristics. Patients with LE had a significantly higher body mass index, more advanced disease and a higher number of lymph nodes removed. Genetic associations were identified for four genes (i.e., lymphocyte cytosolic protein 2 (rs315721), neuropilin-2 (rs849530), protein tyrosine kinase (rs158689), vascular cell adhesion molecule 1 (rs3176861)) and three haplotypes (i.e., Forkhead box protein C2 (haplotype A03), neuropilin-2 (haplotype F03), vascular endothelial growth factor-C (haplotype B03)) involved in lymphangiogensis and angiogenesis. These genetic associations suggest a role for a number of lymphatic and angiogenic genes in the development of LE following breast cancer treatment.
Introduction
Lymphedema (LE) following treatment for breast cancer is the most common form of LE in the industrialized world [1]. LE is caused by a reduction in lymph transport capacity associated with the cancer and its treatment (e.g., surgery, radiation therapy (RT), chemotherapy (CTX)) and results in the accumulation of protein-rich fluid in the interstitial space. LE results in disfigurement, discomfort, and functional impairments. In addition, LE may precipitate cellulitis and lymphangitis [2].
The exact prevalence of LE in breast cancer survivors is unknown with estimates ranging from 3% to 87% [3]. This wide variation is due to differences in measurement methods, inconsistencies in diagnostic criteria, as well as variations in sample characteristics, timing of measurements, and duration of follow-up. However, as noted by Armer [3], if the incidence of LE is conservatively set at 20%, more than 500,000 breast cancer survivors in the United States are affected by this condition.
One of the major goals of LE research is the identification of women at greatest risk for the development of this condition following breast cancer treatment. Risk factors evaluated in the majority of studies done to date focused on patient, disease, and treatment characteristics. However, in four large scale prospective cohort studies [4]–[7], the factors associated with increased risk for LE were not concordant. In a study of 1,287 women (104 were diagnosed with LE) [5], risk factors for LE included: higher stage of disease, increased number of lymph nodes removed, removal of tumor positive nodes, receipt of adjuvant CTX, higher body mass index (BMI), and poorer health status. In a second study of 997 women (133 had LE) [4], risk factors for LE included: being African American, being better educated, higher stage of disease, and removal of at least one lymph node. In a population-based sample of 631 women (237 had LE) [6], hazard ratios for the development of LE were increased following axillary lymph node dissection (ALND) and receipt of CTX. Finally, in a sample of breast cancer patients (n = 2,431) from the Women's Healthy Eating and Living Well Study [7], women with LE (28.5% of the sample) were diagnosed at a younger age, had a higher BMI, had a larger tumor size, had more lymph nodes removed, were more likely to have a mastectomy with radiation therapy (RT) and were more likely to have CTX. While all four studies evaluated a large number of risk factors, the diagnosis of LE relied on patient self-report [5]–[7] or medical record data [4].
While the phenotypic characterization of risk for LE following breast cancer treatment warrants additional investigation, recent evidence suggests that some of the variation in the occurrence and time to onset of LE may be related to genomic determinants [8], [9]. These two studies evaluated candidate genes that are known to play a role in lymphatic development [10]–[15] or were identified in studies of primary LE which are Mendelian inherited disorders. In a study that evaluated hepatocyte growth factor (HGF) and the high affinity hepatocyte growth factor receptor (MET) in 59 women with breast cancer related LE [9], mutations leading to truncation or missense changes in evolutionarily conserved residues of HGF and MET were identified. In a case control study of 188 women (80 diagnosed with LE) [8], mutations in GJC2 that encodes for connexin 47 were identified in women with LE. Taken together, these findings suggest that additional studies are warranted to determine the phenotypic and genotypic factors associated with increased risk for LE following breast cancer treatment. Therefore, the purposes of this study were to evaluate for differences in phenotypic and genotypic characteristics in women who did and did not develop LE following breast cancer treatment.
Methods
Study Samples and Procedures
Demographic, clinical, and genomic data from a cross-sectional study (i.e., LE Study (NR0101282)) and a longitudinal study (i.e., Breast Symptoms Study (CA107091 and CA118658)) were combined for these analyses. Both studies used the same subjective and objective measures. Both studies were approved by the UCSF Committee on Human Research and the CTSI Clinical Research Center Advisory Committee.
LE Study
The LE study used a cross-sectional design to evaluate for differences in phenotypic and genotypic characteristics in women with (n = 74) and without LE (n = 71). Women who were ≥18 years of age, ≥6 months post-treatment for unilateral breast cancer, and with or without upper extremity LE were recruited. Women were excluded for bilateral breast cancer, current upper extremity infection, lymphangitis, preexisting LE, current breast cancer, or contraindications to bioimpedance spectroscopy (BIS) testing. Women were recruited through the National Lymphedema Network website, San Francisco Bay area hospitals, and breast cancer or LE support groups and conferences. Women were evaluated in the Clinical Research Center at UCSF. After obtaining written informed consent, women completed the study questionnaires. Following the completion of the questionnaires, the research staff performed the objective measurements: height, weight, and BIS. A blood sample was drawn for genomic analyses.
Breast Symptoms Study
The Breast Symptoms Study used a prospective, longitudinal design to evaluate neuropathic pain and LE following breast cancer surgery. Women were recruited from Breast Care Centers located in a Comprehensive Cancer Center, two public hospitals, and four community practices. Patients were eligible to participate if they were: adult women (≥18 years) who would undergo breast cancer surgery on one breast; able to read, write, and understand English; agreed to participate, and gave written informed consent. Patients were excluded if they were: having breast cancer surgery on both breasts and/or had distant metastasis at the time of diagnosis. A total of 516 patients were approached to participate, 410 were enrolled in the study (response rate 79.4%), and 398 completed the preoperative assessment. The major reasons for refusal were: too busy, overwhelmed with the cancer diagnosis, or insufficient time available to do the baseline assessment prior to surgery. During the patient's preoperative visit, a clinician explained the study, determined the patient's willingness to participate, and introduced the patient to the research nurse. The research nurse met with the woman, determined eligibility, and obtained written informed consent prior to surgery.
After obtaining written informed consent, the patient completed the enrollment questionnaires prior to surgery. Following the completion of the questionnaires, the research nurse performed the objective measurements: height, weight, and BIS. A blood sample was drawn for genomic analyses. Patients were contacted two weeks after surgery to schedule the first post-surgical appointment. The research nurse met with the patients either in their home or in the Clinical Research Center at 1, 2, 3, 4, 5, 6, 8, 10, and 12 months after surgery. In the second through fifth years of the study, patients were seen every four months. During each of the study visits, the women completed the study questionnaires and had the objective measures done by the research nurse.
Subjective Measures
A demographic questionnaire was used to obtain information on age, marital status, education, ethnicity, employment status, living situation, and financial status. Functional status was evaluated using the Karnofsky Performance Status (KPS) scale that has well established validity and reliability [16], [17]. Patients rated their functional status using the KPS scale that ranged from 30 (I feel severely disabled and need to be hospitalized) to 100 (I feel normal; I have no complaints or symptoms). Patients were asked to indicate if they exercised on a regular basis (yes/no). Clinical information was obtained from patient interviews and medical record reviews.
Self-Administered Comorbidity Questionnaire (SCQ) is a short and easily understood instrument that was developed to measure comorbidity in clinical and health service research settings [18]. The questionnaire consists of 13 common medical conditions that were simplified into language that could be understood without any prior medical knowledge. Patients were asked to indicate if they had the condition using a “yes/no” format. If they indicated that they had a condition, they were asked if they received treatment for it (yes/no; proxy for disease severity) and did it limit their activities (yes/no; indication of functional limitations). Patients were given the option to add two additional conditions not listed on the instrument. For each condition, a patient can receive a maximum of 3 points. Because there are 13 defined medical conditions and 2 optional conditions, the maximum score totals 45 points if the open-ended items are used and 39 points if only the closed-ended items are used. The SCQ has well-established validity and reliability and has been used in studies of patients with a variety of chronic conditions [18]–[22].
Objective Measures
Bioimpedance Spectroscopy (BIS) of LE
BIS measurements, of the affected and unaffected arm, were done using the procedures described by Cornish and colleagues [23]–[25]. Patients were instructed not to exercise or take a sauna within 8 hours of the assessment. In addition, they were asked to refrain from drinking alcohol for 12 hours prior to the assessment. BIS measurements were taken using a single channel BIS device (i.e., SFB7 device; ImpediMed, San Diego, CA in the LE study or the Quantum X Bioelectrical Impedance Device; RJL Systems, Clinton Township, MI in the Breast Symptoms Study). Women removed all jewelry and their skin was prepped with an alcohol wipe prior to surface electrode placement. Patients lay supine on a massage table with their arms 30 degrees from the body and legs not touching for at least 10 minutes prior to the BIS measurements. Electrodes were placed on the dorsum of the wrists adjacent to the ulnar styloid process, the dorsum of the hands just proximal to the third metacarpophalangeal joint, anterior to the ankle joints between the malleoli, and over the dorsum of the feet over the third metatarsal bone just proximal to the third metatarsophalangeal joint. Two ‘measurement’ electrodes were placed at either end of the 40 cm length over which the circumference measurements were made and the ‘drive’ electrodes were placed 8 to 10 cm distal to these measurement electrodes. These electrode sites were chosen, for the segmental measurement of the arm, in preference to the standard shoulder to wrist sites so that direct comparisons could be made between the volumes measured by the circumference method and by the BIS method. Two readings of resistance were obtained from the affected and unaffected arms and averaged for subsequent analyses.
While cases and non-cases of LE were known in the LE study, for the Breast Symptoms Study, LE cases were determined based on the procedures of Cornish and colleagues [23]–[25], using all of the data obtained from each woman during her participation in the study. A woman was defined as a LE case if the resistance ratio for the untreated arm/treated arm prior to surgery was >1.139 or >1.066 for those women who had surgery on the dominant or nondominant side, respectively at any of the BIS assessments.
Methods of Analysis for Phenotypic Data
Data were analyzed using SPSS Version 19 [26]. Descriptive statistics and frequency distributions were generated on the sample characteristics. Independent sample t-tests, Chi-square analyses, and Mann Whitney U tests were done to evaluate for differences in demographic, clinical, and genotypic characteristics between patients with and without LE. Logistic regression analyses were performed to evaluate the association between phenotypic characteristics and LE group membership.
Methods of Analysis for Genomic Data
Gene Selection
Candidate genes for secondary LE include genes that cause monogenic (i.e., primary) forms of LE or genes that cause primary LE in animal models [27]–[31]. While the genetic causes of primary LE might be due to ablative mutations or variations in these candidate genes, secondary LE may be caused by modest functional variations that do not result in critical loss of gene function. Candidate genes for primary LE include angiopoeitin-2 (ANGPT2), elastin microfibril interfacer (EMILIN1), Forkhead box protein C2 (FOXC2), hepatocye growth factor (HGF), lymphocyte cytosolic protein 2 (LCP2), lymphatic vessel endothelial hyaluronan receptor 1 (LYVE1, XLKD1), hepatocyte growth factor receptor (MET), neuropilin-2 (NRP2), Prospero-related homeobox 1 (PROX1), ROR orphan receptor C (RORC), SpSRY-box 17 (SOX17), protein tyrosine kinase (SYK), vascular cell adhesion molecule 1 (VCAM1), and vascular endothelial growth factor-B (VEGFB), -C (VEGFC), -D (VEGFD), -receptor 2 (VEGFR2), and -receptor 3 (VEGFR3).
Blood collection and genotyping
Genomic DNA was extracted from archived buffy coats using the PUREGene DNA Isolation System (Invitrogen, Carlsbad, CA). Of the 543 patients recruited for this study, DNA was recovered from the archive buffy coat of 407 patients (i.e., 110 with and 297 without LE) who provided a blood sample. No differences were found in any demographic and clinical characteristics between patients who did and did not choose to participate in the studies or between those patients who did and did not provide a blood sample for genomic analyses.
Genotyping was performed blinded to LE status and positive and negative controls were included. DNA was quantitated with a Nanodrop Spectrophotometer (ND-1000) and normalized to a concentration of 50 ng/μL (diluted in 10 mM Tris/1 mM EDTA). Samples were genotyped using the GoldenGate genotyping platform (Illumina, San Diego, CA) and processed according to the standard protocol using GenomeStudio (Illumina, San Diego, CA). Signal intensity profiles and resulting genotype calls for each single nucleotide polymorphism (SNP) were visually inspected by two blinded reviewers. Disagreements were adjudicated by a third reviewer.
SNP Selection
A combination of tagging SNPs and literature driven SNPs (i.e., SNPs reported as being associated with LE) were selected for analysis. Tagging SNPs were required to be common (i.e., estimated to have a minor allele frequency ≥.05) in public databases (e.g., HapMap). In order to ensure robust genetic association analyses, quality control filtering of SNPs was performed. SNPs with call rates of <95% or Hardy-Weinberg p-values of <.001 were excluded.
As shown in Table 1, a total of 157 SNPs among the 17 candidate genes (ANGPT2: 25 SNPs, FOXC2: 3 SNPs; HGF: 3 SNPs; LCP2: 11 SNPs; LYVE1: 1 SNP; MET: 15 SNPs; NRP2: 32 SNPs; PROX1: 3 SNPs; RORC: 4 SNPs; SOX17:1 SNP; SYK: 19 SNPs; VCAM1: 7 SNPs; VEGFB: 1 SNP; VEGFC: 8 SNPs; VEGFD: 4 SNPs; VEGFR2: 12 SNPs, VEGFR3: 8 SNPs) passed all quality control filters and were included in the genetic association analyses. The one SNP used to evaluate genetic variation in EMILIN1 did not pass the quality control filters. Potential functional roles of SNPs associated with LE were examined using PUPASuite 2.0 [32], a comprehensive search engine that predicts a series of functional effects (i.e., non-synonymous changes, altered transcription factor binding sites, exonic splicing enhancing or silencing, splice site alterations, microRNA target alterations).
Table 1. Lymphatic and angiogenic genes and single nucleotide polymorphisms analyzed for lymphedema versus no lymphedema.
| Gene | SNP | Position | Chr | MAF | Alleles | Chi Square | p-value | Model |
| ANGPT2 | rs2916716 | 6344643 | 8 | .287 | T>A | .380 | .827 | A |
| ANGPT2 | rs2442468 | 6351358 | 8 | .493 | G>C | .073 | .964 | A |
| ANGPT2 | rs2515409 | 6352330 | 8 | .146 | T>C | .131 | .937 | A |
| ANGPT2 | rs2515413 | 6353028 | 8 | .205 | T>C | 1.464 | .481 | A |
| ANGPT2 | rs2442636 | 6354369 | 8 | .446 | T>C | .346 | .841 | A |
| ANGPT2 | rs2442631 | 6356184 | 8 | .428 | G>A | .096 | .953 | A |
| ANGPT2 | rs1982386 | 6358884 | 8 | .314 | G>A | .542 | .762 | A |
| ANGPT2 | rs2515462 | 6371007 | 8 | .287 | G>A | 2.660 | .264 | A |
| ANGPT2 | rs6990020 | 6373612 | 8 | .380 | T>C | FE | .040 | D |
| ANGPT2 | rs2515466 | 6373694 | 8 | .231 | G>A | .230 | .892 | A |
| ANGPT2 | rs2442608 | 6374028 | 8 | .447 | A>G | .985 | .611 | A |
| ANGPT2 | rs734701 | 6375655 | 8 | .329 | T>C | 1.570 | .456 | A |
| ANGPT2 | rs2515477 | 6376048 | 8 | .131 | C>T | .904 | .636 | A |
| ANGPT2 | rs12674822 | 6376624 | 8 | .440 | G>T | .865 | .649 | A |
| ANGPT2 | rs2515483 | 6379677 | 8 | .353 | G>C | .644 | .725 | A |
| ANGPT2 | rs17552444 | 6381587 | 8 | .269 | A>G | .731 | .694 | A |
| ANGPT2 | rs11989215 | 6383317 | 8 | .338 | A>G | .371 | .831 | A |
| ANGPT2 | rs11989242 | 6383428 | 8 | .478 | G>A | .567 | .753 | A |
| ANGPT2 | rs11137037 | 6383590 | 8 | .326 | A>C | 3.009 | .222 | A |
| ANGPT2 | rs17623313 | 6383749 | 8 | .431 | C>T | .150 | .928 | A |
| ANGPT2 | rs13269021 | 6384171 | 8 | .270 | G>T | .176 | .916 | A |
| ANGPT2 | rs1823375 | 6384406 | 8 | .349 | C>G | 1.537 | .464 | A |
| ANGPT2 | rs13255574 | 6386085 | 8 | .204 | C>T | .208 | .901 | A |
| ANGPT2 | rs2922869 | 6387201 | 8 | .349 | A>G | 1.543 | .462 | A |
| ANGPT2 | rs2515488 | 6390414 | 8 | .458 | A>C | 2.811 | .245 | A |
| HapA04 | .119 | .942 | ||||||
| HapA06 | .533 | .766 | ||||||
| HapB01 | .093 | .955 | ||||||
| HapB04 | .203 | .903 | ||||||
| HapC01 | 3.938 | .140 | ||||||
| HapC04 | 2.438 | .295 | ||||||
| HapD01 | 1.570 | .456 | ||||||
| HapD02 | 1.087 | .581 | ||||||
| HapD03 | .797 | .671 | ||||||
| HapE01 | 1.816 | .403 | ||||||
| HapE02 | .682 | .711 | ||||||
| HapE03 | .860 | .650 | ||||||
| HapF01 | 3.375 | .185 | ||||||
| HapF07 | .158 | .924 | ||||||
| HapG01 | .677 | .713 | ||||||
| HapG02 | 1.392 | .499 | ||||||
| HapG03 | .295 | .863 | ||||||
| HapH02 | 1.523 | .467 | ||||||
| HapH03 | .202 | .904 | ||||||
| FOXC2 | rs34221221 | 85157931 | 16 | .452 | T>C | 2.830 | .243 | A |
| FOXC2 | rs11640590 | 85159945 | 16 | 0.000 | C>A | All patients were homozygous for CC | ||
| FOXC2 | rs1035550 | 85160208 | 16 | .087 | C>T | 3.934 | .140 | A |
| HapA01 | 2.830 | .243 | ||||||
| HapA03 | 6.214 | .045 | ||||||
| HGF | rs5745692 | 81196202 | 7 | .033 | G>C | n/a | n/a | n/a |
| HGF | rs757830 | 81200320 | 7 | .221 | T>C | 1.998 | .368 | A |
| HGF | rs5745616 | 81236292 | 7 | .222 | G>A | 1.315 | .518 | A |
| LCP2 | rs3789184 | 169611615 | 5 | .494 | T>C | 2.794 | .247 | A |
| LCP2 | rs572192 | 169621175 | 5 | .410 | A>G | FE | .047 | D |
| LCP2 | rs10475933 | 169622825 | 5 | .285 | C>T | .409 | .815 | A |
| LCP2 | rs2338872 | 169625132 | 5 | .366 | A>G | 1.492 | .474 | A |
| LCP2 | rs315745 | 169630285 | 5 | .459 | T>C | .431 | .806 | A |
| LCP2 | rs2271146 | 169634968 | 5 | .466 | G>T | .119 | .942 | A |
| LCP2 | rs2338873 | 169641269 | 5 | .141 | G>A | 3.164 | .206 | A |
| LCP2 | rs315721 | 169647616 | 5 | .305 | A>G | FE | .005 | D |
| LCP2 | rs182618 | 169655691 | 5 | .278 | A>G | 4.040 | .133 | A |
| LCP2 | rs6866733 | 169655807 | 5 | .197 | C>T | FE | .026 | D |
| LCP2 | rs315730 | 169656902 | 5 | .434 | A>T | .644 | .725 | A |
| HapA03 | .510 | .775 | ||||||
| HapA04 | 4.304 | .116 | ||||||
| LYVE1 | rs17318858 | 10536263 | 11 | .176 | T>C | .547 | .761 | A |
| MET | rs714180 | 116106238 | 7 | .488 | G>A | .981 | .612 | A |
| MET | rs38841 | 116107162 | 7 | .362 | A>G | .001 | 1.000 | A |
| MET | rs39747 | 116108275 | 7 | .441 | T>C | 2.782 | .249 | A |
| MET | rs38845 | 116109038 | 7 | .456 | G>A | 4.550 | .103 | A |
| MET | rs39748 | 116114666 | 7 | .442 | G>C | 1.177 | .555 | A |
| MET | rs38849 | 116119775 | 7 | .249 | G>C | 2.414 | .299 | A |
| MET | rs2237710 | 116124588 | 7 | .289 | T>G | 1.666 | .435 | A |
| MET | rs38850 | 116124885 | 7 | .246 | G>A | 2.491 | .288 | A |
| MET | rs11762213 | 116126518 | 7 | .053 | G>A | 1.434 | .488 | A |
| MET | rs2299437 | 116128724 | 7 | .244 | G>A | 1.244 | .537 | A |
| MET | rs38857 | 116152649 | 7 | .275 | C>T | 1.954 | .376 | A |
| MET | rs2402118 | 116215809 | 7 | .338 | C>A | 1.915 | .384 | A |
| MET | rs193686 | 116218663 | 7 | .287 | T>C | 1.569 | .456 | A |
| MET | rs2023748 | 116223258 | 7 | .447 | G>A | 2.968 | .227 | A |
| MET | rs1621 | 116224842 | 7 | .318 | A>G | 1.909 | .385 | A |
| HapA01 | 2.427 | .297 | ||||||
| HapA03 | 4.016 | .134 | ||||||
| HapB02 | 2.545 | .280 | ||||||
| HapC02 | 3.296 | .192 | ||||||
| HapC06 | 1.704 | .427 | ||||||
| HapC07 | 2.003 | .367 | ||||||
| NRP2 | rs1517527 | 206252547 | 2 | .408 | C>T | 1.126 | .569 | A |
| NRP2 | rs6711044 | 206252910 | 2 | .479 | T>C | .413 | .813 | A |
| NRP2 | rs1400733 | 206253168 | 2 | .432 | C>G | .329 | .848 | A |
| NRP2 | rs3806577 | 206254395 | 2 | .412 | A>G | .291 | .865 | A |
| NRP2 | rs861078 | 206260223 | 2 | .351 | A>G | 1.320 | .517 | A |
| NRP2 | rs849530 | 206264049 | 2 | .482 | T>G | FE | .042 | R |
| NRP2 | rs950219 | 206268858 | 2 | .308 | G>A | 3.601 | .165 | A |
| NRP2 | rs3771052 | 206269672 | 2 | .293 | G>A | 2.566 | .272 | A |
| NRP2 | rs849556 | 206271503 | 2 | .305 | G>A | 1.858 | .395 | A |
| NRP2 | rs863707 | 206284428 | 2 | .445 | T>G | .678 | .712 | A |
| NRP2 | rs849525 | 206301486 | 2 | .466 | G>A | .635 | .728 | A |
| NRP2 | rs3771033 | 206304055 | 2 | .326 | G>A | 3.654 | .161 | A |
| NRP2 | rs849523 | 206304181 | 2 | .279 | C>T | 1.299 | .522 | A |
| NRP2 | rs1983343 | 206304908 | 2 | .343 | A>G | 1.258 | .533 | A |
| NRP2 | rs849584 | 206309961 | 2 | .318 | G>T | 2.925 | .232 | A |
| NRP2 | rs3771021 | 206318233 | 2 | .405 | C>T | .723 | .697 | A |
| NRP2 | rs849563 | 206318747 | 2 | .187 | A>C | FE | .012 | R |
| NRP2 | rs1996412 | 206320830 | 2 | .473 | A>G | 4.227 | .121 | A |
| NRP2 | rs2241156 | 206323209 | 2 | .358 | G>C | 1.564 | .457 | A |
| NRP2 | rs2241155 | 206323311 | 2 | .358 | C>T | .293 | .864 | A |
| NRP2 | rs3771016 | 206323784 | 2 | .418 | G>A | .005 | .998 | A |
| NRP2 | rs3771010 | 206331839 | 2 | .477 | G>C | .219 | .896 | A |
| NRP2 | rs867344 | 206335660 | 2 | .346 | C>T | .783 | .676 | A |
| NRP2 | rs3771004 | 206339132 | 2 | .305 | G>A | .006 | .997 | A |
| NRP2 | rs16837637 | 206339499 | 2 | .402 | G>A | .417 | .812 | A |
| NRP2 | rs16837641 | 206343114 | 2 | .371 | G>A | FE | .009 | R |
| NRP2 | rs2241153 | 206344368 | 2 | .383 | G>T | 1.748 | .417 | A |
| NRP2 | rs2160328 | 206350582 | 2 | .374 | C>T | 2.150 | .341 | A |
| NRP2 | rs4675542 | 206353492 | 2 | .213 | G>C | 1.940 | .379 | A |
| NRP2 | rs10932125 | 206360545 | 2 | .456 | G>C | 3.922 | .141 | A |
| NRP2 | rs3755232 | 206361208 | 2 | .253 | A>G | 3.096 | .213 | A |
| NRP2 | rs15994 | 206370542 | 2 | .377 | C>G | 1.106 | .575 | A |
| HapA01 | 5.793 | .055 | ||||||
| HapA06 | 2.709 | .258 | ||||||
| HapB01 | 3.427 | .180 | ||||||
| HapB04 | .631 | .730 | ||||||
| HapB05 | 1.340 | .512 | ||||||
| HapC01 | .163 | .922 | ||||||
| HapC02 | .367 | .832 | ||||||
| HapC03 | 3.431 | .180 | ||||||
| HapD01 | .137 | .934 | ||||||
| HapD06 | .430 | .807 | ||||||
| HapE01 | 3.083 | .214 | ||||||
| HapE02 | .372 | .830 | ||||||
| HapE03 | .006 | .997 | ||||||
| HapF01 | 8.937 | .011 | ||||||
| HapF03 | 1.574 | .455 | ||||||
| HapF06 | 1.426 | .490 | ||||||
| PROX1 | rs340874 | 212225879 | 1 | .499 | A>G | 1.919 | .383 | A |
| PROX1 | rs340839 | 212228443 | 1 | .483 | C>T | 2.961 | .227 | A |
| PROX1 | rs726334 | 212246741 | 1 | .284 | C>T | .726 | .696 | A |
| RORC | rs9826 | 150045523 | 1 | .371 | A>G | .097 | .953 | A |
| RORC | rs939595 | 150050312 | 1 | .381 | C>A | .232 | .891 | A |
| RORC | rs7540530 | 150057482 | 1 | .466 | G>A | .420 | .811 | A |
| RORC | rs11204894 | 150059798 | 1 | .225 | G>T | 3.952 | .139 | A |
| HapA01 | .420 | .811 | ||||||
| HapA03 | .624 | .732 | ||||||
| HapA06 | 4.005 | .135 | ||||||
| SOX17 | rs12541742 | 55533707 | 10 | .210 | C>T | FE | .008 | D |
| SYK | rs1319677 | 92605820 | 9 | .484 | T>C | .171 | .918 | A |
| SYK | rs1333633 | 92607393 | 9 | .416 | T>C | .835 | .659 | A |
| SYK | rs3789889 | 92611162 | 9 | .339 | A>G | 2.008 | .366 | A |
| SYK | rs290237 | 92614471 | 9 | .204 | A>G | .034 | .983 | A |
| SYK | rs4744505 | 92619818 | 9 | .391 | G>A | .140 | .932 | A |
| SYK | rs2065583 | 92632383 | 9 | .151 | G>C | 1.174 | .556 | A |
| SYK | rs290213 | 92635628 | 9 | .143 | A>G | 1.116 | .572 | A |
| SYK | rs1870660 | 92637826 | 9 | .163 | C>G | .682 | .711 | A |
| SYK | rs1864202 | 92641776 | 9 | .261 | C>A | 2.573 | .276 | A |
| SYK | rs17489214 | 92642959 | 9 | .186 | G>A | .342 | .843 | A |
| SYK | rs2035073 | 92649240 | 9 | .328 | T>C | .093 | .955 | A |
| SYK | rs11787537 | 92656440 | 9 | .185 | G>A | .576 | .750 | A |
| SYK | rs10993726 | 92660569 | 9 | .214 | C>T | .678 | .713 | A |
| SYK | rs9695993 | 92663585 | 9 | .123 | A>C | .742 | .690 | A |
| SYK | rs290229 | 92674234 | 9 | .259 | C>T | 3.746 | .154 | A |
| SYK | rs10761395 | 92682718 | 9 | .300 | T>C | 2.009 | .366 | A |
| SYK | rs290254 | 92691706 | 9 | .408 | G>C | .078 | .962 | A |
| SYK | rs158689 | 92697582 | 9 | .456 | A>T | FE | .039 | R |
| SYK | rs1049164 | 92698027 | 9 | .178 | G>A | 2.849 | .241 | A |
| HapA01 | .204 | .903 | ||||||
| HapA04 | .688 | .709 | ||||||
| HapB01 | .617 | .735 | ||||||
| HapB02 | .161 | .923 | ||||||
| HapB03 | .029 | .985 | ||||||
| HapC01 | 1.608 | .448 | ||||||
| HapD01 | 9.769 | .008 | ||||||
| HapD03 | .093 | .955 | ||||||
| HapE01 | 3.746 | .154 | ||||||
| HapE02 | .024 | .988 | ||||||
| HapE03 | 2.009 | .366 | ||||||
| HapF01 | 5.000 | .082 | ||||||
| HapF03 | 5.254 | .072 | ||||||
| VCAM1 | rs1409419 | 100955984 | 1 | .498 | T>C | 1.095 | .578 | A |
| VCAM1 | rs3176860 | 100959807 | 1 | .412 | A>G | .880 | .644 | A |
| VCAM1 | rs3176861 | 100959909 | 1 | .201 | C>T | FE | .036 | D |
| VCAM1 | rs3917012 | 100968247 | 1 | .350 | T>G | 3.381 | .184 | A |
| VCAM1 | rs3181088 | 100971296 | 1 | .167 | C>T | 1.520 | .468 | A |
| VCAM1 | rs3176877 | 100975983 | 1 | .396 | T>A | 2.366 | .306 | A |
| VCAM1 | rs3176879 | 100976415 | 1 | .061 | A>G | 1.579 | .454 | A |
| HapA01 | 1.579 | .454 | ||||||
| HapA05 | .825 | .662 | ||||||
| HapB01 | 8.241 | .016 | ||||||
| HapB02 | 2.366 | .306 | ||||||
| VEGFB | rs3741403 | 63756105 | 11 | .433 | G>A | .553 | .759 | A |
| VEGFC | rs1485762 | 177844725 | 4 | .310 | C>T | 5.000 | .082 | A |
| VEGFC | rs7664413 | 177845701 | 4 | .206 | C>T | 6.364 | .041 | A |
| VEGFC | rs6828869 | 177847127 | 4 | .451 | C>G | .329 | .848 | A |
| VEGFC | rs1485766 | 177847878 | 4 | .469 | A>C | .219 | .896 | A |
| VEGFC | rs3775202 | 177848205 | 4 | .494 | A>G | 2.226 | .329 | A |
| VEGFC | rs3775195 | 177858104 | 4 | .238 | C>A | 1.112 | .573 | A |
| VEGFC | rs3775194 | 177860871 | 4 | .407 | C>G | .482 | .786 | A |
| VEGFC | rs1485765 | 177864946 | 4 | .156 | A>G | 1.453 | .484 | A |
| HapA01 | .297 | .862 | ||||||
| HapA05 | 6.173 | .046 | ||||||
| HapB01 | 2.463 | .292 | ||||||
| HapB03 | 7.194 | .027 | ||||||
| HapB04 | 1.254 | .534 | ||||||
| VEGFD | rs6527518 | 15276100 | X | .435 | G>T | .009 | .996 | A |
| VEGFD | rs6418686 | 15297927 | X | .329 | T>C | .116 | .944 | A |
| VEGFD | rs4830939 | 15309204 | X | .324 | G>A | .182 | .913 | A |
| VEGFD | rs6632528 | 15312319 | X | .258 | T>C | .264 | .876 | A |
| HapA01 | .001 | .999 | ||||||
| HapA02 | .474 | .789 | ||||||
| HapA03 | .325 | .850 | ||||||
| VEGFR2 | rs12642307 | 55646938 | 4 | .256 | T>C | 2.524 | .283 | A |
| VEGFR2 | rs1531289 | 55649989 | 4 | .288 | G>A | .111 | .946 | A |
| VEGFR2 | rs7671745 | 55651593 | 4 | .329 | G>A | .557 | .757 | A |
| VEGFR2 | rs6828477 | 55661558 | 4 | .398 | T>C | .801 | .670 | A |
| VEGFR2 | rs2168945 | 55662240 | 4 | .327 | T>G | 1.008 | .604 | A |
| VEGFR2 | rs1870377 | 55667731 | 4 | .254 | T>A | 3.543 | .170 | A |
| VEGFR2 | rs2034965 | 55672557 | 4 | .278 | G>A | .923 | .630 | A |
| VEGFR2 | rs11941492 | 55672967 | 4 | .243 | C>T | 2.307 | .316 | A |
| VEGFR2 | rs10020464 | 55673827 | 4 | .330 | C>T | FE | .025 | D |
| VEGFR2 | rs11133360 | 55677509 | 4 | .455 | T>C | FE | .032 | R |
| VEGFR2 | rs1531290 | 55681319 | 4 | .437 | A>G | .836 | .658 | A |
| VEGFR2 | rs12502008 | 55685799 | 4 | .397 | G>T | 1.751 | .417 | A |
| HapA01 | .386 | .825 | ||||||
| HapA02 | .604 | .739 | ||||||
| HapA03 | .111 | .946 | ||||||
| HapB02 | 3.331 | .189 | ||||||
| HapB03 | 1.096 | .578 | ||||||
| HapB04 | .716 | .699 | ||||||
| HapC01 | 4.155 | .125 | ||||||
| HapC03 | .278 | .870 | ||||||
| HapC04 | 7.730 | .021 | ||||||
| VEGFR3 | rs2242216 | 179974097 | 5 | .430 | G>A | .137 | .934 | A |
| VEGFR3 | rs400330 | 179974268 | 5 | .375 | T>C | .274 | .872 | A |
| VEGFR3 | rs307823 | 179984714 | 5 | .271 | A>G | 1.617 | .446 | A |
| VEGFR3 | rs3797102 | 179987794 | 5 | .392 | T>C | .255 | .880 | A |
| VEGFR3 | rs2290983 | 179991569 | 5 | .453 | T>C | .708 | .702 | A |
| VEGFR3 | rs10085109 | 179993410 | 5 | .484 | C>G | n/a | n/a | n/a |
| VEGFR3 | rs11748431 | 180001347 | 5 | .237 | G>A | 2.704 | .259 | A |
| VEGFR3 | rs307814 | 180006854 | 5 | .387 | C>T | .179 | .914 | A |
| HapA01 | .276 | .871 | ||||||
| HapA03 | .137 | .934 | ||||||
| HapB02 | FE | .643 | ||||||
| HapB03 | 3.693 | .158 | ||||||
| HapB04 | .453 | .797 | ||||||
| HapC01 | .556 | .757 | ||||||
| HapC02 | 3.163 | .206 | ||||||
| HapC03 | .490 | .783 | ||||||
Abbreviations: A = Additive model; ANGPT2 = angiopoeitin-2; Chr = chromosome; D = Dominant model; FOXC2 – forkhead box C2; HGF = hepatocyte growth factor; LCP2 = lymphocyte cytosolic protein 2; LYVE1 = lymphatic vessel endothelial hyaluronan receptor 1 (XLKD1); MAF = minor allele frequency; MET = hepatocyte growth factor receptor; n/a = not assayed because SNP violated Hardy-Weinberg expectations (p<.001); NRP2 – neuropilin-2; PROX1 = prospero-related homeobox 1; R = Recessive model; RORC = ROR orphan receptor C; SNP = single nucleotide polymorphism; SOX17 = SpSRY-box 17; SYK = protein tyrosine kinase; VCAM1 = vascular cell adhesion molecule 1; VEGFB = vascular endothelial growth factor B; VEGFC = vascular endothelial growth factor C; VEGFD = vascular endothelial growth factor D; VEGFR2 = vascular endothelial growth factor receptor 2; VEGFR3 = vascular endothelial growth factor receptor 3.
Statistical Analyses
Allele and genotype frequencies were determined by gene counting. Hardy-Weinberg equilibrium was assessed by the Chi-square or Fisher Exact tests. Measures of linkage disequilibrium ((LD) i.e., D' and r2) were computed from the participants' genotypes with Haploview 4.2. LD-based haplotype block definition was based on D' confidence interval [33].
For SNPs that were members of the same haploblock, haplotype analyses were conducted in order to localize the association signal within each gene and to determine if haplotypes improved the strength of the association with the phenotype. Haplotypes were constructed using the program PHASE version 2.1 [34]. In order to improve the stability of haplotype inference, the haplotype construction procedure was repeated five times using different seed numbers with each cycle. Only haplotypes that were inferred with probability estimates of ≥.85, across the five iterations, were retained for downstream analyses. Haplotypes were evaluated assuming a dosage model (i.e., analogous to the additive model).
Ancestry informative markers (AIMS) were used to minimize confounding due to population stratification [35]–[37]. Homogeneity in ancestry among participants was verified by principal component analysis [38], using Helix Tree (Golden Helix, Bozeman, MT). Briefly, the number of principal components (PCs) was sought which distinguished the major racial/ethnic groups in the sample by visual inspection of scatter plots of orthogonal PCs (i.e., PC 1 versus PC2, PC2 versus PC3). This procedure was repeated until no discernible clustering of patients by their self-reported race/ethnicity was possible (data not shown). One hundred and six AIMs were included in the analysis. The first three PCs were selected to adjust for potential confounding due to population substructure (i.e., race/ethnicity) by including the three covariates in all regression models.
For association tests, three genetic models were assessed for each SNP: additive, dominant, and recessive. Barring trivial improvements (i.e., delta <10%), the genetic model that best fit the data, by maximizing the significance of the p-value, was selected for each SNP. Logistic regression analysis, that controlled for significant covariates, as well as genomic estimates of and self-reported race/ethnicity, was used to evaluate the association between genotype and LE group membership. A backwards stepwise approach was used to create the most parsimonious model. Genetic model fit and both unadjusted and covariate-adjusted odds ratios were estimated using STATA version 9 [39].
As was done in our previous studies [40]–[42], based on recommendations in the literature [43], [44], the implementation of rigorous quality controls for genomic data, the non-independence of SNPs/haplotypes in LD, and the exploratory nature of the analyses, adjustments were not made for multiple testing. In addition, significant SNPs identified in the bivariate analyses were evaluated further using regression analyses that controlled for differences in phenotypic characteristics, potential confounding due to population stratification, and variation in other SNPs/haplotypes within the same gene. Only those SNPs that remained significant were included in the final presentation of the results. Therefore, the significant independent associations reported are unlikely to be due solely to chance. Unadjusted (bivariate) associations are reported for all SNPs passing quality control criteria in Table 1 to allow for subsequent comparisons and meta-analyses.
Results
Differences in demographic and clinical characteristics
As shown in Table 2, no differences were found between patients with and without LE for the majority of the demographic and clinical characteristics. Patients with LE had a significantly higher BMI and a lower KPS score, and were more likely to report lung disease. In addition, patients with LE had a higher number of lymph nodes removed, a higher number of positive nodes, more advanced disease at the time of diagnosis, were less likely to have had a SLNB, and were more likely to have had an ALND, had received CTX prior to or following surgery, and had received RT following surgery.
Table 2. Differences in demographic and clinical characteristics between patients with (n = 155) and without (n = 387) lymphedema.
| Characteristic | No Lymphedema | Lymphedema | Statistics |
| Mean (SD) | Mean (SD) | ||
| Age (years) | 54.9 (11.1) | 56.2 (10.8) | NS |
| Education (years) | 16.0 (2.7) | 15.8 (2.8) | NS |
| Age at menopause (years) | 47.8 (7.2) | 46.7 (9.1) | NS |
| Body mass index (kg/m2) | 26.1 (5.6) | 28.2 (6.7) | p = .001 |
| Karnofsky Performance Status score | 93.3 (9.7) | 91.1 (11.1) | p = .028 |
| Comorbidity score | 4.0 (2.9) | 4.5 (3.3) | NS |
| Number of nodes removed | 5.8 (6.3) | 10.9 (9.0) | p<.0001 |
| Number of positive nodes | 0.7 (1.7) | 1.7 (3.4) | p = .009 |
Abbreviations: kg = kilograms, m2 – meter squared, NS = not significant, SD = standard deviation.
Regression analyses for phenotypic predictors of LE
As shown in Table 3, two regression analyses were done to evaluate the associations between phenotypic characteristics and LE group membership. In the first regression analysis, that included BMI, stage of disease, SLNB status, number of lymph nodes removed, receipt of CTX, and receipt of RT predicted 13.8% of the variance in LE group membership (p<0.0001). The odds of developing LE increased significantly for women who had a higher BMI, had a higher stage of disease at diagnosis, had a higher number of lymph nodes removed, had received CTX prior to or following surgery, and had received RT. The odds of having LE decrease significantly in women who had a SLNB.
Table 3. Multiple logistic regression analyses for phenotypic predictors of the development of lymphedema.
| Regression analysis WITHOUT the inclusion of genomic and self-reported race/ethnicity | |||||
| Predictor | Odds Ratio | Standard Error | 95% CI | Z | p-value |
| BMI | 1.06 | 0.018 | 1.025, 1.097 | 3.39 | 0.001 |
| Stage of disease Stage 0 versus I Stage 0 versus II Stage 0 versus III and IV | 3.23 3.18 3.62 | 1.590 1.622 2.151 | 1.234, 8.479 1.171, 8.640 1.129, 11.603 | 2.39 2.27 2.16 | 0.017 0.023 0.030 |
| SLNB | 0.42 | 0.116 | 0.243, 0.719 | −3.16 | 0.002 |
| Number of nodes removed | 1.06 | 0.017 | 1.028, 1.093 | 3.70 | <.0001 |
| Any chemotherapy | 1.74 | 0.416 | 1.086, 2.779 | 2.30 | 0.021 |
| Any radiation therapy | 1.94 | 0.452 | 1.224, 3.060 | 2.83 | 0.005 |
| Overall model fit: χ2 = 85.32, p<0.0001, R2 = 0.1380 | |||||
Abbreviations: AIMS = ancestry informative markers; BMI = body mass index; CI = confidence interval; SLNB = sentinel lymph node biopsy.
In the second regression analysis that added genomic estimates of and self-reported race/ethnicity in addition to the characteristics listed above, the overall model explained 17.4% of the variance in LE group membership (p<0.0001). While BMI, stage of disease, SLNB status, and number of lymph nodes removed remained significant, when genomic and self-reported race/ethnicity were included in the model, the receipt of CTX and RT were no longer significant predictors of LE group membership.
Candidate gene analyses for the development of LE
As summarized in Table 1, no associations with the occurrence of LE were found in the SNPs evaluated for HGF, LYVE1, MET, PROX1, RORC, VEGFB, VEGFD, and VEGFR3. However, the genotype frequency was significantly different between those who did and did not develop LE for thirteen SNPs and seven haplotypes spanning nine genes (i.e., ANGPT2, FOXC2, LCP2, NRP2, SOX 17, SYK, VCAM1, VEGFC, and VEGFR2). For the SNP in ANGPT2 (rs6990020), a dominant model fit the data best (p = .040). One haplotype (HapA03, p = .045) was identified in FOXC2. For the three SNPs (rs572192, rs315721, rs6866733) identified in LCP2, a dominant model fit the data best (p = .047, p = .005, p = .026, respectively). Three SNPs (rs849530, rs849563, rs16837641) and one haplotype (HapF01, p = .011) were identified in NRP2. For all three SNPs, a recessive model fit the data best (p = .042, p = .012, p = .009, respectively). For the SNP in SOX17 (rs12541742), a dominant model fit the data best (p = .008). One SNP (rs158689) and 1 haplotype (HapD01, p = .008) were identified in SYK. For rs158689, a recessive model fit the data best (p = .039). One SNP (rs3176861) and one haplotype (HapB01, p = .016) were identified in VCAM1. For rs3176861, a dominant model fit the data best (p = .036). One SNP (rs7664413) and two haplotypes (HapA05, p = .046; HapB03, p = .027) were identified in VEGFC. For rs7664413, an additive model fit the data best (p = .041). Two SNPs (rs10020464, rs11133360) and one haplotype (HapC04, p = .021) were identified for VEGFR2. For rs10020464, a dominant model fit the data best (p = .025) and for rs11133360 a recessive model fit the data best (p = .032).
Regression analyses of FOXC2, LCP2, NRP2, SYK, VCAM1, and VECFC genotypes and haplotypes and the development of LE
In order to better estimate the magnitude (i.e., odds ratio, OR) and precision (95% confidence interval, CI) of genotype on the development of LE, multivariate logistic regression models were fit. As shown in Table 4, in addition to genotype, the phenotypic variables included in the regression models were ethnicity (i.e., White, Black, Asian, Hispanic/Mixed ethnic background/Other), BMI, stage of disease, having a SLNB, and number of lymph nodes removed. Receipt of CTX and RT, while not significant after the inclusion of genomic estimates of and self-reported race/ethnicity, were retained in all of the regression models for face validity.
Table 4. Multiple logistic regression analyses for FOXC2. LCP2, NRP2, SYK, VCAM1, and VEGF-C genotypes and halotypes to predict the development of lymphedema.
| Predictor | Odds Ratio | Standard Error | 95% CI | Z | p-value | ||||||||
| FOXC2 haplotype | 0.63 | 0.125 | 0.430, 0.931 | −2.32 | 0.020 | ||||||||
| BMI | 1.06 | 0.022 | 1.016, 1.104 | 2.72 | 0.006 | ||||||||
| Stage of disease Stage 0 versus I Stage 0 versus II Stage 0 versus III and IV | 3.43 4.10 6.71 | 2.053 2.619 4.975 | 1.060, 11.085 1.175, 14.333 1.571, 28.687 | 2.06 2.21 2.57 | 0.040 0.027 0.010 | ||||||||
| SLNB | 0.40 | 0.138 | 0.203, 0.784 | −2.67 | 0.008 | ||||||||
| Number of nodes removed | 1.08 | 0.022 | 1.042, 1.127 | 4.01 | <.0001 | ||||||||
| Any chemotherapy | 1.22 | 0.380 | 0.664, 2.246 | 0.64 | 0.521 | ||||||||
| Any radiation therapy | 1.35 | 0.406 | 0.752, 2.436 | 1.01 | 0.313 | ||||||||
| Overall model fit: χ2 = 83.44, p<0.0001, R2 = 0.1860 | |||||||||||||
| LCP2 genotype | 0.50 | 0.132 | 0.298, 0.838 | −2.63 | 0.009 | ||||||||
| BMI | 1.05 | 0.022 | 1.012, 1.097 | 2.54 | 0.011 | ||||||||
| Stage of disease Stage 0 versus I Stage 0 versus II Stage 0 versus III and IV | 3.01 3.56 5.94 | 1.788 2.238 4.365 | 0.937, 9.643 1.040, 12.203 1.407, 25.079 | 1.85 2.02 2.42 | 0.064 0.043 0.015 | ||||||||
| SLNB | 0.40 | 0.137 | 0.204, 0.784 | −2.67 | 0.008 | ||||||||
| Number of nodes removed | 1.08 | 0.022 | 1.040, 1.125 | 3.93 | <.0001 | ||||||||
| Any chemotherapy | 1.25 | 0.389 | 0.679, 2.298 | 0.71 | 0.475 | ||||||||
| Any radiation therapy | 1.33 | 0.398 | 0.744, 2.392 | 0.97 | 0.334 | ||||||||
| Overall model fit: χ2 = 84.86, p<0.0001, R2 = 0.1891 | |||||||||||||
| NRP2 genotype | 0.38 | 0.143 | 0.185, 0.799 | −2.56 | 0.010 | ||||||||
| BMI | 1.06 | 0.023 | 1.016, 1.105 | 2.68 | 0.007 | ||||||||
| Stage of disease Stage 0 versus I Stage 0 versus II Stage 0 versus III and IV | 2.79 3.41 6.01 | 1.647 2.150 4.430 | 0.874, 8.875 0.990, 11.733 1.420, 25.477 | 1.73 1.94 2.44 | 0.083 0.052 0.015 | ||||||||
| SLNB | 0.39 | 0.136 | 0.203, 0.775 | −2.71 | 0.007 | ||||||||
| Number of nodes removed | 1.08 | 0.021 | 1.040, 1.122 | 4.00 | <0.0001 | ||||||||
| Any chemotherapy | 1.18 | 0.369 | 0.635, 2.174 | 0.51 | 0.607 | ||||||||
| Any radiation therapy | 1.20 | 0.363 | 0.667, 2.171 | 0.61 | 0.539 | ||||||||
| Overall model fit: χ2 = 85.11, p<0.0001, R2 = 0.1897 | |||||||||||||
| NRP2 haplotype | 0.54 | 0.114 | 0.358, 0.817 | −2.92 | 0.003 | ||||||||
| BMI | 1.05 | 0.022 | 1.011, 1.099 | 2.48 | 0.013 | ||||||||
| Stage of disease Stage 0 versus I Stage 0 versus II Stage 0 versus III and IV | 3.03 3.81 5.49 | 1.805 2.433 4.081 | 0.941, 9.738 1.088, 13.321 1.281, 23.561 | 1.86 2.09 2.29 | 0.063 0.036 0.022 | ||||||||
| SLNB | 0.40 | 0.137 | 0.201, 0.780 | −2.68 | 0.007 | ||||||||
| Number of nodes removed | 1.09 | 0.021 | 1.046, 1.132 | 4.21 | <0.0001 | ||||||||
| Any chemotherapy | 1.11 | 0.348 | 0.602, 2.054 | 0.34 | 0.734 | ||||||||
| Any radiation therapy | 1.30 | 0.391 | 0.723, 2.346 | 0.88 | 0.379 | ||||||||
| Overall model fit: χ2 = 86.91, p<0.0001, R2 = 0.1937 | |||||||||||||
| SYK genotype | 3.43 | 1.131 | 1.797, 6.546 | 3.74 | <0.0001 | ||||||||
| BMI | 1.06 | 0.023 | 1.017, 1.106 | 2.76 | 0.006 | ||||||||
| Stage of disease Stage 0 versus I Stage 0 versus II Stage 0 versus III and IV | 3.05 3.10 5.50 | 1.821 1.968 4.078 | 0.946, 9.832 0.891, 10.760 1.286, 23.523 | 1.87 1.78 2.30 | 0.062 0.075 0.022 | ||||||||
| SLNB | 0.40 | 0.141 | 0.201, 0.797 | −2.61 | 0.009 | ||||||||
| Number of nodes removed | 1.10 | 0.022 | 1.054, 1.142 | 4.53 | <0.0001 | ||||||||
| Any chemotherapy | 1.44 | 0.459 | 0.773, 2.692 | 1.15 | 0.249 | ||||||||
| Any radiation therapy | 1.45 | 0.437 | 0.803, 2.616 | 1.23 | 0.218 | ||||||||
| Overall model fit: χ2 = 91.90, p<0.0001, R2 = 0.2049 | |||||||||||||
| VCAM1 genotype | 0.55 | 0.158 | 0.309, 0.963 | −2.09 | 0.037 | ||||||||
| BMI | 1.06 | 0.022 | 1.015, 1.102 | 2.68 | 0.007 | ||||||||
| Stage of disease Stage 0 versus I Stage 0 versus II Stage 0 versus III and IV | 2.85 3.78 5.92 | 1.710 2.413 4.385 | 0.879, 9.239 1.080, 13.214 1.388, 25.275 | 1.75 2.08 2.40 | 0.081 0.038 0.016 | ||||||||
| SLNB | 0.41 | 0.141 | 0.206, 0.801 | −2.60 | 0.009 | ||||||||
| Number of nodes removed | 1.08 | 0.022 | 1.043, 1.128 | 4.07 | <0.0001 | ||||||||
| Any chemotherapy | 1.18 | 0.365 | 0.643, 2.161 | 0.53 | 0.596 | ||||||||
| Any radiation therapy | 1.30 | 0.387 | 0.724, 2.327 | 0.88 | 0.381 | ||||||||
| Overall model fit: χ2 = 82.35, p<0.0001, R2 = 0.1836 | |||||||||||||
| VEGFC haplotype | 0.64 | 0.145 | 0.409, 0.997 | −1.97 | 0.049 | ||||||||
| BMI | 1.05 | 0.022 | 1.011, 1.097 | 2.46 | 0.014 | ||||||||
| Stage of disease Stage 0 versus I Stage 0 versus II Stage 0 versus III and IV | 2.94 3.74 5.42 | 1.740 2.351 3.962 | 0.920, 9.380 1.091, 12.820 1.291, 22.716 | 1.82 2.10 2.31 | 0.069 0.036 0.021 | ||||||||
| SLNB | 0.42 | 0.145 | 0.215, 0.826 | −2.51 | 0.012 | ||||||||
| Number of nodes removed | 1.09 | 0.021 | 1.044, 1.128 | 4.16 | <0.0001 | ||||||||
| Any chemotherapy | 1.24 | 0.387 | 0.675, 2.287 | 0.70 | 0.487 | ||||||||
| Any radiation therapy | 1.31 | 0.391 | 0.729, 2.354 | 0.90 | 0.366 | ||||||||
| Overall model fit: χ2 = 81.85, p<0.0001, R2 = 0.1825 | |||||||||||||
For each model, the first three principal components identified from the analysis of ancestry informative markers as well as self-report race/ethnicity (i.e., White, Black, Asian/Pacific Islander, Hispanic/Mixed ethnic background/Other) were retained in all models to adjust for potential confounding due to race or ethnicity (data not shown). Predictors evaluated in each model included genotype (FOXC2 haplotype A03 composed of the rs34221221 “C” allele and the rs1035550 “C” allele; LCP2 rs315721: AA versus AG+GG; NRP2 rs849530: TT+TG versus GG; NPR2 haplotype F01 composed of the rs849530 “G” allele, the rs950219 “G” allele, and the rs3771052 “G” allele; SYK rs158689: AA+AT versus TT; VCAM1 rs3176861: CC versus CT + TT; VEGFC haplotype B03 composed of the rs3775202 “G” allele and the rs3775195 “C” allele), BMI (kilograms/meter squared), stage of disease, SLNB, number of lymph nodes removed, receipt of chemotherapy prior to or following surgery, and receipt of radiation therapy following surgery.
Abbreviations: BMI = body mass index; CI = confidence interval; FOXC2 = Forkhead box protein C2; LCP2 = Lymphocyte cytosolic protein 2; NRP = neuropilin-2; SLNB = sentinel lymph node biopsy; SYK = spleen tyrosine kinase; VCAM1 = vascular cell adhesion molecule 1; VEGFC = vascular endothelial growth factor-C.
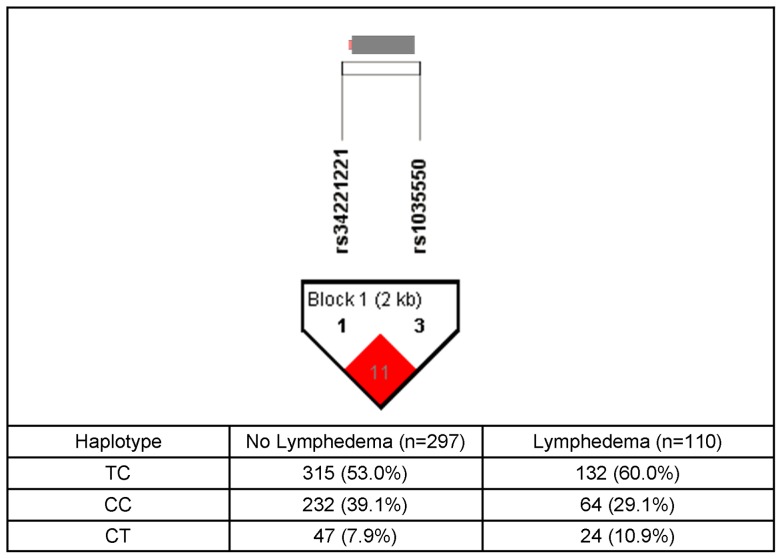
The only genetic associations that remained significant in the multivariate logistic regression analyses were for FOXC2 haplotype A03, LCP2 rs315721, NRP2 rs849530, NRP2 haplotype F01, SYK rs158689, VCAM1 rs3176861, and VEGFC haplotype B03 (see Table 4 and Figures 1, 2, 3, and 4). In the regression analysis for the FOXC2 haplotype A3, which was composed of two SNPs (i.e., rs34221221, rs1035550), the overall model explained 18.6% of the variance in the odds of developing LE. Each additional dose of the FOXC2 haplotype A03 was associated with 37.0% decrease in the odds of developing LE. Figure 1 displays the FOXC2 LD-based heatmap and haplotype analysis.
Figure 1. FOXC2 Gene Structure and Linkage Disequilibrium.
An ideogram of forkhead box C2 (FOXC2) is presented above the white bar that represents the physical distance along human chromosome 16 (chr16: 85,158,358–85,160,040; genome assembly 36.3, NM_005251.2). Exons are represented as thick bars. Reference sequence identifiers (rsID) for each single nucleotide polymorphism (SNP) are plotted both in terms of their physical distance (i.e., the white bar at the top of the figure) and equidistantly to render the pairwise linkage disequilibrium (LD) estimates that were calculated and visualized with Haploview 4.2. The gene structure for FOXC2 was rendered with FancyGene 1.4. The correlation statistic (r2 and D') is provided in the heatmap. LD-based haplotype block definition was based on the D' confidence interval method. The haploblock is indicated in a bolded triangle and its component SNPs are rendered in bold font. Pairwise D' value (range: 0–1, inclusive) was rendered in color, with darker red diamond representing D' value approaching 1.0. When the r2 value (range of 0–100, inclusive) is not equal to 0 or 100, it is provided in a given diamond. The 2-SNP haplotype associated with LE is composed of one rare and one common allele of two SNPs located in the immediate early promoter (rs34221221; rare “C” allele) and immediately downstream of the FOXC2 coding region (rs1035550; common “C” allele).
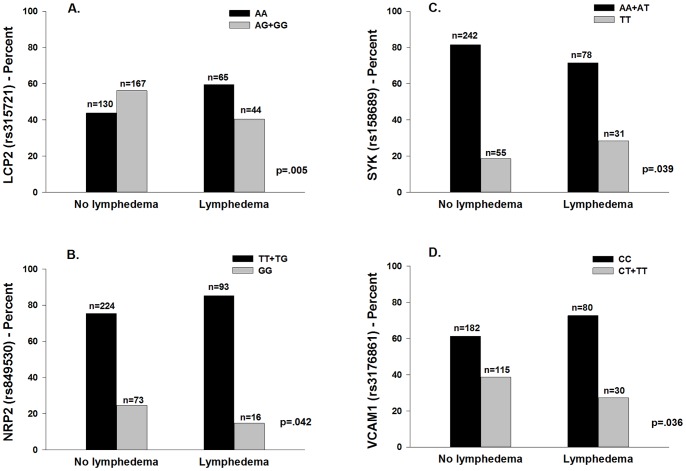
Figure 2. Differences between the lymphedema and no lymphedema groups.
A – Differences between the lymphedema and no lymphedema groups in the percentages of patients who were homozygous for the common allele (AA) or heterozygous or homozygous for the rare allele (AG+GG) for rs315721 in lymphocyte cytosolic protein 2 (LCP2). B – Differences between the lymphedema and no lymphedema groups in the percentages of patients who were homozygous or heterozygous for the common allele (TT+TG) or homozygous for the rare allele (GG) for rs849530 in neuropilin-2 (NRP2). C – Differences between the lymphedema and no lymphedema groups in the percentages of patients who were homozygous or heterozygous for the common allele (AA+AT) or homozygous for the rare allele (TT) for rs158689 in protein tyrosine kinase (SYK). D – Differences between the lymphedema and no lymphedema groups in the percentages of patients who were homozygous for the common allele (CC) or heterozygous or homozygous for the rare allele (CT+TT) for rs3176861 in vascular cell adhesion molecule 1 (VCAM1).
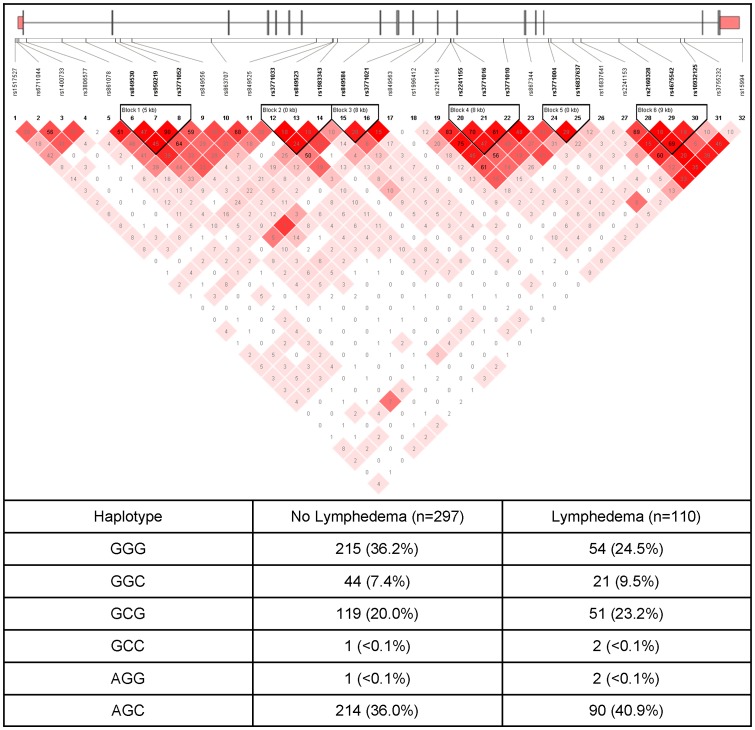
Figure 3. NRP2 Gene Structure and Linkage Disequilibrium.
An ideogram of neuropilin 2 (NRP2) is presented above the white bar that represents the physical distance along human chromosome 2 (chr2: 206,255,469–206,371,102; genome assembly 36.3, NM_201266.1). Exons are represented as thick bars. Gray lines connecting the exons represent introns. Reference sequence identifiers (rsID) for each single nucleotide polymorphism (SNP) are plotted both in terms of their physical distance (i.e., the white bar at the top of the figure) and equidistantly to render the pairwise linkage disequilibrium (LD) estimates that were calculated and visualized with Haploview 4.2. The gene structure for NRP2 was rendered with FancyGene 1.4. The correlation statistics (r2 and D') are provided in the heatmap. LD-based haplotype block definition was based on the D' confidence interval method. The haploblock is indicated in a bolded triangle and its component SNPs are rendered in bold font. Pairwise D' values (range: 0–1, inclusive) were rendered in color, with darker red diamonds representing D' values approaching 1.0. When the r2 values (range of 0–100, inclusive) are not equal to 0 or 100, they are provided in a given diamond. The 3-SNP haplotype associated with LE consists of one rare and two common alleles of three SNPs (rs849530 “G” rare allele, rs950219 “G” common allele, rs3771052 “G” common allele) located in intron 1 of the gene.
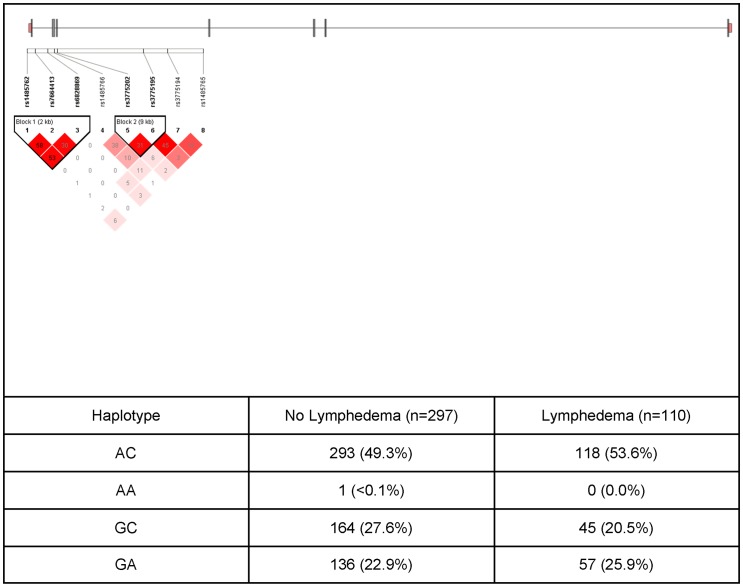
Figure 4. VEGFC Gene Structure and Linkage Disequilibrium.
An ideogram of vascular endothelial growth factor C (VEGFC) is presented above the white bar that represents the physical distance along human chromosome 4 (chr4: 177,841,685–177,950,889; genome assembly 36.3, NM_005429.2). Exons are represented as thick bars. Gray lines connecting the exons represent introns. Reference sequence identifiers (rsID) for each single nucleotide polymorphism (SNP) are plotted both in terms of their physical distance (i.e., the white bar at the top of the figure) and equidistantly to render the pairwise linkage disequilibrium (LD) estimates that were calculated and visualized with Haploview 4.2. The gene structure for VEGFC was rendered with FancyGene 1.4. The correlation statistics (r2 and D') are provided in the heatmap. LD-based haplotype block definition was based on the D' confidence interval method. The haploblock is indicated in a bolded triangle and its component SNPs are rendered in bold font. Pairwise D' values (range: 0–1, inclusive) were rendered in color, with darker red diamonds representing D' values approaching 1.0. When the r2 values (range of 0–100, inclusive) are not equal to 0 or 100, they are provided in a given diamond. The 2-SNP haplotype associated with LE consists of one rare and one common allele of two SNPs (rs3775202 “G” rare allele, rs3775195 “C” common allele) located in intron 4 of the gene. Of note, the strong linkage disequilibrium estimates observed in public databases (i.e., HapMap) resulted in the selection of 8 SNPs that tagged the entire coding region of the VEGFC gene.
In the regression analysis for LCP2 rs315721, the overall model explained 18.9% of the variance in the odds of developing LE. Carrying one or two doses of the rare allele (i.e., AA versus AG + GG) was associated with a 50.0% decrease in the odds of developing LE (Figure 2A).
In the regression analysis for NRP2 rs849530, the overall model explained 19.0% of the variance in the odds of developing LE. Carrying two doses of the rare allele (i.e., TT+TG versus GG) was associated with 62.0% decrease in the odds of developing LE (Figure 2B). In the regression analysis for the NRP2 haplotype F01, which was composed of three SNPs (i.e., rs849530, rs950219, rs3771052), the overall model explained 19.4% of the variance in the odds of developing LE. Each additional dose of the NRP2 haplotype F01 was associated with 46.0% decrease in the odds of developing LE. Figure 3 displays the NRP2 LD-based heatmap and haplotype analysis.
In the regression analysis for SYK rs158689, the overall model explained 20.1% of the variance in the odds of developing LE. Carrying two doses of the rare allele (i.e., AA + AT versus TT) was associated with 3.43-fold increase in the odds of developing LE (Figure 2C).
In the regression analysis for VCAM1 rs3176861, the overall model explained 18.4% of the variance in the odds of developing LE. Carrying one or two doses of the rare allele (i.e., CC versus CT + TT) was associated with a 45.0% decrease in the odds of developing LE (Figure 2D).
In the regression analysis for the VEGFC haplotype B03, which was composed of two SNPs (i.e., rs3775202, rs3775195), the overall model explained 18.3% of the variance in the odds of developing LE. Each additional dose of the VEGFC haplotype B03 was associated with 36.0% decrease in the odds of developing LE. Figure 4 displays the VEGFC LD-based heatmap and haplotype analysis.
Discussion
This study is the first to evaluate phenotypic and genotypic predictors of LE in a large cohort of women who had LE diagnosed using BIS rather than self-report. Based on an extensive evaluation of demographic, disease, and treatment characteristics, the factors associated with an increased risk of LE in the bivariate analyses were higher BMI, poorer functional status, having lung disease, a higher stage of disease, increased number of lymph nodes removed, increased number of positive lymph nodes, not having a SLND, having had an ALND, and receipt of CTX or RT. All of these risk factors are consistent with previous reports.
In the initial multivariate analyses, poorer functional status, having lung disease, number of positive nodes, and having had an ALND were not retained in the final model (Table 3). However, when genomic estimates of and self-reported race/ethnicity were added to the multivariate logistic regression analysis, receipt of CTX and RT were no longer significant predictors of LE group membership. This final phenotypic model explained only 17.4% of the variance in LE group membership. These findings suggest that complex interactions may exist between phenotypic characteristics and the development of LE. Future studies of LE risk need to evaluate a comprehensive list of phenotypic characteristics as well as interactions among these characteristics.
In the past few years, the complex array of molecular events that regulate the development and maintenance of lymphatic system, as well as contribute to its malfunction have begun to be elucidated in animals and humans [10], [11], [13]. In this study, candidate genes were selected that were identified in previous animal and human studies to play a role in lymphatic morphogenesis (Table 1). While not all of the candidate genes were associated with the development of LE, the significant associations that were identified provide new information on genomic risk factors and potential therapeutic targets. Of note, each of the SNPs explained between 1.0% (VCAM1 rs3176861) to 3.1% (SYK rs158689) of the variance in the development of LE.
FOXC2 is a transcription factor that appears to be important for the normal development and maintenance of both venous and lymphatic vessels [45]. In adults, FOXC2 is highly expressed in developing lymphatic vessels as well as in lymphatic valves [46], [47]. While early lymphatic development will proceed normally in the absence of FOXC2, the collecting lymphatic vessels that are formed lack valves and the lymphatic capillaries acquire an ectopic coverage of basement membrane components and smooth muscle cells [46]. FOXC2 is the transcription factor associated with lymphedema-distichiasis (LD, OMIM #153400) a monogenic disorder that is characterized by late onset LE, a double row of eyelashes, and varicose veins [48]–[50]. Therefore, common functional polymorphisms that result in modest alterations in function or expression of the FOXC2 transcription factor may be associated with the development of secondary LE following breast cancer treatment. The FOXC2 A03 haplotype consists of one rare and one common allele in two SNPs located in the immediate early promoter (rs34221221; rare “C” allele) and immediately downstream from the FOXC2 coding region (rs1035550, common “C” allele). SNP rs34221221 occurs in the immediate early promoter of the FOXC2 gene (−514) which is a highly conserved region of the gene. While functional studies need to be done, it is reasonable to suggest that this polymorphism alters transcription factor binding and subsequent gene expression. SNP rs103550 lies downstream from the FOXC2 coding region of the gene and is not likely to have a functional role. Functional studies of this two-SNP haplotype are warranted if the association with LE is replicated in an independent sample.
LCP2 functions in lymphatic vessel development by modulation of the hematopoetic signaling pathway that mediates the separation of the two major vascular networks (i.e., blood, lymphatic) [51]. SYK acts on LCP2 as part of a central signaling pathway that regulates separation of these two vascular networks. Variation in either locus could result in their altered interaction with upstream (i.e., SYK) or downstream (i.e., LCP2) members of the signaling cascade. Mice with mutations in SYK develop arterio-venous shunts and abnormal lymphatic-venous connections [51], [52]. In addition, genetic ablation of SYK causes the accumulation of leukocytes that is associated with lymphatic proliferation and lymphatic vessel dilation which results in the formation of shunts between the blood and lymphatic vessels (F. Kiefer, personal communication cited in Tammela and Alitalo [12]). While LPC2 rs35721 is located in the intronic region of the gene and has no known function, it could be in LD with a functional variation. Although SYK rs158689 is located in an intron, it is predicted to disrupt a putative CCCTC-binding factor (CTCF) termed insulator elements. These insulator elements play a vital role in the regulation of gene expression by limiting the boundary of heterochromatin and by restricting transcriptional factor access [53]. Functional studies are needed to determine if the rare “T” allele of rs158689 alters the function of the putative insulator element and SYK gene expression.
Lymphatic vessels participate in inflammatory responses by promoting lymphocyte transport to draining lymph nodes [54]. VCAM1 plays a role in the migration of lymphocytes into lymphoid organs. Compared to healthy controls, cytokines levels are increased in lymphatic fluid from patients with obstructive LE. These elevated levels of cytokines may be due to their local production by infiltrating immune cells [55]. The elevated levels of these cytokines would be expected to contribute to the chronic inflammation that is typically observed in tissues with poor lymphatic drainage. Allelic variations in VCAM1 could influence the rate of lymphocyte homing to lymph nodes. Like other SNPs in this study, rs3176861, located in the intron of the VCAM gene, has no known function. However, it could be in LD with a functional variation.
NRP1 and NRP2 are transmembrane proteins involved in a number of physiologic processes [56]–[59]. NRP2 can bind to members of the VEGF family of growth factors and influence lymphagiogenesis. NRP2 is expressed in a subset of lymphatic vessels and acts as a co-receptor for VEGFC [60]. VEGFC binds to and activates VEGFR3 and VEGFR2 receptors on the lymphatic epithelium [61]–[65]. In one study [66], Nrp2 knockout mice did not exhibit defects in blood vessels. However, lymphatic development was abnormal, including abnormal patterning and marked reduction in small lymphatic vessels and capillaries. These findings suggest that NRP2 has a role in VEGFC mediated VEGFR3 signaling and lymphangiogenesis. The NRP2 haplotype consists of one rare and two common alleles in three SNPs (rs849530 “G” rare allele, rs950219 “G” common allele, rs3771052 “G” common allele) located in intron 1 of the gene. These SNPs lie in a conserved region of the NRP2 gene. However, this haplotype has no known function which suggest that this haplotype is a surrogate for an unmeasured SNP(s) in LD with the haplotype. The VEGFC haplotype consists of one rare and one common allele in two SNPs (rs3775202 “G” rare allele, rs3775195 “C” common allele) located in intron 4 of the gene. These SNPs lie in a conserved region of the VEGFC gene. However, this haplotype has no known or predicted function which suggests that it is a surrogate for an unmeasured SNP(s) in LD with this haplotype.
It is interesting to note that no associations were found in SNPs for HGF, LYVE1, MET, PROX1, RORC, VEGFB, VEGFC, and VEGFR3. One possible reason for this lack of association is that these genes are involved primarily during embryogenesis in the development of the lymphatic system and are not involved in the mechanisms that underlie the development of secondary LE [10], [11], [13]. However, recent evidence suggests that the VEGF pathway is involved in lymphangiogenesis associated with inflammation [12], [67]. An alternative explanation for the lack of association is that the occurrence of the minor allele was too infrequent in this sample to identify significant differences between patients with and without LE. Evidence to support this hypothesis is found in Table 1, where p-values for some of these genes approached statistical significance.
Several study limitations need to be acknowledged. While the sample size was relatively large, larger samples may identify additional candidate gene associations. In terms of the genetic analyses, additional studies are needed to confirm the associations found in this study. Future studies can evaluate additional candidate genes or perform genome wide association studies to uncover novel molecular pathways. Once candidate genes are confirmed, then DNA sequencing may need to be performed to find the causal variant(s). Despite these limitations, findings from this study suggest a role for a number of lymphatic and angiogenic candidate genes in the development of secondary LE following breast cancer treatment.
Funding Statement
This study was funded by grants from the National Cancer Institute (CA107091 and CA118658) and the National Institute of Nursing Research (NR0101282). Dr. Miaskowski is an American Cancer Society Clinical Research Professor. Dr. Bradley Aouizerat was funded through the National Institutes of Health (NIH) Roadmap for Medical Research Grant (KL2 RR624130). This project was supported by NIH/NCRR UCSF-CTSI Grant Number UL1 RR024131. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Maby-El Hajjami H, Petrova TV (2008) Developmental and pathological lymphangiogenesis: from models to human disease. Histochem Cell Biol 130: 1063–1078. [DOI] [PubMed] [Google Scholar]
- 2. Stamatakos M, Stefanaki C, Kontzoglou K (2011) Lymphedema and breast cancer: a review of the literature. Breast Cancer 18: 174–180. [DOI] [PubMed] [Google Scholar]
- 3. Armer JM (2010) Research on risk assessment for secondary lymphedema following breast cancer treatment. Cancer Epidemiol Biomarkers Prev 19: 2715–2717. [DOI] [PubMed] [Google Scholar]
- 4. Kwan ML, Darbinian J, Schmitz KH, Citron R, Partee P, et al. (2010) Risk factors for lymphedema in a prospective breast cancer survivorship study: the Pathways Study. Arch Surg 145: 1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahmed RL, Schmitz KH, Prizment AE, Folsom AR (2011) Risk factors for lymphedema in breast cancer survivors, the Iowa Women's Health Study. Breast Cancer Res Treat 130: 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Norman SA, Localio AR, Kallan MJ, Weber AL, Torpey HA, et al. (2010) Risk factors for lymphedema after breast cancer treatment. Cancer Epidemiol Biomarkers Prev 19: 2734–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominick SA, Madlensky L, Natarajan L, Pierce JP (2012) Risk factors associated with breast cancer-related lymphedema in the WHEL Study. J Cancer Surviv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Finegold D, Baty C, Knickelbein K, Perschke S, Noon S, et al. (2012) Connexin 47 mutations increase risk for secondary lymphedema following breast cancer treatment. Clin Cancer Res 18: 232–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Finegold DN, Schacht V, Kimak MA, Lawrence EC, Foeldi E, et al. (2008) HGF and MET mutations in primary and secondary lymphedema. Lymphat Res Biol 6: 65–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schulte-Merker S, Sabine A, Petrova TV (2011) Lymphatic vascular morphogenesis in development, physiology, and disease. J Cell Biol 193: 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y, Oliver G (2010) Current views on the function of the lymphatic vasculature in health and disease. Genes Dev 24: 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tammela T, Alitalo K (2010) Lymphangiogenesis: Molecular mechanisms and future promise. Cell 140: 460–476. [DOI] [PubMed] [Google Scholar]
- 13. Butler MG, Isogai S, Weinstein BM (2009) Lymphatic development. Birth Defects Res C Embryo Today 87: 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rockson SG (2009) The unique biology of lymphatic edema. Lymphat Res Biol 7: 97–100. [DOI] [PubMed] [Google Scholar]
- 15. Jurisic G, Detmar M (2009) Lymphatic endothelium in health and disease. Cell Tissue Res 335: 97–108. [DOI] [PubMed] [Google Scholar]
- 16.Karnofsky D (1977) Performance scale; Kennealey GT, Mitchell MS, editors. New York: Plenum Press.
- 17. Karnofsky D, Abelmann WH, Craver LV, Burchenal JH (1948) The use of nitrogen mustards in the palliative treatment of carcinoma. Cancer 1: 634–656. [Google Scholar]
- 18. Sangha O, Stucki G, Liang MH, Fossel AH, Katz JN (2003) The Self-Administered Comorbidity Questionnaire: a new method to assess comorbidity for clinical and health services research. Arthritis Rheum 49: 156–163. [DOI] [PubMed] [Google Scholar]
- 19. MacLean CD, Littenberg B, Kennedy AG (2006) Limitations of diabetes pharmacotherapy: results from the Vermont Diabetes Information System study. BMC Fam Pract 7: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cieza A, Geyh S, Chatterji S, Kostanjsek N, Ustun BT, et al. (2006) Identification of candidate categories of the International Classification of Functioning Disability and Health (ICF) for a Generic ICF Core Set based on regression modelling. BMC Med Res Methodol 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smith SK, Zimmerman S, Williams CS, Zebrack BJ (2009) Health status and quality of life among non-Hodgkin lymphoma survivors. Cancer 115: 3312–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brunner F, Bachmann LM, Weber U, Kessels AG, Perez RS, et al. (2008) Complex regional pain syndrome 1 – the Swiss cohort study. BMC Musculoskelet Disord 9: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hayes S, Cornish B, Newman B (2005) Comparison of methods to diagnose lymphoedema among breast cancer survivors: 6-month follow-up. Breast Cancer Res Treat 89: 221–226. [DOI] [PubMed] [Google Scholar]
- 24. Cornish BH, Chapman M, Hirst C, Mirolo B, Bunce IH, et al. (2001) Early diagnosis of lymphedema using multiple frequency bioimpedance. Lymphology 34: 2–11. [PubMed] [Google Scholar]
- 25. Cornish BH, Chapman M, Thomas BJ, Ward LC, Bunce IH, et al. (2000) Early diagnosis of lymphedema in postsurgery breast cancer patients. Ann N Y Acad Sci 904: 571–575. [DOI] [PubMed] [Google Scholar]
- 26.SPSS (2010) IBM SPSS for Windows (Version 19). Chicago, Illinois: SPSS, Inc. [Google Scholar]
- 27. Ferrell RE, Kimak MA, Lawrence EC, Finegold DN (2008) Candidate gene analysis in primary lymphedema. Lymphat Res Biol 6: 69–76. [DOI] [PubMed] [Google Scholar]
- 28. Irrthum A, Devriendt K, Chitayat D, Matthijs G, Glade C, et al. (2003) Mutations in the transcription factor gene SOX18 underlie recessive and dominant forms of hypotrichosis-lymphedema-telangiectasia. Am J Hum Genet 72: 1470–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ghalamkarpour A, Holnthoner W, Saharinen P, Boon LM, Mulliken JB, et al. (2009) Recessive primary congenital lymphoedema caused by a VEGFR3 mutation. J Med Genet 46: 399–404. [DOI] [PubMed] [Google Scholar]
- 30. Traboulsi EI, Al-Khayer K, Matsumoto M, Kimak MA, Crowe S, et al. (2002) Lymphedema-distichiasis syndrome and FOXC2 gene mutation. Am J Ophthalmol 134: 592–596. [DOI] [PubMed] [Google Scholar]
- 31. Saito Y, Nakagami H, Morishita R, Takami Y, Kikuchi Y, et al. (2006) Transfection of human hepatocyte growth factor gene ameliorates secondary lymphedema via promotion of lymphangiogenesis. Circulation 114: 1177–1184. [DOI] [PubMed] [Google Scholar]
- 32. Conde L, Vaquerizas JM, Dopazo H, Arbiza L, Reumers J, et al. (2006) PupaSuite: finding functional single nucleotide polymorphisms for large-scale genotyping purposes. Nucleic Acids Res 34: W621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. (2002) The structure of haplotype blocks in the human genome. Science 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- 34. Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T (2008) A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Human Mutat 29: 648–658. [DOI] [PubMed] [Google Scholar]
- 36. Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, et al. (2003) Control of confounding of genetic associations in stratified populations. Am J Hum Genet 72: 1492–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tian C, Gregersen PK, Seldin MF (2008) Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet 17: R143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 39.StataCorp (2005) Stata Statistical Software: Release 9. College Station, Texas: Stata Corporation. [Google Scholar]
- 40. Illi J, Miaskowski C, Cooper B, Levine JD, Dunn L, et al. (2012) Association between pro- and anti-inflammatory cytokine genes and a symptom cluster of pain, fatigue, sleep disturbance, and depression. Cytokine 58: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCann B, Miaskowski C, Koetters T, Baggott C, West C, et al. (2012) Associations between pro- and anti-inflammatory cytokine genes and breast pain in women prior to breast cancer surgery. J Pain 13: 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miaskowski C, Cooper BA, Dhruva A, Dunn LB, Langford DJ, et al. (2012) Evidence of associations between cytokine genes and subjective reports of sleep disturbance in oncology patients and their family caregivers. PLoS One 7: e40560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rothman KJ (1990) No adjustments are needed for multiple comparisons. Epidemiology 1: 43–46. [PubMed] [Google Scholar]
- 44. Hattersley AT, McCarthy MI (2005) What makes a good genetic association study? Lancet 366: 1315–1323. [DOI] [PubMed] [Google Scholar]
- 45. Mellor RH, Brice G, Stanton AW, French J, Smith A, et al. (2007) Mutations in FOXC2 are strongly associated with primary valve failure in veins of the lower limb. Circulation 115: 1912–1920. [DOI] [PubMed] [Google Scholar]
- 46. Petrova TV, Karpanen T, Norrmen C, Mellor R, Tamakoshi T, et al. (2004) Defective valves and abnormal mural cell recruitment underlie lymphatic vascular failure in lymphedema distichiasis. Nat Medicine 10: 974–981. [DOI] [PubMed] [Google Scholar]
- 47. Dagenais SL, Hartsough RL, Erickson RP, Witte MH, Butler MG, et al. (2004) Foxc2 is expressed in developing lymphatic vessels and other tissues associated with lymphedema-distichiasis syndrome. Gene Expr Patterns 4: 611–619. [DOI] [PubMed] [Google Scholar]
- 48. Fang J, Dagenais SL, Erickson RP, Arlt MF, Glynn MW, et al. (2000) Mutations in FOXC2 (MFH-1), a forkhead family transcription factor, are responsible for the hereditary lymphedema-distichiasis syndrome. Am J Hum Genet 67: 1382–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bell R, Brice G, Child AH, Murday VA, Mansour S, et al. (2001) Analysis of lymphoedema-distichiasis families for FOXC2 mutations reveals small insertions and deletions throughout the gene. Hum Genet 108: 546–551. [DOI] [PubMed] [Google Scholar]
- 50. Connell F, Brice G, Mortimer P (2008) Phenotypic characterization of primary lymphedema. Ann N Y Acad Sci 1131: 140–146. [DOI] [PubMed] [Google Scholar]
- 51. Abtahian F, Guerriero A, Sebzda E, Lu MM, Zhou R, et al. (2003) Regulation of blood and lymphatic vascular separation by signaling proteins SLP-76 and Syk. Science 299: 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sebzda E, Hibbard C, Sweeney S, Abtahian F, Bezman N, et al. (2006) Syk and Slp-76 mutant mice reveal a cell-autonomous hematopoietic cell contribution to vascular development. Dev Cell 11: 349–361. [DOI] [PubMed] [Google Scholar]
- 53. Dhruva A, Paul SM, Cooper BA, Lee K, West C, et al. (2012) A longitudinal study of measures of objective and subjective sleep disturbance in patients with breast cancer before, during, and after radiation therapy. J Pain Symptom Manage 44: 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kerjaschki D, Regele HM, Moosberger I, Nagy-Bojarski K, Watschinger B, et al. (2004) Lymphatic neoangiogenesis in human kidney transplants is associated with immunologically active lymphocytic infiltrates. J Am Soc Nephrol 15: 603–612. [DOI] [PubMed] [Google Scholar]
- 55. Olszewski WL (2003) Pathophysiological aspects of lymphedema of human limbs: I. Lymph protein composition. Lymphat Res Biol 1: 235–243. [DOI] [PubMed] [Google Scholar]
- 56. Chen C, Li M, Chai H, Yang H, Fisher WE, et al. (2005) Roles of neuropilins in neuronal development, angiogenesis, and cancers. World J Surg 29: 271–275. [DOI] [PubMed] [Google Scholar]
- 57. Bagri A, Tessier-Lavigne M, Watts RJ (2009) Neuropilins in tumor biology. Clin Cancer Res 15: 1860–1864. [DOI] [PubMed] [Google Scholar]
- 58. Staton CA, Kumar I, Reed MW, Brown NJ (2007) Neuropilins in physiological and pathological angiogenesis. J Pathol 212: 237–248. [DOI] [PubMed] [Google Scholar]
- 59. Karpanen T, Heckman CA, Keskitalo S, Jeltsch M, Ollila H, et al. (2006) Functional interaction of VEGF-C and VEGF-D with neuropilin receptors. FASEB J 20: 1462–1472. [DOI] [PubMed] [Google Scholar]
- 60. Karkkainen MJ, Saaristo A, Jussila L, Karila KA, Lawrence EC, et al. (2001) A model for gene therapy of human hereditary lymphedema. Proc Natl Acad Sci U S A 98: 12677–12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Joukov V, Pajusola K, Kaipainen A, Chilov D, Lahtinen I, et al. (1996) A novel vascular endothelial growth factor, VEGF-C, is a ligand for the Flt4 (VEGFR-3) and KDR (VEGFR-2) receptor tyrosine kinases. EMBO J 15: 1751. [PMC free article] [PubMed] [Google Scholar]
- 62. Lee J, Gray A, Yuan J, Luoh SM, Avraham H, et al. (1996) Vascular endothelial growth factor-related protein: a ligand and specific activator of the tyrosine kinase receptor Flt4. Proc Natl Acad Sci U S A 93: 1988–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Orlandini M, Marconcini L, Ferruzzi R, Oliviero S (1996) Identification of a c-fos-induced gene that is related to the platelet-derived growth factor/vascular endothelial growth factor family. Proc Natl Acad Sci U S A 93: 11675–11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yamada Y, Nezu J, Shimane M, Hirata Y (1997) Molecular cloning of a novel vascular endothelial growth factor, VEGF-D. Genomics 42: 483–488. [DOI] [PubMed] [Google Scholar]
- 65. Achen MG, Jeltsch M, Kukk E, Makinen T, Vitali A, et al. (1998) Vascular endothelial growth factor D (VEGF-D) is a ligand for the tyrosine kinases VEGF receptor 2 (Flk1) and VEGF receptor 3 (Flt4). Proc Natl Acad Sci U S A 95: 548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yuan L, Moyon D, Pardanaud L, Breant C, Karkkainen MJ, et al. (2002) Abnormal lymphatic vessel development in neuropilin 2 mutant mice. Development 129: 4797–4806. [DOI] [PubMed] [Google Scholar]
- 67. Alitalo K (2011) The lymphatic vasculature in disease. Nat Med 17: 1371–1380. [DOI] [PubMed] [Google Scholar]