Abstract
Purpose
The combination of systemic cisplatin with local and regional radiotherapy as primary treatment of head and neck squamous cell carcinoma (HNSCC) leads to cure in approximately half of the patients. The addition of cisplatin has significant effects on outcome, but despite extensive research the mechanism underlying cisplatin response is still not well understood.
Methods
We examined 19 HNSCC cell lines with variable cisplatin sensitivity. We determined the TP53 mutational status of each cell line and investigated the expression levels of 11 potentially relevant genes by quantitative real-time PCR. In addition, we measured cisplatin accumulation and retention, as well as the level of platinum-DNA adducts.
Results
We found that the IC50 value was significantly correlated with the platinum-DNA adduct levels that accumulated during four hours of cisplatin incubation (p = 0.002). We could not find a significant correlation between cisplatin sensitivity and any of the other parameters tested, including the expression levels of established cisplatin influx and efflux transporters. Furthermore, adduct accumulation did not correlate with mRNA expression of the investigated influx pumps (CTR1 and OCT3) nor with that of the examined DNA repair genes (ATR, ATM, BRCA1, BRCA2 and ERCC1).
Conclusion
Our findings suggest that the cisplatin-DNA adduct level is the most important determinant of cisplatin sensitivity in HNSCC cells. Imaging with radio-labeled cisplatin might have major associations with outcome.
Introduction
Head and neck squamous cell carcinomas (HNSCCs) arise in the mucosal linings of the upper aerodigestive tract. Early stage tumors are treated with surgery or radiotherapy and these treatment modalities are combined to treat more advanced tumors. Since recent years chemoradiation protocols are increasingly applied to treat locally advanced carcinomas, consisting of systemic application of cisplatin (cis-diammine-dichloroplatinum II) combined with locoregional radiotherapy. The addition of cisplatin causes a significant increase in survival rates [1] and has become standard treatment. Despite these promising results, only 50% of patients are cured by chemoradiation. The tumors that do not respond well might better have been treated by surgery combined with postoperative radiotherapy. Therefore, biomarkers for personalized therapy are urgently awaited, and these might be found among factors that determine cisplatin and radiation response. It is conceivable that cells acquire resistance to cisplatin during treatment, but also intrinsic resistance of chemo-naive cells might be important. It is necessary to fully understand the mechanism of cisplatin response in order to find ways to personalize treatment, to sensitize tumor cells to this anticancer drug or to protect normal cells.
Several mechanisms of cisplatin resistance have been described previously (reviewed in [2], [3]), and it has become clear that the cellular processing of platinum provides important insights into the molecular mechanisms involved in cisplatin response. With the observation that the kinetics of uptake and efflux of cisplatin appears to follow a parallel pattern with those of copper [4], [5], CTR1 was identified as an important cisplatin influx transporter [4], [6]–[9]. ATP7A [4], [10], ATP7B [4], [11]–[13] as well as members of the organic cation transporter (OCT) family [14] are reported to play an important role in cisplatin efflux. Altered expression of these proteins has also been correlated to poor outcome in several tumor types, including HNSCC [15], [16].
Once inside the cell, cisplatin looses the chloride groups as a result of the low cellular chloride concentration, a process called aquation. This hydrolyzed form is more reactive to cellular targets of which the nuclear DNA is biologically most relevant. Cisplatin exerts its cytotoxic effect by the formation of intrastrand and interstrand DNA adducts, resulting in cell cycle arrest and eventually cell death. Cells may survive the toxic DNA damage by increased DNA repair activities that lead to the removal of cisplatin adducts. Nucleotide excision repair (NER) is one of the pathways known to remove cisplatin lesions from the DNA [17]. ERCC1 is one of the key factors in the NER system, and the expression of this endonuclease was identified as a prognostic marker in several types of cancer, including HNSCC [18], [19]. Unrepaired cisplatin-DNA adducts eventually result in stalled replication forks during the process of DNA replication in the S-phase of the cell cycle. The cell is then forced into an S-phase arrest to allow time for DNA repair, for example via translesion synthesis or homologous recombination. Members of pathways involved in these processes are described to be extremely important in the process of crosslink repair, and deficiencies in two important players, BRCA1 and BRCA2, sensitize cells to cisplatin treatment [20], [21]. The Fanconi anemia (FA) pathway also proved to be essential in the repair of cisplatin induced DNA damage, since patients with defects in one of the 15 known FA genes show cellular hypersensitivity to DNA cross-linking agents, including cisplatin [22]. Also, ATM and ATR, the proteins that are responsible for the detection of the initial DNA damage and for the initiation of the actual DNA repair process, are likely essential to establish a good DNA repair response.
Furthermore, the TP53 gene, an important player in cellular homeostasis, has also been associated with cisplatin response. It was demonstrated that mutated and inactivated TP53 influences cisplatin sensitivity [23]–[25]. This gene is of special interest concerning its frequently mutated status in human tumors, including HNSCC [26]–[28]. In HNSCC cells the G1 and G2 cell cycle checkpoints can be impaired by TP53 mutations, loss of CDKN2A (p16INK4a ) or gain of CCND1 (Cyclin D1). This might result in the entrance of tumor cells into S-phase despite the presence of cisplatin adducts, leading to cell death.
Despite the importance of the aforementioned genes in the pharmacodynamics and anti-tumor effect of cisplatin, the actual mechanism of cisplatin response in HNSCC is still not understood. Many genes seem to be important in this response, but their relevance may vary from one tumor model to another and affect cisplatin activity at different levels (influx, efflux, metabolism or DNA repair). In the present study we characterized cisplatin sensitivity and the cellular characteristics of 19 HNSCC cell lines, the largest panel tested as far as we know. We investigated the expression levels of several genes that are implicated in cisplatin toxicity and measured cisplatin accumulation and retention rates as well as cisplatin-DNA adduct levels in these cell lines. Finally, we investigated potential correlations between the different parameters studied and the cellular sensitivity to cisplatin.
Materials and Methods
Cell Lines and Chemicals
HNSCC cell lines used were all cultured in Dulbecco's modified Eagle's medium (DMEM), 5% fetal calf serum (FCS) and 2 mM L-glutamine, in a humidified atmosphere of 5% CO2 at 37°C. Cell lines of sporadic tumors were established as described previously [29] and are indicated as VU-SCC-040 (formerly known as 92VU040), VU-SCC-094 (formerly 93VU094), VU-SCC-096 (formerly 93VU096), VU-SCC-120 (formerly 93VU120), VU-SCC-147 (formerly 93VU147) and VU-SCC-OE. Three HNSCC cell lines were established from patients carrying a mutation in one of the Fanconi anemia genes [30]; VU-SCC-1131 (formerly VU1131, with mutations in FANCC), VU-SCC-1365 (formerly VU1365, with mutations in FANCA) and OHSU-0974 (with mutations in FANCA). Cell line VU-SCC-9917 was newly established from a sporadic oral cavity tumor, after written informed consent was obtained. The studies involving clinical specimens were according to the Dutch legislations on research with human material. This study was approved by the Institutional Review Board (Medisch Ethische Toetsingscommissie VUmc).
Cell lines UM-SCC-6, UM-SCC-11B, UM-SCC-14A, UM-SCC-14B, UM-SCC-14C, UM-SCC-22A, UM-SCC-22B and UM-SCC-38 were obtained from Dr. T. Carey (University of Michigan) [31]. The commercially available cell line FaDu was obtained from the American Type Culture Collection. All cell lines have been genetically characterized by microsatellite markers to allow authentication. All cell lines were tested for human papillomavirus (HPV) and only cell line VU-SCC-147 is HPV-positive [32].
Cisplatin was obtained from Teva Pharmachemie (Haarlem, The Netherlands) in a concentration of 1 mg/ml.
IC50 Analysis
Cells were plated in 96 well plates in concentrations that allow the cells to grow till 70% confluence after 5 days (ranging from 1,000–5,000 per well, optimized per cell line). After 24 hours, cells were treated with 18 different concentrations of cisplatin (0–666 µM). Plates were incubated for 72 hours at 37°C/5% CO2. Cell viability was determined by 2 hours incubation with 20 µl of a 1∶1 dilution of CellTiter-Blue (Promega, Leiden, The Netherlands) with culture medium. Fluorescence was measured using an Infinite 200 microplate reader (Tecan, Männedorf, Switzerland).
TP53 Mutation Analysis
Direct dideoxynucleotide sequencing of the evolutionary conserved regions of TP53 (exons 5–9) was performed on all 19 HNSCC cell lines as described previously [28]. In case mutations were not found in these exons, the remaining coding exons (exons 2–4, 10 and 11) were sequenced in addition. Scored mutations were classified according to criteria described by Lindenbergh-van der Plas et al. [28], dividing the tested cell lines into two distinctive groups. Mutations were assigned “truncating” in case of a deletion or insertion. Mutations introducing a stop codon or a splice site change resulting in a frame shift were also called “truncating”. All other mutations leading to an amino acid change were considered “missense”.
Quantitative Real-time PCR
Cell pellets were made from cultures grown to 70% confluence. RNA was extracted using TRIzol (Invitrogen, Bleiswijk, The Netherlands), and quality control tests were performed using the Nanodrop (Thermo Fisher Scientific, Landsmeer, The Netherlands). Complementary DNA was synthesized from 250 ng of RNA template using AMV reverse transcriptase (Promega, Leiden, The Netherlands) and a custom designed reverse primer specific for the gene of interest. Amplification of the cDNA was performed on the ABI/Prism 7500 Sequence Detector System (Taqman-PCR, Applied Biosystems, Nieuwerkerk aan den IJssel, The Netherlands) with 1× Power SYBR Green PCR Master Mix (Applied Biosystems, Nieuwerkerk aan den IJssel, The Netherlands) and custom designed primers for each gene of interest. The primer sequences are displayed in Table S1. For each sample the cycle number at which the amount of amplified target crossed a pre-determined threshold (the Ct-value) was determined. To correct for differences in RNA input and quality, beta-glucuronidase (GUSB) was used as a housekeeping gene [33] for each RNA sample. The mRNA expression was calculated relative to that of GUSB using the Delta/Delta Ct method [34].
Cisplatin Accumulation
Cells were seeded in triplicate in flasks. When 70% confluence was reached, 2 flasks were incubated with 75 µM cisplatin, and the medium of the third flask was refreshed. After four hours, the cells of one cisplatin-incubated flask (the accumulation flask) and the control flask were harvested. The other cisplatin-incubated flask was washed twice with PBS and incubated with fresh medium for another 3 hours, in order to analyze retention of the platinum compound. After counting, cells were resuspended in 0.5 ml 2 M NaOH for overnight incubation at 55°C. Subsequently, 1 ml 1 M HCl was added. Platinum was measured using flameless atomic absorption spectrometry (FAAS) using a Perkin Elmer AS-800 with a THGA graphite furnace (Perkin Elmer,LLC, Norfalk, USA). The instrument response was calibrated across the range 0.2 to 3.0 µM for the whole cell lysate determinations and from 50 to 250 pmol per extract for the DNA adduct determinations (the DNA isolation procedure is specified below). With the graphite furnace 20 µl of individual samples was dried at 110–130°C, ashing was performed at 1300°C and atomization at 2200°C with a read time of 5 seconds.
Cisplatin-DNA Adducts
Cells were treated as described above for the accumulation and retention experiments. Cells were harvested in culture medium, centrifuged and cell pellets were stored at −20°C until further use. The QiaAmp DNA mini kit (Qiagen, Venlo, The Netherlands) was used to extract genomic DNA from the cell pellets, and the quality was controlled by OD260/280 nm analysis on a Nanodrop (Thermo Fisher Scientific, Landsmeer, The Netherlands). Resulting DNA samples were analyzed for cisplatin content using FAAS as described above.
Results
Growth Inhibition
Nineteen head and neck squamous cell carcinoma (HNSCC) cell lines were subjected to treatment with 18 different concentrations of cisplatin, ranging from 0.01 to 666 µM. Examples of resulting curves are shown in Fig. 1. The half-maximal inhibitory concentrations (IC50) and the tumor characteristics of the cell lines are displayed in Table S2. Cell lines VU-SCC-1131 and VU-SCC-1365 were among the most sensitive cell lines with IC50 values of 0.15 µM and 0.80 µM, respectively. This was not unexpected as both these cell lines were established from patients carrying a mutation in one of the Fanconi anemia (FA) genes. On the other hand, cell line OHSU-0974, also established from a FA patient, did not show an extremely sensitive phenotype (IC50 value 2.99 µM). Cell line UM-SCC-38 showed the highest IC50 value with 7.57 µM.
Figure 1. The half-maximal inhibitory concentration of cisplatin.

Nineteen HNSCC cell lines were continuously treated with 18 different concentrations of cisplatin during 72 h. Cell viability was determined and plotted against the cisplatin concentration. Cell lines VU-SCC-094 and UM-SCC-38 are presented as examples of a relatively sensitive and a relatively resistant cell line, respectively. Measurements were performed in at least three independent triplicate experiments. Error bars represent the standard error of the mean.
There was no significant correlation between the stage of the tumor from which the cell lines were derived and the IC50 value (p = 0.297, Spearman’s rho = −0.289). Cell lines UM-SCC-14B, UM-SCC-14C and UM-SCC-22B were left out of this analysis since these were not established from independent primary tumors. When we grouped the cell lines according to their tumor stage (early stage tumors (T1–T2) or late stage (T3–T4)), a significant correlation with cisplatin sensitivity was not found either (p = 0.614, Spearman’s rho = −0.142).
TP53 Mutation Status
We investigated the TP53 mutation status in all 19 cell lines (Table S2). Two cell lines (UM-SCC-6 and VU-SCC-040) harbored wild-type TP53, and the other 17 cell lines (88%) showed mutations in the TP53 gene. Nine cell lines (UM-SCC-14A, UM-SCC-14B, UM-SCC-14C, UM-SCC-22A, UM-SCC-22B, VU-SCC-094, VU-SCC-9917, VU-SCC-OE and OHSU-0974) had truncating TP53 mutations according to the classification described by Lindenbergh-van der Plas et al. [28]. These mutations included one deletion, two frameshift mutations, 2 splice site mutations and four nonsense mutations that resulted in a stop codon. Cell lines UM-SCC-14A, UM-SCC-14B, UM-SCC-14C, UM-SCC-22A and UM-SCC-22B each harbor two TP53 mutations; one missense and one truncating mutation. For further analyses, we only counted the truncating mutation since this mutation has most impact on TP53 functioning. The other eight (47%) cell lines showed one or more missense TP53 mutations. The TP53 mutation status (wild type, truncating or missense) did not significantly correlate to cisplatin sensitivity (p = 0.128, Spearman’s rho = 0.600), even when UM-SCC-14B, UM-SCC-14C and UM-SCC-22B were eliminated from the analysis (p = 0.611, Spearman’s rho = 0.138).
Expression of Genes Related to Cisplatin Sensitivity
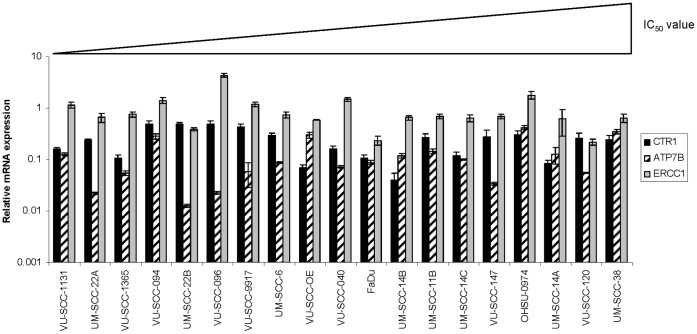
The toxic effect of cisplatin may be dependent on the transport of the compound into the cell, the activation of platinum inside the cell and also the efflux out of the cell (reviewed in [2]). Even when the drug is already bound to the DNA, the toxic effect can be abolished by removal of the formed crosslink. Several genes have been described to mediate cisplatin influx (CTR1, OCT1, OCT2 and OCT3), efflux (ATP7A, ATP7B) or are involved in DNA crosslink repair (ATR, ATM, BRCA1, BRCA2 and ERCC1). We used quantitative real-time PCR to investigate the expression of these genes in our cell line panel (Table S3). The expression patterns of CTR1, ATP7B and ERCC1 are presented in Fig. 2. The expression of both OCT1 and OCT2 could often not be determined due to very low mRNA expression levels. Therefore, these genes were eliminated from further analyses. The mRNA expression levels of the remaining nine genes were correlated to each other as well as to the IC50 values, the clinical characteristics of the cell lines and the TP53 mutation status. Unfortunately, we could not find a significant correlation between the IC50 value for cisplatin and the expression of any of the genes tested. Even when we excluded the cell lines with a known mutation in one of the FA genes (VU-SCC-1131, VU-SCC-1365 and OHSU-0974), no correlations were found. Furthermore, the type of TP53 mutation (wild type, truncating or missense) also showed no correlation with the expression of the nine genes tested. Remarkably, there was a highly significant correlation (Spearman’s rho = 0.95, p<0.001) between BRCA1 and BRCA2 expression in the 19 cell lines (Table 1), suggesting that these genes are co-regulated in HNSCC, similar to what has been shown in mammary epithelial cells [35]. The relative mRNA expression of BRCA1 ranged from 0.31 till 6.5 and for BRCA2 from 0.20 till 5,0. The expression of BRCA1 and BRCA2 also correlated with the expression of ERCC1, the gene that is held responsible for cisplatin-DNA adduct repair via the NER pathway.
Figure 2. Relative mRNA expression levels of CTR1, ATP7B and ERCC.
The mRNA expression level of eleven genes were determined in our HNSCC cell line panel. Genes were selected on basis of their involvement in cisplatin toxicity. CTR1 is believed to mediate cisplatin influx, ATP7B is a known cisplatin efflux pump and ERCC1 is a DNA repair protein. Results are medians of triplicate experiments and are presented relative to the expression of the beta-glucuronidase (GUSB) housekeeping gene. Error bars represent standard deviations.
Table 1. Spearman’s rho correlations of the mRNA expression levels of the nine genes examined in 19 HNSCC cell lines.
| CTR1 | OCT3 | ATP7A | ATP7B | ATM | ATR | BRCA1 | BRCA2 | ERCC1 | ||
| CTR1 | N | 19 | 19 | 19 | 13 | 19 | 19 | 19 | 19 | |
| Correlation | −0.026 | 0.426 | 0.28 | 0.487 | 0.366 | 0.586 | 0.534 | 0.426 | ||
| p | 0.915 | 0.069 | 0.246 | 0.091 | 0.123 | 0.008 | 0.019 | 0.069 | ||
| OCT3 | N | 19 | 19 | 13 | 19 | 19 | 19 | 19 | ||
| Correlation | 0.214 | 0.126 | −0.187 | 0.356 | −0.034 | −0.011 | 0.282 | |||
| p | 0.379 | 0.606 | 0.541 | 0.135 | 0.889 | 0.966 | 0.243 | |||
| ATP7A | N | 19 | 13 | 19 | 19 | 19 | 19 | |||
| Correlation | 0.202 | 0.011 | 0.521 | 0.497 | 0.398 | 0.582 | ||||
| p | 0.408 | 0.972 | 0.022 | 0.03 | 0.091 | 0.009 | ||||
| ATP7B | N | 13 | 19 | 19 | 19 | 19 | ||||
| Correlation | −0.56 | 0.253 | 0.253 | 0.298 | 0.075 | |||||
| p | 0.046 | 0.297 | 0.296 | 0.215 | 0.759 | |||||
| ATM | N | 13 | 13 | 13 | 13 | |||||
| Correlation | 0.28 | 0.033 | 0.016 | 0.385 | ||||||
| p | 0.354 | 0.915 | 0.957 | 0.194 | ||||||
| ATR | N | 19 | 19 | 19 | ||||||
| Correlation | 0.458 | 0.367 | 0.396 | |||||||
| p | 0.048 | 0.123 | 0.093 | |||||||
| BRCA1 | N | 19 | 19 | |||||||
| Correlation | 0.953 | 0.494 | ||||||||
| p | 0.000 | 0.032 | ||||||||
| BRCA2 | N | 19 | ||||||||
| Correlation | 0.517 | |||||||||
| p | 0.023 | |||||||||
| ERCC1 | N | |||||||||
| Correlation | ||||||||||
| p |
Bold Correlation is significant at the 0.01 level (2-tailed). Italics Correlation is significant at the 0.05 level (2-tailed).
Cisplatin Accumulation and Retention
The cellular cisplatin accumulation was determined after incubating the cells with 75 µM cisplatin during four hours (Table 2). This concentration is not toxic in this short incubation period and allows reliable measurements. The cisplatin accumulation ranged from 91 pmol/106 cells for cell line UM-SCC-14C to 923 pmol/106 cells for cell line UM-SCC-6. Cisplatin retention was determined by incubating the cells with fresh medium during three hours post cisplatin incubation. The maximum efflux was found in cell line VU-SCC-040 (80%, Table 2). There were also three cell lines (UM-SCC-14B, UM-SCC-22A and OHSU-0974) that did not show any cisplatin efflux during the drug-free period.
Table 2. Cisplatin accumulation after four hours of cisplatin exposure and the percentage of cisplatin retention after three hours in drug free medium.
| HNSCC cell line | IC50 (µM) | Cisplatin accumulation (pmol cisplatin/106 cells) | Retention (%) |
| UM-SCC-6 | 1.12±0.15 | 922±56 | 70.4 |
| UM-SCC-11B | 2.48±0.16 | 153±6 | 79.6 |
| UM-SCC-14A | 3.27±0.51 | 337±12 | 69.6 |
| UM-SCC-14B | 2.35±0.70 | 242±62 | 100 |
| UM-SCC-14C | 2.60±0.17 | 91±13 | 85.9 |
| UM-SCC-22A | 0.69±0.32 | 97±30 | 100 |
| UM-SCC-22B | 0.93±0.31 | 452±9 | 74.2 |
| UM-SCC-38 | 7.57±1.25 | 373±22 | 52.6 |
| VU-SCC-040 | 1.81±0.45 | 267±10 | 20.0 |
| VU-SCC-094 | 0.91±0.13 | ND | ND |
| VU-SCC-096 | 0.99±0.46 | 588±29 | 56.5 |
| VU-SCC120 | 4.03±0.92 | 176±14 | 87.9 |
| VU-SCC-147 | 2.97±0.39 | ND | ND |
| VU-SCC-9917 | 0.99±0.12 | 258±16 | 95.5 |
| VU-SCC-OE | 1.66±0.25 | 252±51 | 98.4 |
| VU-SCC-1131 | 0.15±0.04 | 334±114 | 82.3 |
| VU-SCC-1365 | 0.80±0.18 | 275±76 | 97.3 |
| OHSU-0974 | 2.99±0.23 | 275±54 | 100 |
| FaDu | 1.83±0.10 | 285±90 | 61.2 |
Retention (%) represents the remaining cellular cisplatin concentration after three hours in drug free medium. Values are the means of at least duplicate experiments ± SEM. ND; not determined.
Intriguingly, the large range in accumulation did not correlate to the IC50 value (p = 0.589, Spearman’s rho = −0.141) nor to any of the mRNA expression levels, among which were the assigned influx pumps CTR1 and OCT3. Elimination of cell lines with a known FA defect from the analysis did not change this observation. The percentage retention also did not show a significant correlation to any of the characteristics of the cell lines, including the mRNA expression of efflux pumps ATP7A and ATP7B. Remarkably, we found a significantly inverse correlation between the cisplatin accumulation and retention (p = 0.015, Spearman’s rho = −0.580), indicating that cells with high cisplatin accumulation also show a strong efflux.
Cisplatin-DNA Adducts
DNA isolated from cell cultures incubated with 75 µM cisplatin for four hours was subjected to flameless atomic absorption spectrometry (FAAS) in order to determine the level of platinum adducts in the DNA. The resulting data are displayed in Table 3. The platinum content ranged from 0.085 pmol/µg DNA in cell line UM-SCC-14C to 1.235 pmol/ug DNA in VU-SCC-1365 cells.
Table 3. Platinum-DNA adducts after four hours of cisplatin exposure and the percentage of adduct retention after three hours in drug free medium.
| HNSCC cell line | IC50 (µM) | Platinum-DNA adducts (pmol platinum/µg DNA) |
| UM-SCC-6 | 1.12±0.15 | 0.375±0.09 |
| UM-SCC-11B | 2.48±0.16 | 0.525±0.08 |
| UM-SCC-14A | 3.27±0.51 | 0.405±0.13 |
| UM-SCC-14B | 2.35±0.70 | 0.975±0.16 |
| UM-SCC-14C | 2.60±0.17 | 0.085±0.03 |
| UM-SCC-22A | 0.69±0.32 | 0.99±0.00 |
| UM-SCC-22B | 0.93±0.31 | 0.88±0.16 |
| UM-SCC-38 | 7.57±1.25 | 0.33±0.06 |
| VU-SCC-040 | 1.81±0.45 | 0.48±0.08 |
| VU-SCC-094 | 0.91±0.13 | ND |
| VU-SCC-096 | 0.99±0.46 | 1.12±0.26 |
| VU-SCC120 | 4.03±0.92 | 0.27±0.05 |
| VU-SCC-147 | 2.97±0.39 | ND |
| VU-SCC-9917 | 0.99±0.12 | 0.73±0.03 |
| VU-SCC-OE | 1.66±0.25 | 0.395±0.03 |
| VU-SCC-1131 | 0.15±0.04 | 0.835±0.09 |
| VU-SCC-1365 | 0.80±0.18 | 1.235±0.08 |
| OHSU-0974 | 2.99±0.23 | 0.525±0.23 |
| FaDu | 1.83±0.10 | 0.365±0.04 |
Values are the means of duplicate experiments ± SEM. ND; not determined.
The most remarkable finding in our study is that the level of cisplatin-DNA adducts showed a significantly inverse correlation with the IC50 value (p = 0.002, Spearman’s rho = −0.703), even when we excluded the three cell lines with a known mutation in one of the FA genes (p = 0.006, Spearman’s rho = −0.697). This indicates that cells with a low IC50 value for cisplatin contain significantly more DNA adducts compared to cells that are more resistant to cisplatin (Figure 3).
Figure 3. Inverse correlation between IC50 value and accumulation of platinum-DNA adducts.
The level of DNA-bound platinum was determined in 17 HNSCC cell lines that were incubated with cisplatin. A significant inverse correlation was found between the platinum-DNA adducts and the IC50 value. Measurements were performed in duplicate and error bars represent standard deviations.
Discussion
Multiple studies have been performed to find key factors determining the cellular sensitivity to cisplatin. Until now, numerous factors have been identified, but there seems to be no concordance. We evaluated the sensitivity to cisplatin as well as the accumulation and retention of cisplatin and platinum-DNA adducts in a large panel of established HNSCC cell lines with known clinical characteristics. Also the expression levels of several genes known to be involved in cisplatin toxicity were determined. The only parameter that showed a significant correlation to cisplatin sensitivity was the platinum-DNA adduct level. Remarkably, neither the intracellular accumulation nor the retention showed a correlation with either IC50 or DNA-bound cisplatin. Apparently, it seems the DNA-bound fraction that determines cisplatin response in head-and-neck cancer more than something else. This suggests that either nuclear transport of cisplatin and/or the level of DNA repair differs between cells, resulting in a different adduct level. It further suggests that assessment of cisplatin-DNA adducts may predict treatment outcome in patients. This has been shown previously but only in small series [36], [37]. At that time the data were obtained by laborious postlabeling techniques to assess adduct levels, but the current highly sensitive FAAS equipment may be much more suited for clinical studies. The remaining major obstacle is the demand for tumor biopsies after cisplatin infusion, which is only possible for easy accessible tumors.
We determined the cisplatin sensitivity in 19 HNSCC cell lines by treating the cells with 18 different cisplatin concentrations during 72 hours. Since pharmacokinetics play an important role in drug availability in patients, it is hard to compare the cisplatin concentrations used in this study to the cisplatin levels that are reached clinically. Patients are nowadays treated with 100 mg/m2 cisplatin every three weeks, usually with a one hour intravenous infusion. This administration results in a peak concentration of cisplatin in the blood at a few minutes after stopping the infusion. The peak concentration of free cisplatin has been analyzed and varies between 3.5 and 6 µg/ml [38], which corresponds to approximately 12–18 µM. Furthermore, the total plasma platinum concentration rapidly decreases already within the first two hours after infusion [39]. The cisplatin concentrations used in this study vary from 0.01–666 µM, which corresponds to 3 ng/ml−0.2 mg/ml. This means that we totally cover the range of peak cisplatin concentrations measured for patients. Nonetheless, it is difficult to compare the fixed cisplatin concentration in vitro with the rapidly decreasing concentration in patients.
Several studies reported an association between cisplatin sensitivity and TP53 mutations in solid cancer cell lines in vitro [23], [40]–[42], but with conflicting results which might be a consequence of the small sample sizes and different origins of the cell lines tested. However, in the present large panel of HNSCC cell lines we could not find a correlation between the type of TP53 mutation and the corresponding IC50 value. However, our cell line panel contained only two wild type cell lines, and therefore we were unable to determine whether wild type TP53 significantly altered the sensitivity to cisplatin as compared to cell lines harboring a mutated form of the tumor suppressor gene. Nevertheless, we observed that the two HNSCC cell lines with wild type TP53 were among the most sensitive cell lines tested. Also in vivo studies searching for a correlation between TP53 mutation status and outcome after cisplatin treatment in multiple cancer types led to opposing results [43]–[45]. In case of HNSCC, it is known that approximately 80% of the tumors has an incorrect functioning of the TP53 pathway by either TP53 mutation or HPV infection [46], [47], implicating that tumors with wild type TP53 are rare. This already makes it difficult to imagine that TP53 mutation status could explain the response to cisplatin.
The influence of copper transporters on the import (CTR1 [6], [7], [9]) and efflux of cisplatin (ATP7A and/or ATP7B [11]–[13], [16]) has been observed in a variety of human cancer cell lines. In our data, we could not find a significant correlation between the mRNA expression of CTR1, ATP7A and ATP7B and cisplatin sensitivity. Since the mRNA level does not necessarily reflect the protein level or pump activity within a cell, we might have missed the effect of these genes on the cellular sensitivity to cisplatin. Similarly, we did not find a correlation between CTR1, ATP7A and ATP7B expression and the cisplatin accumulation and retention. On the other hand, mutations that do not influence the expression level of the copper transporters but that do have an effect on the pump activity and the efficiency of cisplatin transport were not detectable with our quantitative real-time PCR approach, although this might have been reflected in an altered cisplatin accumulation, retention or DNA adduct formation. In addition, the subcellular localization of the ATP7A and ATP7B proteins could not be determined in our experimental setup because of very low transcript levels, but might be of importance in the process of cisplatin efflux [48].
Although defects in DNA repair mechanisms are essential for cisplatin induced toxicity, we could not find clear associations between the expression levels of ERCC1, BRCA1 and BRCA2 and cisplatin sensitivity. Again, this might be a result of the detection method we employed. The BRCA1 and BRCA2 genes are described to be often mutated or transcriptionally silenced by promoter methylation, although this has not been observed in HNSCC [49]. Certain polymorphisms in ERCC1 have been described to be associated with cisplatin sensitivity in multiple types of cancer. These polymorphisms do not alter the expression level of the gene, but might modify the functional activity of the protein.
When comparing the expression levels of the genes tested in our HNSCC panel, we found several significant correlations. Some correlations were explainable when looking at the gene functions. We found a very strong correlation between the expression of BRCA1 and BRCA2, which might point at a possible co-regulation of these genes. It has been shown in prostate cancer and in breast cancer that BRCA1 and BRCA2 are indeed co-regulated in response to DNA damage. This phenomenon was also observed in untreated mammary epithelial cells [35]. Furthermore, the expression level of several other genes with distinct functions within a cell (like ATP7A and BRCA1) showed a significant correlation, but with the current literature at hand no likely explanation for these correlations could be suggested.
From these results we can conclude that the mechanism of cisplatin response seems a relatively simple process in HNSCC. Cells with high IC50 values harbor less platinum-DNA adducts. This suggests that the DNA adduct level resulting from cisplatin incubation is the most important determinant of cisplatin sensitivity. Therefore, imaging of radio-labeled cisplatin in patients might be an excellent indicator for treatment outcome. A genome-wide functional siRNA screen might be a useful tool to determine all the genes that specifically determine the cisplatin response and consequently the adduct accumulation level.
Supporting Information
Primer sequences for quantitative real-time PCR.
(DOC)
Inhibitory cisplatin concentrations (IC50 values) and TP53 mutation status of 19 HNSCC cell lines.
(DOC)
Relative mRNA expression of the indicated genes in 19 HNSCC cell lines.
(XLS)
Acknowledgments
We thank Nienke Losekoot for technical assistance.
Funding Statement
The authors have no support or funding to report.
References
- 1. Bernier J, Domenge C, Ozsahin M, Matuszewska K, Lefebvre JL, et al. (2004) Postoperative irradiation with or without concomitant chemotherapy for locally advanced head and neck cancer. N. Engl. J. Med. 350: 1945–1952. [DOI] [PubMed] [Google Scholar]
- 2. Kelland L (2007) The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 7: 573–584. [DOI] [PubMed] [Google Scholar]
- 3. Wang D, Lippard SJ (2005) Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 4: 307–320. [DOI] [PubMed] [Google Scholar]
- 4. Katano K, Kondo A, Safaei R, Holzer A, Samimi G, et al. (2002) Acquisition of resistance to cisplatin is accompanied by changes in the cellular pharmacology of copper. Cancer Res 62: 6559–6565. [PubMed] [Google Scholar]
- 5. Safaei R, Katano K, Samimi G, Naerdemann W, Stevenson JL, et al. (2004) Cross-resistance to cisplatin in cells with acquired resistance to copper. Cancer Chemother. Pharmacol. 53: 239–246. [DOI] [PubMed] [Google Scholar]
- 6. Holzer AK, Samimi G, Katano K, Naerdemann W, Lin X, et al. (2004) The copper influx transporter human copper transport protein 1 regulates the uptake of cisplatin in human ovarian carcinoma cells. Mol. Pharmacol. 66: 817–823. [DOI] [PubMed] [Google Scholar]
- 7. Ishida S, Lee J, Thiele DJ, Herskowitz I (2002) Uptake of the anticancer drug cisplatin mediated by the copper transporter Ctr1 in yeast and mammals. Proc. Natl. Acad. Sci. U. S. A 99: 14298–14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin X, Okuda T, Holzer A, Howell SB (2002) The copper transporter CTR1 regulates cisplatin uptake in Saccharomyces cerevisiae. Mol. Pharmacol. 62: 1154–1159. [DOI] [PubMed] [Google Scholar]
- 9. Song IS, Savaraj N, Siddik ZH, Liu P, Wei Y, et al. (2004) Role of human copper transporter Ctr1 in the transport of platinum-based antitumor agents in cisplatin-sensitive and cisplatin-resistant cells. Mol. Cancer Ther. 3: 1543–1549. [PubMed] [Google Scholar]
- 10. Samimi G, Safaei R, Katano K, Holzer AK, Rochdi M, et al. (2004) Increased expression of the copper efflux transporter ATP7A mediates resistance to cisplatin, carboplatin, and oxaliplatin in ovarian cancer cells. Clin. Cancer Res 10: 4661–4669. [DOI] [PubMed] [Google Scholar]
- 11. Katano K, Safaei R, Samimi G, Holzer A, Tomioka M, et al. (2004) Confocal microscopic analysis of the interaction between cisplatin and the copper transporter ATP7B in human ovarian carcinoma cells. Clin. Cancer Res 10: 4578–4588. [DOI] [PubMed] [Google Scholar]
- 12. Komatsu M, Sumizawa T, Mutoh M, Chen ZS, Terada K, et al. (2000) Copper-transporting P-type adenosine triphosphatase (ATP7B) is associated with cisplatin resistance. Cancer Res. 60: 1312–1316. [PubMed] [Google Scholar]
- 13. Nakayama K, Miyazaki K, Kanzaki A, Fukumoto M, Takebayashi Y (2001) Expression and cisplatin sensitivity of copper-transporting P-type adenosine triphosphatase (ATP7B) in human solid carcinoma cell lines. Oncol. Rep. 8: 1285–1287. [DOI] [PubMed] [Google Scholar]
- 14. Noordhuis P, Laan AC, van de BK, Losekoot N, Kathmann I, et al. (2008) Oxaliplatin activity in selected and unselected human ovarian and colorectal cancer cell lines. Biochem. Pharmacol. 76: 53–61. [DOI] [PubMed] [Google Scholar]
- 15. Higashimoto M, Kanzaki A, Shimakawa T, Konno S, Naritaka Y, et al. (2003) Expression of copper-transporting P-type adenosine triphosphatase in human esophageal carcinoma. Int. J. Mol. Med. 11: 337–341. [PubMed] [Google Scholar]
- 16. Miyashita H, Nitta Y, Mori S, Kanzaki A, Nakayama K, et al. (2003) Expression of copper-transporting P-type adenosine triphosphatase (ATP7B) as a chemoresistance marker in human oral squamous cell carcinoma treated with cisplatin. Oral Oncol. 39: 157–162. [DOI] [PubMed] [Google Scholar]
- 17. Wang Z, Wu X, Friedberg EC (1993) Nucleotide-excision repair of DNA in cell-free extracts of the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A 90: 4907–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Handra-Luca A, Hernandez J, Mountzios G, Taranchon E, Lacau-St-Guily J, et al. (2007) Excision repair cross complementation group 1 immunohistochemical expression predicts objective response and cancer-specific survival in patients treated by Cisplatin-based induction chemotherapy for locally advanced head and neck squamous cell carcinoma. Clin. Cancer Res. 13: 3855–3859. [DOI] [PubMed] [Google Scholar]
- 19. Jun HJ, Ahn MJ, Kim HS, Yi SY, Han J, et al. (2008) ERCC1 expression as a predictive marker of squamous cell carcinoma of the head and neck treated with cisplatin-based concurrent chemoradiation. Br. J. Cancer 99: 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burkitt K, Ljungman M (2007) Compromised Fanconi anemia response due to BRCA1 deficiency in cisplatin-sensitive head and neck cancer cell lines. Cancer Lett. 253: 131–137. [DOI] [PubMed] [Google Scholar]
- 21. Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, et al. (1999) BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 59: 3547–3551. [PubMed] [Google Scholar]
- 22. de Winter JP, Joenje H (2009) The genetic and molecular basis of Fanconi anemia. Mutat. Res. 668: 11–19. [DOI] [PubMed] [Google Scholar]
- 23. Branch P, Masson M, Aquilina G, Bignami M, Karran P (2000) Spontaneous development of drug resistance: mismatch repair and p53 defects in resistance to cisplatin in human tumor cells. Oncogene 19: 3138–3145. [DOI] [PubMed] [Google Scholar]
- 24. Reles A, Wen WH, Schmider A, Gee C, Runnebaum IB, et al. (2001) Correlation of p53 mutations with resistance to platinum-based chemotherapy and shortened survival in ovarian cancer. Clin. Cancer Res. 7: 2984–2997. [PubMed] [Google Scholar]
- 25. Siddik ZH, Mims B, Lozano G, Thai G (1998) Independent pathways of p53 induction by cisplatin and X-rays in a cisplatin-resistant ovarian tumor cell line. Cancer Res. 58: 698–703. [PubMed] [Google Scholar]
- 26. Hollstein M, Sidransky D, Vogelstein B, Harris CC (1991) p53 mutations in human cancers. Science 253: 49–53. [DOI] [PubMed] [Google Scholar]
- 27. Levine AJ, Momand J, Finlay CA (1991) The p53 tumour suppressor gene. Nature 351: 453–456. [DOI] [PubMed] [Google Scholar]
- 28. Lindenbergh-van der Plas M, Brakenhoff RH, Kuik DJ, Buijze M, Bloemena E, et al. (2011) Prognostic significance of truncating TP53 mutations in head and neck squamous cell carcinoma. Clin. Cancer Res. 17: 3733–3741. [DOI] [PubMed] [Google Scholar]
- 29. Hermsen MA, Joenje H, Arwert F, Welters MJ, Braakhuis BJ, et al. (1996) Centromeric breakage as a major cause of cytogenetic abnormalities in oral squamous cell carcinoma. Genes Chromosomes. Cancer 15: 1–9. [DOI] [PubMed] [Google Scholar]
- 30. van Zeeburg HJ, Snijders PJ, Pals G, Hermsen MA, Rooimans MA, et al. (2005) Generation and molecular characterization of head and neck squamous cell lines of fanconi anemia patients. Cancer Res. 65: 1271–1276. [DOI] [PubMed] [Google Scholar]
- 31. Lin CJ, Grandis JR, Carey TE, Gollin SM, Whiteside TL, et al. (2007) Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck 29: 163–188. [DOI] [PubMed] [Google Scholar]
- 32. Steenbergen RD, Hermsen MA, Walboomers JM, Joenje H, Arwert F, et al. (1995) Integrated human papillomavirus type 16 and loss of heterozygosity at 11q22 and 18q21 in an oral carcinoma and its derivative cell line. Cancer Res. 55: 5465–5471. [PubMed] [Google Scholar]
- 33. Aerts JL, Gonzales MI, Topalian SL (2004) Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. Biotechniques 36: 84–1. [DOI] [PubMed] [Google Scholar]
- 34. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 35. Rajan JV, Wang M, Marquis ST, Chodosh LA (1996) Brca2 is coordinately regulated with Brca1 during proliferation and differentiation in mammary epithelial cells. Proc. Natl. Acad. Sci. U. S. A 93: 13078–13083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoebers FJ, Pluim D, Verheij M, Balm AJ, Bartelink H, et al. (2006) Prediction of treatment outcome by cisplatin-DNA adduct formation in patients with stage III/IV head and neck squamous cell carcinoma, treated by concurrent cisplatin-radiation (RADPLAT). Int. J. Cancer 119: 750–756. [DOI] [PubMed] [Google Scholar]
- 37. Welters MJ, Braakhuis BJ, Jacobs-Bergmans AJ, Kegel A, Baan RA, et al. (1999) The potential of plantinum-DNA adduct determination in ex vivo treated tumor fragments for the prediction of sensitivity to cisplatin chemotherapy. Ann. Oncol. 10: 97–103. [DOI] [PubMed] [Google Scholar]
- 38. Andersson A, Fagerberg J, Lewensohn R, Ehrsson H (1996) Pharmacokinetics of cisplatin and its monohydrated complex in humans. J. Pharm. Sci. 85: 824–827. [DOI] [PubMed] [Google Scholar]
- 39. Urien S, Lokiec F (2004) Population pharmacokinetics of total and unbound plasma cisplatin in adult patients. Br. J. Clin. Pharmacol. 57: 756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bradford CR, Zhu S, Ogawa H, Ogawa T, Ubell M, et al. (2003) P53 mutation correlates with cisplatin sensitivity in head and neck squamous cell carcinoma lines. Head Neck 25: 654–661. [DOI] [PubMed] [Google Scholar]
- 41.Hoffmann TK, Sonkoly E, Hauser U, van LA, Whiteside TL et al.. (2008) Alterations in the p53 pathway and their association with radio- and chemosensitivity in head and neck squamous cell carcinoma. Oral Oncol. [DOI] [PubMed]
- 42. Mandic R, Schamberger CJ, Muller JF, Geyer M, Zhu L, et al. (2005) Reduced cisplatin sensitivity of head and neck squamous cell carcinoma cell lines correlates with mutations affecting the COOH-terminal nuclear localization signal of p53. Clin. Cancer Res 11: 6845–6852. [DOI] [PubMed] [Google Scholar]
- 43. Andrews GA, Xi S, Pomerantz RG, Lin CJ, Gooding WE, et al. (2004) Mutation of p53 in head and neck squamous cell carcinoma correlates with Bcl-2 expression and increased susceptibility to cisplatin-induced apoptosis. Head Neck 26: 870–877. [DOI] [PubMed] [Google Scholar]
- 44. Bauer JA, Trask DK, Kumar B, Los G, Castro J, et al. (2005) Reversal of cisplatin resistance with a BH3 mimetic, (-)-gossypol, in head and neck cancer cells: role of wild-type p53 and Bcl-xL. Mol. Cancer Ther. 4: 1096–1104. [DOI] [PubMed] [Google Scholar]
- 45. Cabelguenne A, Blons H, de W, I, Carnot F, Houllier AM, et al. (2000) p53 alterations predict tumor response to neoadjuvant chemotherapy in head and neck squamous cell carcinoma: a prospective series. J. Clin. Oncol. 18: 1465–1473. [DOI] [PubMed] [Google Scholar]
- 46. Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, et al. (2011) Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333: 1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Leemans CR, Braakhuis BJ, Brakenhoff RH (2011) The molecular biology of head and neck cancer. Nat. Rev. Cancer 11: 9–22. [DOI] [PubMed] [Google Scholar]
- 48. Kalayda GV, Wagner CH, Buss I, Reedijk J, Jaehde U (2008) Altered localisation of the copper efflux transporters ATP7A and ATP7B associated with cisplatin resistance in human ovarian carcinoma cells. BMC. Cancer 8: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Snyder ER, Ricker JL, Chen Z, Waes CV (2007) Variation in cisplatinum sensitivity is not associated with Fanconi Anemia/BRCA pathway inactivation in head and neck squamous cell carcinoma cell lines. Cancer Lett. 245: 75–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences for quantitative real-time PCR.
(DOC)
Inhibitory cisplatin concentrations (IC50 values) and TP53 mutation status of 19 HNSCC cell lines.
(DOC)
Relative mRNA expression of the indicated genes in 19 HNSCC cell lines.
(XLS)