Abstract
The purpose of the present work was to determine the incidence and clinical implications of somatic EZH2 mutations in 714 patients with de novo acute myelogenous leukemia by sequencing the entire coding region. EZH2 mutations were identified in 13/714 (1.8%) of AML patients were found to be more common in males (P = 0.033). The presence of EZH2 mutations was significantly associated with lower blast percentage (21–30%) in bone marrow (P<0.0001) and -7/del(7q) (P = 0.025). There were no differences in the incidence of mutation in 13 genes, ASXL1, CBL, c-KIT, DNMT3A, FLT3, IDH1, IDH2, MLL, NPM1, NRAS, RUNX1, TET2, and WT1, between patients with and without EZH2 mutations. No difference in complete remission, event-free survival, or overall survival was observed between patients with and without EZH2 mutation (P>0.05). Overall, these results showed EZH2 mutation in de novo acute myeloid leukemia as a recurrent genetic abnormality to be associated with lower blast percentage in BM and -7/del(7q).
Introduction
Acute myeloid leukemia (AML) is a heterogeneous group of neoplastic disorders with considerable variability with regard to clinical course and response to treatment, as well as in their cytogenetic and molecular genetic features. Major advances in the understanding of the mechanisms for AML have been made by the characterization and the study of acquired cytogenetic abnormalities, especially reciprocal translocations and inversions observed in AML. Cytogenetic abnormalities are been considered the most important prognostic parameters in AML. Seven recurrent balanced chromosomal abnormalities, specifically t(15;17)(q22;q12), t(8;21)(q22;q22), inv(16)(p13.1q22) or t(16;16)(p13.1;q22), t(9;11)(p22;q23), t(6;9)(p23;q34), inv(3)(q21q26.2) or t(3;3)(q21;q26.2), and t(1;22)(p13;q13), are recognized as the cytogenetic hallmarks of genetically defined disease entities in the WHO classification criteria, which were revised in 2008. [1] Nevertheless, approximately half of patients with de novo AML lack typical clonal karyotypic abnormalities. [2]–[3] In recent years, genome-wide profiling of DNA copy-number variations and candidate gene sequencing have described abundant recurrent genetic mutations in patients with AML, such as FLT3, c-KIT, NPM1, WT1, ASXL1, DNMT3A, IDH1, IDH2, TET2, CBL, RUNX1, PHF6, and CEBPA. [4] Remarkably, several genes involved in the epigenetic regulation of transcription, including TET2, ASXL1, IDH1, IDH2, and DNMT3A, have been found to be mutated in AML, myelodisplastic syndrome (MDS) and other myeloproliferative neoplasmas (MPN). [5]–[12].
EZH2, located in 7q36.1, is another important gene associated with epigenetic regulation of transcription. EZH2 encodes the catalytic component of the polycomb repressive complex 2 (PRC2), which is responsible for the methylation of lysine 27 on the N-terminal tail of histone H3 (H3K27), which influences stem cell renewal by epigenetic modification. [13]–[14] Macro- and microdeletions involving EZH2 have been detected in about 10% of MDS patients [15], and a few MDS patients have shown loss-of-heterozygosity caused by acquired uniparental disomy. [16]–[17] Somatic mutations of EZH2 were recently identified in lymphoma and myeloid neoplasmas. In lymphoma, somatic mutations affecting codon 641 of EZH2 were detected in 7% of follicular lymphomas and 22% of diffuse large cell B-cell lymphomas of germinal center origin, resulting in enhanced lymphomagenesis. [18] In myeloid neoplasms, mutations were found throughout the EZH2 and have been described in 10–13% of poor-prognosis myelodysplasia-myeloproliferative neoplasms (MDS/MPN), 13% of myelofibrosis (MF), and 6% of MDS. [16]–[17] However, the prevalence and clinical significance of somatic EZH2 mutations in patients with acute myelogenous leukemia (AML) remains largely unknown.
In this study, we investigated the prevalence and prognostic value of somatic EZH2 mutations in 714 patients with de novo AML by PCR amplification of the entire coding region of EZH2 followed by direct bidirectional DNA sequencing. Patients were also assessed for the presence of mutations in 13 genes, including ASXL1, CBL, c-KIT, DNMT3A, FLT3, IDH1, IDH2, MLL, NPM1, NRAS, RUNX1, TET2, and WT1.
Methods
Patients
From January 2005 to December 2010, a total of 714 patients who had been newly diagnosed with AML at Jiangsu Institute of Hematology (JIH) were enrolled in the present study. Patients with antecedent hematologic diseases, especially agnogenic hematocytopenia, and those experiencing MDS, MDS/MPN, or therapy-related AML were excluded. In order to exclude AMLs attributable to MDS, all the samples from patients with karyotypes -7, 7q-, -5, 5q-, 20q-, or BM-blast less than 30% were examined retrospectively, and no evidence of morphologic myelodysplasia or atypical localization of immature progenitor (ALIP) was found. Diagnosis and classification of AML were defined according to the French-American-British classification (FAB) system and were revised using the World Health Organization (WHO 2008) classification system. The main characteristics of the patients in this study and the entire group are summarized in Table S1. This study was approved by the Ethics Committee of the First Affiliated Hospital of Soochow University. Written informed consent was provided for sample preservation and genetic analysis from every subject. For minors (56 patients, age 8–17 y) written informed consent of the parent or guardian was also obtained. Genomic DNA was extracted from frozen bone marrow mononuclear cells (BMMCs) after Ficoll gradient centrifugation using standard procedures.
For acute promyelocytic leukemia (APL) patients with t (15;17), all-trans-retinoic-acid- and arsenic-trioxide-based treatments were used in induction and consolidation therapy. Patients with non-APL AML were induced with standard first-line treatment, specifically the DA-like regimen, which consisted of daunorubicin (45 mg/m2, d1–3) and cytarabine (100–150 mg/m2, d1–7). With regard to the consolidation therapy, high-dose cytarabine based chemotherapy was performed on young patients. In addition, 27 patients received allogeneic hematopoietic stem cell transplantation (allo-HSCT) or autologous HSCT.
Mutational Analysis of EZH2
EZH2 mutations were analyzed by PCR amplification of the entire coding region of 20 EZH2 exons followed by direct bidirectional DNA sequencing as previously described. [16]–[17] Patients were also assessed for the presence of mutations of ASXL1 [19], CBL [19], c-KIT [20], DNMT3A [21], FLT3 [19], IDH1 [19], IDH2 [19], MLL [22], NPM1 [19], NRAS [19], RUNX1 [19], TET2 [19], and WT1 [19] as previously reported. Abnormal sequencing results were confirmed by at least 2 repeated analyses.
Statistical Analysis
Patient characteristics were analyzed by chi-square (x 2) or Fisher exact tests for univariate analysis. Kaplan-Meier analysis was used to evaluate patient survival. The log-rank test was used to compare survival difference. Binary logistic regression and COX model was used for the multivariate analysis of CR, EFS and OS, respectively. P-values less than 0.05 were deemed significant. All calculations were performed using the SPSS software package (version 13.0).
Results and Discussion
EZH2 Mutations in de novo AML
In this study, a total of 14 EZH2 mutations were documented in 13 patients with de novo AML. These involved exons 2, 9, 13, 17, and 18 (one case each) and exons 4, 12, 14, and 20 (2, 2, 3, and 2 cases, respectively). In this way, the frequency of somatic EZH2 mutations was 13/714 (1.8%) in de novo AML patients analyzed. The EZH2 mutations detected in the present study are listed in Table 1 and depicted graphically in Figure S1. Nonsense, frameshift, and missense mutations accounted for 7.1% (1/14), 28.6% (4/14), and 64.3% (9/14) of all of EZH2 mutations, respectively, showing a heterozygous pattern. Mutation of EZH2 Y641 was not found in this group of patients. Four out of fourteen mutations were located in the conserved catalytic SET domain (amino acids 618–731), which is essential to the methyltransferase activity of EZH2. We analyzed the genomic DNA of 3 EZH2-mutated cases upon diagnosis and again during remission. Genomic DNA confirmed the somatic origin of EZH2 mutations (frame-shift mutations: Tyr297fs, Glu731fs, Cys534fs; missense mutation: His516Asn).
Table 1. Characteristics of 13 AML patients with EZH2 mutations.
| No. cases | Sex | Age | BM blast (%) | Karyotype | Mutation(s) | Exon | Type of mutation | Protein level | Status | Other mutations |
| 1 | M | 34 | 96.7 | 46, XY [20] | c.65_66delAG | 2 | frame-shift | p.Glu22fs | N | NPM1: A-type |
| IDH2: c.419>A/p.Arg140Gln | ||||||||||
| NRAS: c.G35>T/p.GLY12Val | ||||||||||
| 2 | M | 50 | 96.2 | 46,XY, t(15;17)(q22;q12) [20] | c.278A>G | 4 | missense | p.Asn93Ser | N | |
| 3 | M | 50 | 56 | 46, XY [20] | c.1978T>G | 17 | missense | p.Leu660Val | N | NPM1: B-type |
| IDH2: c.419>A/p.Arg140Gln | ||||||||||
| TET2: c.767C>G/p.A256G | ||||||||||
| 4 | M | 41 | 51.5 | 46, XY [20] | c.278A>G | 4 | missense | p.Asn93Ser | N | ASXL1: c.2254dupG/p.Ala752fs |
| IDH1: c.C394T/p.R132C | ||||||||||
| 5 | F | 12 | 26.5 | 48,XX,-7,+8,t(11;12)(q12;p13),+mar1,+mar2 [20] | c.1615C>T | 14 | nonsense | p.Gln539X | N | |
| 6 | M | 22 | 46 | 46, XY [20] | c.2003C>T | 18 | missense | p.Ala668Val | N | |
| 7 | M | 37 | 91.5 | 46,XY,t(15;17)(q22;q12) [20] | c.2186_2187insGCCCCG | 20 | frame-shift | p.Ile730fs | N | |
| 8 | M | 42 | 25.9 | 45,X,-Y,t(8;21)(q22;q22) [20] | c.891delT+insGGCATA | 9 | frame-shift | p.Tyr297fs; | N | C-KIT: c.85G>T/p.D816Y |
| c.2190_2191dupATC | 20 | frame-shift | p.Glu731fs | MLL-PTD: mutated | ||||||
| 9 | M | 36 | 89.5 | 47,XY,-8,+i(8q)*2 [20] | c.1600_1601dupT | 14 | missense | p.Cys534fs | N | FLT3-ITD: mutated |
| RUNX1: c.415C>T/p.Arg140X | ||||||||||
| 10 | F | 57 | 63 | 46, XY [20] | c.1445A>G | 12 | missense | p.Lys482Arg | N | |
| 11 | M | 73 | 21 | 46, XY [20] | c.1484G>C | 13 | missense | p.Ser495Thr | N | |
| 12 | M | 53 | 51.3 | 45,X,-Y,t(8;21)(q22;q22) [20] | c.1546C>A | 14 | missense | p.His516Asn | N | C-KIT: c.105T>G/p.N822K |
| ASXL1: c.327G>T/p.E108D | ||||||||||
| 13 | M | 39 | 20.8 | 46,xy,del(7q),der(8)(p?),der(7)i(7q-) [18]/48,idem,der(8)(p?),+19 [2] | c.1394C>A | 12 | missense | p.Ser465Tyr | N |
M, male; F, female; R, reported; N, novel.
Correlation of EZH2 Mutations with Clinical and Cytogenetic Features
A comparison of clinical characteristics of patients with and without EZH2 mutations is shown in Table 2 . Analysis of gender distribution in EZH2-mutated AML patients showed that EZH2 mutations in male patients (11/396; 2.8%) were 4.6 times more common (11/396; 2.8%) than in female patients (2/318; 0.6%) with AML (P = 0.033). Patients with EZH2 mutations showed lower BM blast counts (P<0.0001), than patients without EZH2 mutations. Among de novo AML patients with 20–30% of blast in BM, 14.3% (4/28) of them harbored EZH2 mutations. However, EZH2 mutations were detected in only 1.3% (9/686) of de novo AML patients with >30% of blast in BM. No difference in frequency of EZH2 mutations was observed between adult and pediatric patients (2.4% vs. 1.8%, P>0.05).
Table 2. Comparison of clinical and laboratory features between AML patients with and without EZH2 mutation.
| Patient characteristics | Total no. cases (%) | EZH2 mutated No. (%) | EZH2 wild-type No. (%) | P |
| Median age, y (range) | 43.0 (8.0–83.0) | 41.0 (12.0–73.0) | 43.0 (8.0–83.0) | 0.754 |
| WBC,×109/L, median (range) | 28.9 (0.66–490) | 19.2 (1.4–167.4) | 29.0 (0.7–490.0) | 0.675 |
| Hb, median (range) | 83 (24–164) | 73 (46–92) | 83.0 (24.0–164.0) | 0.189 |
| Plt, ×109/L, median(range) | 35 (3.0–530) | 38.0 (10.0–105.0) | 35.0 (3.0–530) | 0.887 |
| BLAST%, median (range) | 79.95 (11.5–99.4) | 51.5 (18–96.7) | 80.0 (11.5–99.4) | 0.151 |
| Gender | 0.033 | |||
| Male | 396/714 (55.5) | 11 (2.8) | 385 (97.2) | |
| Female | 318/714 (44.5) | 2 (0.6) | 316 (99.4) | |
| Age (y) | 0.505 | |||
| <16 | 41/714 (5.7) | 1 (2.4) | 40 (97.6) | |
| 16–60 | 526/714 (73.7) | 11 (2.1) | 515 (97.9) | |
| >60 | 147/714 (20.6) | 1 (0.7) | 146 (99.3) | |
| WBC count, ×109/L | 0.946 | |||
| <4.0 | 66/475 (13.9) | 1 (1.5) | 65 (98.5) | |
| 4.0–30.0 | 177/475 (37.3) | 3 (1.7) | 174 (98.3) | |
| >30.0 | 232/475 (48.8) | 3 (1.3) | 229 (98.7) | |
| Hb, g/L | 0.165 | |||
| <20 | 57/456 (12.5) | 2 (3.5) | 55 (96.5) | |
| 20–100 | 272/456 (59.6) | 5 (1.8) | 267 (98.2) | |
| >100 | 127/456 (27.9) | 0/7 (0) | 127 (100.0) | |
| PLT count, ×109/L | 0.743 | |||
| <20.0 | 122/454 (26.9) | 1 (0.8) | 121 (99.2) | |
| 20.0–100.0 | 269/454 (59.3) | 5 (1.9) | 264 (98.1) | |
| >100.0 | 63/454 (13.9) | 1 (1.6) | 62 (98.4) | |
| BM-blast (%) | <0.0001 | |||
| <30% | 28/714 (3.9) | 4 (14.3) | 24 (85.7) | |
| ≥30% | 686/714 (96.1) | 9 (1.3) | 677 (98.7) | |
| Karyotype | 0.988 | |||
| normal | 360/698 (51.6) | 6(1.7) | 354 (98.3) | |
| t(8;21) | 59/698 (8.5) | 2(3.4) | 57 (96.6) | |
| t(15;17)/t (11;17) | 93/698 (13.3) | 2(2.2) | 91 (97.8) | |
| inv(16), t(16;16) | 24/698 (3.4) | 0(0) | 24 (100.0) | |
| tri8 | 11/698 (1.6) | 0(0) | 11 (100.0) | |
| complex abnormalities | 74/698 (10.6) | 2(2.7) | 72 (97.3) | |
| hyperdiploid | 8/698 (1.1) | 0(0) | 8 (100.0) | |
| t(9;22) | 17/698 (2.4) | 0(0) | 17 (100.0) | |
| 11q23/MLL | 10/698 (1.4) | 0(0) | 10 (100.0) | |
| others | 38/698 (5.4) | 1(2.6) | 37 (97.4) | |
| Risk statusψ | 0.887 | |||
| Better-risk | 176/698 (25.2) | 4(2.3) | 172 (97.7) | |
| Intermediate-risk | 417/698 (59.7) | 7(1.7) | 410 (98.3) | |
| Poor- risk | 105/698 (15.0) | 2(1.9) | 103 (98.1) | |
| -7,7q- | 0.025 | |||
| with | 26/698(3.7) | 2(7.7) | 24 (92.3) | |
| without | 672/698(96.3) | 11(1.6) | 661 (98.4) |
Risk status: Better-risk: inv(16)/t(16;16), t(8;21),t(15;17); Intermediate-risk: normal, +8, t(9;11), other undefined risk; Poor-risk: complex, −5, 5q−, −7, 7q−, 11q23(non t(9;11)), inv(3), t(3;3), t(6;9), t(9;22).
(A) Structure of the EZH2 protein and location of EZH2 mutations. (B) DNA sequencing chromatograms of AML genomic DNA samples showing 14 mutations in 13 AML patients.
Among 714 AML patients undergoing cytogenetic analysis, karyotypic data were available in 698 cases. The remainder (n = 16) of cases lacked karyotypic data due to lack of metaphase. Out of 698 patients supplied with cytogenetic data, abnormal karyotypes were detected in 338 cases (47.9%). EZH2 status was correlated to cytogenetics as summarized in Table 2 . EZH2 mutations were revealed in patients with normal karyotype in 6 cases, -7/del(7q) accompanied by complex karyotypes in 2 cases, t(15;17)(q22;q12) in 2 cases, t(8;21)(q22;q22) in 2 cases, and i(8q) in 1 case. There was no difference in the incidence of EZH2 mutations among patients with favorable karyotype (4/176, 2.3%), intermediate-risk karyotype (7/417, 1.7%), or unfavorable karyotype (2/105, 1.9%; P = 0.887). Of note, EZH2 mutations were observed much more frequently in the cases with -7/del(7q) (2/26, 7.7%) than in the cases without -7/del(7q) (11/672, 1.6%; P = 0.025). No relationship was observed between EZH2 mutations with other cytogenetic abnormalities.
Association of EZH2 Mutations with Other Molecular Abnormalities
To investigate the interactions of gene mutations in the leukemogenesis of de novo AML, a comprehensive mutational screening of EZH2 and 13 other genes was performed. After excluding known polymorphisms and silent mutations, FLT3-ITD mutations were found in 25.0%, NPM1 in 22.5%, TET2 in 10.2%, IDH2 in 9.7%, N-RAS in 8.2%, FLT3-TKD in 6.5%, C-KIT in 6.1%, DNMT3A in 5.3%, WT1 in 4.6%, ASXL1 in 4.5%, IDH1 in 4.2%, MLL-PTD in 3.3%, RUNX1 in 2.5%, and CBL in 0.8% of all cases examined. No association was found between EZH2 mutations and other molecular abnormalities in the present study.
Impact of EZH2 Mutations on Response to Therapy and Clinical Outcome
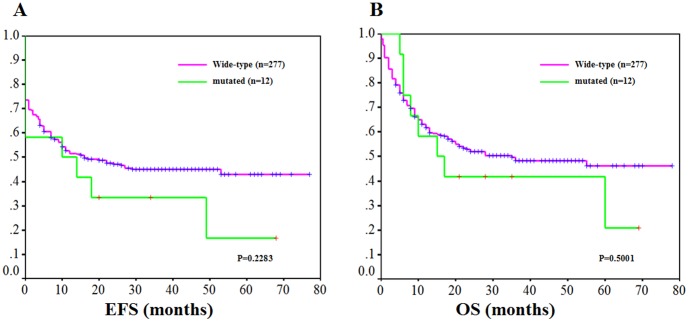
For correlation with clinical outcome, patients with APL were excluded. EZH2 mutations had no influence on achieving complete remission (CR) ( Figure 1 ). The CR rates of cases with and without EZH2 mutations were not significantly different (61.5% vs. 75.0%, P = 0.329). No significant difference in event-free survival (EFS) (3 year rates, 33.1% vs. 45.2%; P = 0.2283) or overall survival (OS) (3 year rates, 41.0% vs. 49.1%; P = 0.5001) with a median follow-up of 46.6 months (range, 1–79 months) were observed.
Figure 1. Kaplan-Meier survival curves according to EZH2 mutation status.
(A) EFS in de novo AML patients according to EZH2 mutations. The green line represents patients with mutated EZH2 (n = 12); and magenta line, patients with unmutated EZH2 (n = 277; P = 0.2283); (B) OS in de novo AML patients according to EZH2 mutations. The green line represents patients with mutated EZH2 (n = 12); and the magenta line represents patients with unmutated EZH2 (n = 277; P = 0.5001).
Multivariate analysis showed FLT3-ITD, NRAS, and risk-status are independent influence factors with regard to CR rate (P<0.001, p = 0.004, P = 0.001 respectively). In a multivariate Cox regression model, mutations of FLT3-ITD, age, risk-status, and N-RAS (only associated with OS) showed independent prognostic significance (EFS: P = 0.002, = 0.002, <0.001; OS: P = 0.001, = 0.005, <0.001, = 0.023).
Collectively, the results of the present study demonstrate that EZH2 mutations could be detected in a substantial proportion of patients with de novo AML and that these mutations occurred more frequently in male patients than in female patients. No difference in CR rates, EFS, or OS was observed between AML patients with and without EZH2 mutation. The presence of EZH2 mutations in AML was found to be closely associated with lower BM blast percentage (20–30%) and -7/del(7q). In the FAB classification of MDS, patients with 21–30% of blasts in BM were defined as having refractory anemia with excess blasts in transformation (RAEB-t). [23] The WHO classification criteria lowered the threshold to 20% for the number of blasts required for the diagnosis of AML. [1] This arbitrary threshold in blast percentage eliminated RAEB-t in MDS and patients in this category were considered to have AML. However, several reports have argued that the biology of RAEB-t is distinct from that of AML and should be considered a subtype of MDS. [24]–[25] In the present study, results showed the frequency of EZH2 mutations in patients with 21–30% of BM blasts to be much higher than that in patients with >30% of blasts in BM (14.3% versus 1.3%, P<0.0001). In addition, EZH2 is located on the distal part of chromosome 7q. EZH2 mutations were observed more frequently in cases with -7/del(7q) than in cases without -7/del(7q) (7.7% versus 1.6%, P = 0.025). It was concluded that somatic mutations of EZH2 might play an important role in pathogenesis of de novo AML patients with -7/del(7q) or with 21–30% of blasts in BM, which is classified as RAEB-t in FAB classification. Because of rarity of EZH2 mutations in de novo AML, the prognostic impact of EZH2 mutations in AML is still uncertain, and will need to be assessed in larger cohorts of patients collected on multi-center co-operative studies, though there no significant difference in EFS or OS was observed between EZH2 mutated patients and wild-type in the present study.
Supporting Information
EZH2 mutations in AML patients.
(TIF)
Clinical characteristics of 714 patients with de novo AML.
(DOC)
Funding Statement
This work was supported in part by grants from National Key Scientific Projects of China (2011CB933501),http: //www.nsfc.gov.cn/Portal0/default152.htm; the Priority Academic Program Development of Jiangsu Higher Education Institutions, Jiangsu Province’s Key Provincial Talents Program, the National Natural Science Foundation of China (81070416), http: //www.ec.js.edu.cn/; Jiangsu Province Natural Science Foundation (BK2010204); Jiangsu Province Natural Science Fund for Distinguished Young Scholars; and Foundation of Jiangsu Province Health Department (H200915), http: //www.most.gov.cn/tztg/index.htm. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (2008) 4th ed. Lyon: International Agency for Research on Cancer (IARC). [Google Scholar]
- 2. Bacher U, Kern W, Schnittger S, Hiddemann W, Schoch C, et al. (2005) Further correlations of morphology according to FAB and WHO classification to cytogenetics in de novo acute myeloid leukemia: a study on 2,235 patients. Ann Hematol 84: 785–791. [DOI] [PubMed] [Google Scholar]
- 3. Grimwade D, Walker H, Harrison G, Oliver F, Chatters S, et al. (2001) The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood 98: 1312–1320. [DOI] [PubMed] [Google Scholar]
- 4. Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, et al. (2012) Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 366: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chou WC, Huang HH, Hou HA, Chen CY, Tang JL, et al. (2010) Distinct clinical and biological features of de novo acute myeloid leukemia with additional sex comb-like 1 (ASXL1) mutations.Blood. 116: 4086–4094. [DOI] [PubMed] [Google Scholar]
- 6. Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, et al. (2010) DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363: 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, et al. (2009) Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thol F, Damm F, Wagner K, Göhring G, Schlegelberger B, et al. (2010) Prognostic impact of IDH2 mutations in cytogenetically normal acute myeloid leukemia. Blood 116: 614–616. [DOI] [PubMed] [Google Scholar]
- 9. Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, et al. (2009) Mutation in TET2 in myeloid cancers. N Engl J Med 360: 2289–2301. [DOI] [PubMed] [Google Scholar]
- 10. Tefferi A, Lasho TL, Abdel-Wahab O, Guglielmelli P, Patel J, et al. (2010) IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia 24: 1302–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walter MJ, Ding L, Shen D, Shao J, Grillot M, et al. (2011) Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia 25: 1153–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stegelmann F, Bullinger L, Schlenk RF, Paschka P, Griesshammer M, et al. (2011) DNMT3A mutations in myeloproliferative neoplasms. Leukemia 25: 1217–1219. [DOI] [PubMed] [Google Scholar]
- 13. Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, et al. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043. [DOI] [PubMed] [Google Scholar]
- 14. Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, et al. (2010) Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A 107: 20980–20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haase D, Germing U, Schanz J, Pfeilstöcker M, Nösslinger T, et al. (2007) New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood 110: 4385–4395. [DOI] [PubMed] [Google Scholar]
- 16. Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, et al. (2010) Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 42 722–726. [DOI] [PubMed] [Google Scholar]
- 17. Nikoloski G, Langemeijer SMC, Kuiper RP, Knops R, Massop M, et al. (2010) Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet 42: 665–667. [DOI] [PubMed] [Google Scholar]
- 18. Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, et al. (2010) Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42: 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rocquain J, Carbuccia N, Trouplin V, Raynaud S, Murati A, et al. (2010) Combined mutations of ASXL1, CBL, FLT3, IDH1,IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias BMC Cancer 2010. 10: 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boissel N, Leroy H, Brethon B, Philippe N, de Botton S, et al. (2006) Incidence and prognostic impact of c-Kit, FLT3, and Ras gene mutations in core binding factor acute myeloid leukemia (CBF-AML). Leukemia. Jun 2006 20 965–970. [DOI] [PubMed] [Google Scholar]
- 21. Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, et al. (2010) DNMT3A Mutations in Acute Myeloid Leukemia. N Engl J Med 2010 363: 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shiah HS, Kuo YY, Tang JL, Huang SY, Yao M, et al. (2002) Clinical and biological implications of partial tandem duplication of the MLL gene in acute myeloid leukemia without chromosomal abnormalities at 11q23. Leukemia 2002 16: 196–202. [DOI] [PubMed] [Google Scholar]
- 23. Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, et al. (1982) Proposals for the classification of the myelodysplastic syndromes. Br J Haematol 51: 189–199. [PubMed] [Google Scholar]
- 24. Huh YO, Jilani I, Estey E, Giles F, Kantarjian H, et al. (2002) More cell death in refractory anemia with excess blasts in transformation than in acute myeloid leukemia. Leukemia 16: 2249–2252. [DOI] [PubMed] [Google Scholar]
- 25. Greenberg P, Anderson J, de Witte T, Estey E, Fenaux P, et al. (2000) Problematic WHO reclassification of myelodysplastic syndromes. Members of the International MDS Study Group. J Clin Oncol 18: 3447–3452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EZH2 mutations in AML patients.
(TIF)
Clinical characteristics of 714 patients with de novo AML.
(DOC)