Abstract
We describe a three-dimensional (3D) confocal imaging technique to characterize and enumerate rare, newly emerging hematopoietic cells located within the vasculature of whole-mount preparations of mouse embryos. However, the methodology is broadly applicable for examining the development and 3D architecture of other tissues. Previously, direct whole-mount imaging has been limited to external tissue layers owing to poor laser penetration of dense, opaque tissue. Our whole-embryo imaging method enables detailed quantitative and qualitative analysis of cells within the dorsal aorta of embryonic day (E) 10.5–11.5 embryos after the removal of only the head and body walls. In this protocol we describe the whole-mount fixation and multimarker staining procedure, the tissue transparency treatment, microscopy and the analysis of resulting images. A typical two-color staining experiment can be performed and analyzed in ~6 d.
INTRODUCTION
Protocol development
Mouse hematopoietic stem cells (HSCs) develop from the major vasculature, some of which is located deep within the trunk region of the midgestation mouse embryo1,2. HSCs emerge through the formation of clusters of hematopoietic cells from endothelial cells, which occurs via an endothelial-to-hematopoietic cell transition. Vascular hematopoietic clusters have historically been visualized in thin sections of the midgestation mouse aorta, vitelline and umbilical arteries after immunostaining with antibodies specific for hematopoietic and vascular markers3. To image an object that is more than 50 µm in thickness or that is located more than 50 µm deep within the midgestation embryo (like the dorsal aorta), sequential sectioning and 2D imaging have been routinely used, and the thin slice images have been reconstructed with software to render a 3D image4. Although this method has been very useful for localizing rare HSCs in some embryonic tissues, it is labor intensive, and furthermore it does not allow for a complete or quantitative assessment of all the relevant cells in the embryo.
For whole-embryo imaging, improvements in the power of the laser light in confocal microscopy have increased the imaging depth of tissues to about 200 µm. However, laser penetration does not reach the depth needed for high-resolution imaging of hematopoietic clusters in the centrally located dorsal aorta, where some of the first HSCs are generated. Light is absorbed, scattered and reflected in the dense and opaque tissue of the midgestation mouse embryo, thus reducing the amount of gathered light and the image quality. To improve light penetration, various tissue transparency techniques were devised using organic solvents such as potassium hydroxide/glycerol, methyl salicylate, carbon disulfide, glycerol, xylene and benzyl alcohol/benzyl benzoate (BABB). BABB was first used to image amphibian eggs and embryos5. Solvents such as BABB clear the opaque tissues of the embryo by replacing water, rendering the refractive index of the embryo the same as the solution. This method was subsequently adapted for use with whole mouse embryos and was found to be compatible with vital dye staining (LysoTracker Red) procedures designed to examine programmed cell death and lysosomal activity6.
We have developed a method by which the whole embryo is immunostained with antibodies specific for hematopoietic clusters and vasculature, and then rendered transparent with BABB, thereby enabling us to reliably image the vasculature and all hematopoietic clusters for quantitative and cartographic analysis (Fig. 1)7–9.
Figure 1.
Representative overview of the protocol for two-color staining.
Applications of the method
The method described here is applicable to imaging large areas of the embryo in order to locate rare cells of interest. It is especially useful for difficult-to-detect cells that are localized within the deepest, most centrally located tissues of the whole mouse embryo. In the example described here, we immunostained all vascular endothelial cells and hematopoietic clusters with a fluorescent antibody specific to CD31, and then combined this with an antibody specific to c-Kit to detect only the hematopoietic clusters. Individual cells in the clusters could be counted, with cluster size ranging from 1 to 100 cells7. Moreover, multicolor immunostaining for a variety of cell surface markers allowed us to describe the developmental heterogeneity of cells within the clusters. In this way, we have shown that aortic hematopoietic clusters contain HSCs, progenitors and mature cells of the hierarchy7.
Here we describe how to optimize whole-mount immunostaining for various types of hematopoietic cells (both rare and abundant); primordial germ cells, another rare type of stem cell deep within the whole midgestation mouse embryo; and mitotic cells. We use this as an example for illustrative purposes; however, the method is broadly applicable. It relies, however, on the specificity of the antibodies and the ability of the immunostaining to withstand the BABB transparency treatment and produce sufficient signal intensity for confocal imaging (Fig. 2).
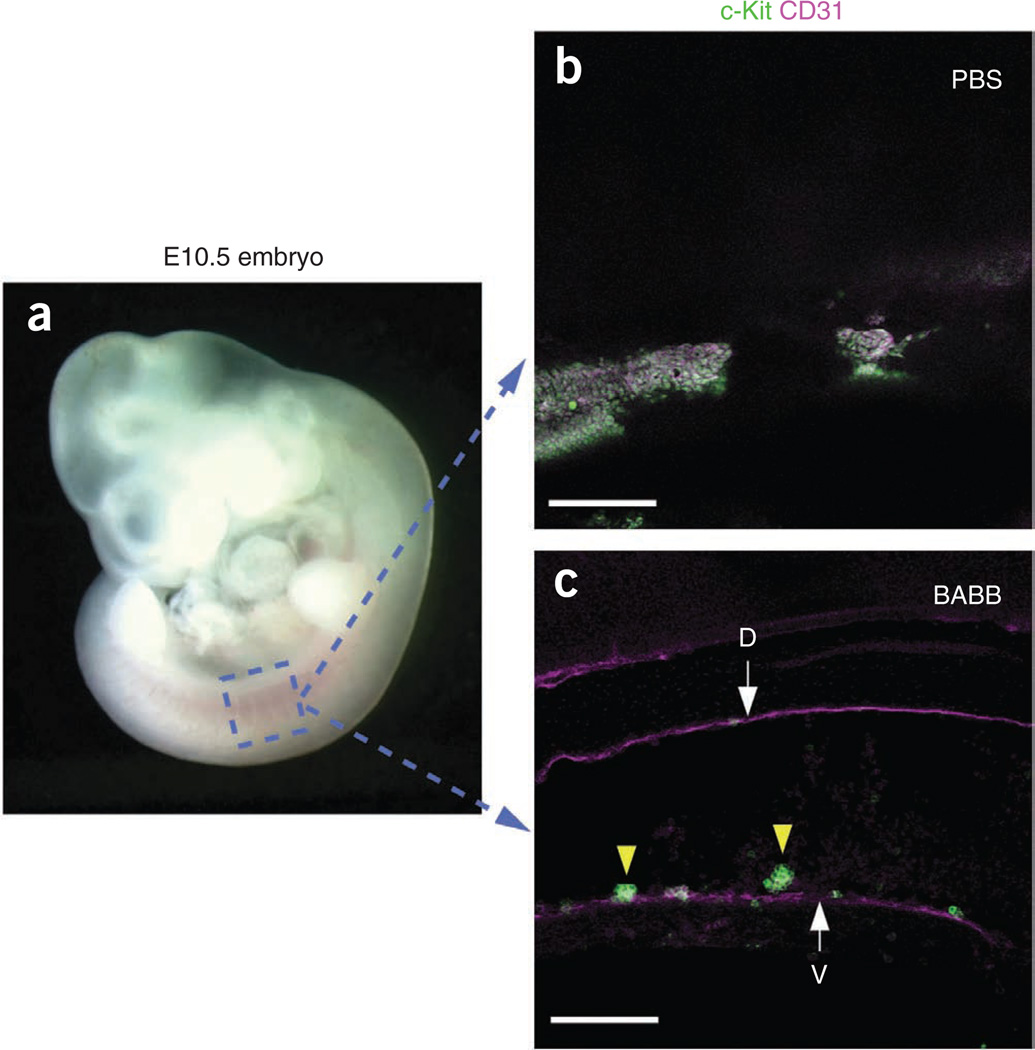
Figure 2.
Increasing tissue transparency using BABB solution. (a) Appearance of E10.5 mouse embryo (before fixation). (b,c) Boxed region was scanned after transferring immunostained embryos into PBS (b) or BABB (c). (b) Although maximum laser power was used, a clear structure was not observed in the embryo immersed in PBS. (c) In contrast, the dorsal aorta and hematopoietic clusters were clearly observed in the embryo immersed in BABB. Scale bars, 100 µm. Arrows indicate ventral (V) and dorsal (D) walls of dorsal aorta. Yellow arrowheads indicate hematopoietic clusters.
Comparison with other methods
Serial sectioning of the trunk region of the chick and mouse embryo has resulted in temporal and spatial maps of hematopoietic clusters in the aorta, vitelline and umbilical arteries10–14, but important quantitative and qualitative information is at risk because of the dislodgement of the vascular adhering hematopoietic cells in the sectioning procedure or the loss of some of the serial sections. In addition, more than 200 sections of 10 µm thickness are required to cover the trunk region of a midgestation mouse embryo. The preparation and analysis of those sections is both labor intensive and relatively expensive, as each section must be incubated with antibody. In comparison, the whole-mount method maintains tissue integrity as well as structure and cell localization, and in our application, it allowed for rapid quantification of aortic clusters. Several embryos can be incubated in a vial with 0.5 ml of solution containing an antibody of interest, thereby saving on reagents. Another important consideration is that the 3D method allows for fine discrimination of cluster localization. Individual histological sections can give the impression that hematopoietic clusters are localized extravascularly. However, using antibodies to endothelial cells enables visualization of the entire embryonic vasculature in fine detail, and revealed that all clusters are on the lumenal side of the vasculature9. We note that a similar whole-mount immunostaining procedure has been used to image hematopoietic cells in the murine yolk sac15. In this procedure, mainly glycerol was used for clearing purposes, with BABB treatment briefly mentioned as an alternative. Although glycerol has an advantage in terms of holding the tissue in place because of its high viscosity, BABB can achieve superior transparency; the tissue-holding problem for BABB has been solved in our procedure by using agarose gel.
Limitations
Although BABB solution markedly enhances the penetration of laser into the embryonic tissue (Fig. 2), there is also a depth limitation for antibody penetration. We have noticed that c-Kit+ hematopoietic clusters within the dorsal aorta at E10.5 cannot be clearly stained without removing the lateral body wall (Fig. 3). The distance from the embryo surface to the dorsal aorta is ~200 µm. After removing the lateral body wall, the distance is reduced to about 120 µm. Therefore, ~150 µm could be an upper limit for the sufficient penetration of antibodies. To analyze older embryos or thick organs, trim them to a thickness of ~300 µm before immunostaining.
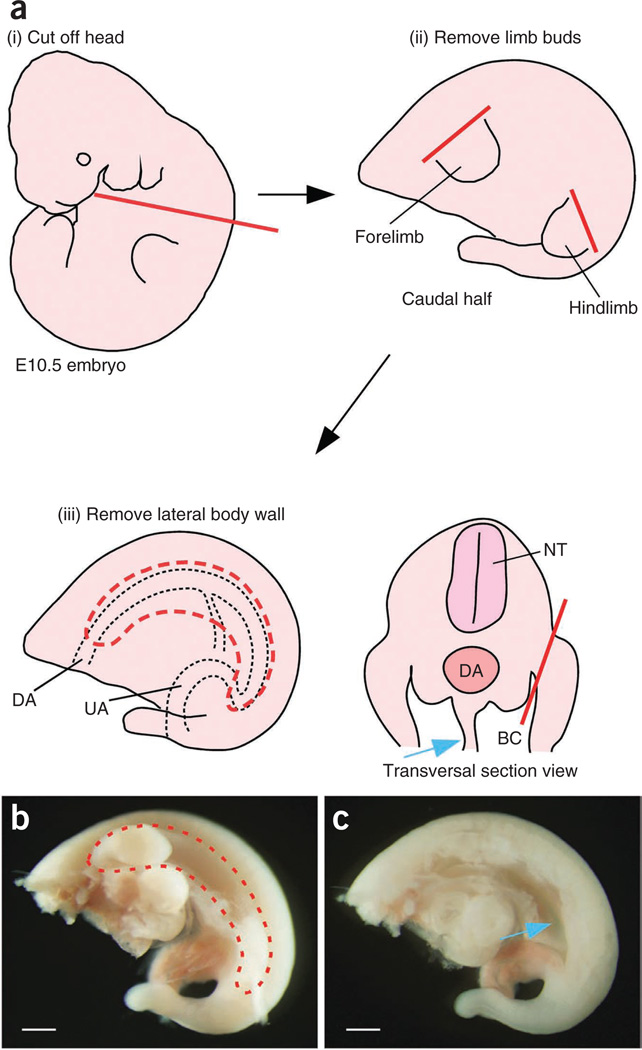
Figure 3.
Embryo trimming. (a–c) Schematic diagram of embryo trimming. First, remove the head (i in a). To remove the lateral body wall (iii in a, left, and red dashed lines in b), an incision is made along the red line (iii in a, right). E10.5 embryo pictures before and after removing the lateral body wall are shown in b and c, respectively. Mesentery (blue arrows) can be seen after removing the lateral body wall. BC, body cavity; DA, dorsal aorta; NT, neural tube; UA, umbilical artery. Scale bars, 500 µm.
Another limitation is that erythrocytes cannot be rendered completely transparent with BABB solution, as they contain a large amount of heme. Therefore, it is difficult to obtain a high-resolution 3D picture from an erythrocyte-rich organ such as the liver.
Experimental design
Lineage-specific markers and fluorescent antibodies
Typically, we have used unlabeled primary antibodies together with species/isotype-specific fluorescent secondary antibodies or biotinylated primary antibodies with labeled streptavidin. Directly labeled antibodies yielded weak signals. Titration of antibodies is performed to optimize signal-to-noise ratio. When secondary antibodies are used, a control sample with only the secondary antibody is included in the experiment. Multimarker analysis requires reoptimization of each of the several antibodies that are used in combination.
Our antibodies were chosen on the basis of our specific interests in HSCs, progenitors and endothelial cells (Table 1). To image the close interactions between hematopoietic clusters and the endothelium, we have routinely used anti-CD31 antibody to image the entire embryonic vasculature. CD31 expression marks all vascular endothelium and hematopoietic cluster cells. To distinctly image hematopoietic clusters, we used c-Kit expression, which marks all hematopoietic cluster cells but not endothelial cells16,17. Other antibodies that we have used with success were specific for GFP, CD41, CD45, Flk1, vascular endothelial cadherin and SSEA-1 (refs. 7–9).
Table 1.
Antibodies and streptavidin list.
| Antibody or streptavidin | Clone | Vendor | Cat. no. | Dilution |
|---|---|---|---|---|
| Primary antibody | ||||
| Rat anti-mouse c-Kit | 2B8 | BD Biosciences | 553352 | 1:500 |
| Biotinylated rat anti-mouse CD31 | MEC13.3 | BD Biosciences | 553371 | 1:500 |
| Mouse anti-mouse SSEA-1 | MC-480 | eBioscience | 14-8813 | 1:500 |
| Mouse anti-phosphohistone H3 (Ser10) | 3H10 | Millipore | 05-806 | 1:1,000 |
| Rabbit anti-GFP | — | MBL | 598 | 1:2,000 |
| Secondary antibody | ||||
| Alexa Fluor 488 goat anti-mouse IgM | — | Invitrogen | A21042 | 1:2,000 |
| Alexa Fluor 647 goat anti-rat IgG (H + L) | — | Invitrogen | A21472 | 1:5,000 |
| Alexa Fluor 555 goat anti-rat IgG (H + L) | — | Invitrogen | A21434 | 1:2,000 |
| Alexa Fluor 488 goat anti-mouse IgG (H + L) | — | Invitrogen | A11029 | 1:1,000 |
| Alexa Fluor 647 goat anti-rabbit IgG (H + L) | — | Invitrogen | A21245 | 1:5,000 |
| Streptavidin | ||||
| Cy3-conjugated streptavidin | — | Jackson ImmunoResearch | 016-160-084 | 1:5,000 |
Autofluorescence from the embryo in the 488-nm channel must also be considered. When examining the subcellular distribution of proteins, or staining of specific cells in the clusters, it is better to use a fluorochrome in the far-red range. The 488-nm channel should be reserved for use with very good primary antibodies (such as CD31). For CD31-biotin, we used Alexa Fluor 488, 555 or Cy3. FITC and Alexa Fluor 647 do not work well with CD31-biotin, yielding grainy images in which the outlines of cells are difficult to discern.
Confocal microscopy
This protocol was designed for use with a single-photon microscope. Thus, it is not necessary to have a multiphoton microscope or special setup. We have used a Zeiss confocal microscope (LSM510 Meta) equipped with three lasers of 488-, 543- and 633-nm wavelengths for fluorochrome excitation and emission. It is also possible to use a Leica confocal microscope (lasers of 488-, 561- and 633-nm wavelengths).
Image analysis
We use Zeiss Image Browser (free software) for creating 3D reconstruction and movies. Other software (e.g., Zeiss Zen, PerkinElmer Volocity, Bitplane Imaris) can also be used. We recommend maximum transparency mode for 3D reconstruction, as the distribution of c-Kit+ hematopoietic clusters within the dorsal aorta can be seen through the half-transparent endothelial layer.
MATERIALS
REAGENTS
Mice (wild-type or reporter/mutant mice)
 Note that all animal care and experimentation should be performed within ethical guidelines and with permission from local regulatory boards.
Note that all animal care and experimentation should be performed within ethical guidelines and with permission from local regulatory boards.Embryos generated from timed matings of adult mice
Phosphate-buffered saline (PBS)
Methanol (Sigma-Aldrich, cat. no. 65543)
Paraformaldehyde (PFA; Sigma-Aldrich, cat. no. P6148)
Streptavidin/biotin blocking kit (Vector Laboratories, cat. no. SP-2002)
Antibodies of choice (Table 1)
Serum (Table 2)
Bovine serum albumin (BSA; Sigma-Aldrich, cat. no. A2153)
Antibodies and streptavidin (Table 1)
Triton X-100 (Sigma-Aldrich, cat. no. T8787)
Powdered skim milk (e.g., Blotting-grade Blocker nonfat dry milk, Bio-Rad, cat. no. 170-6404)
Agarose, low melting point (Sigma-Aldrich, cat. no. A9414)
Benzyl alcohol (Sigma-Aldrich, cat. no. 402834)
Benzyl benzoate (Sigma-Aldrich, cat. no. B6630)
Sodium hydroxide
Phosphate-buffered saline (PBS)
Hydrochloric acid
Table 2.
Serum (all from Jackson ImmunoResearch).
| Species | Code number | Concentration of stock (mg ml−1) |
Dilution |
|---|---|---|---|
| Rabbit | 011-000-001 | 60 | 1 μl per 1 ml |
| Rat | 012-000-001 | 60 | 1 μl per 1 ml |
| Goat | 005-000-001 | 60 | 1 μl per 1 ml |
| Donkey | 017-000-001 | 60 | 1 μl per 1 ml |
EQUIPMENT
Glass scintillation vials (20 ml; Wheaton) or clear vials with screw tops (15 ml; Sigma-Aldrich, cat. no. 27161) or low-adhesion 12-well plates for single-embryo staining and washing
 To save on the volume of antibodies to be used, incubations can be done with the vials standing upright and rocking slowly (750 µl for a 20-ml vial and 500 µl for a 15-ml vial).
To save on the volume of antibodies to be used, incubations can be done with the vials standing upright and rocking slowly (750 µl for a 20-ml vial and 500 µl for a 15-ml vial).Dissection tools (fine-tip forceps, scissors)
Insulin needles (e.g., BD Ultra-Fine needle, 29 G)
Nonsterile graduated plastic pipette (3 ml)
Small Petri dish
Rocking platform
Pipettes and pipette tips (e.g., Eppendorf)
Eppendorf tubes (1.5-ml, Eppendorf)
FastWell (Grace Bio-Labs, cat. no. FW20, http://www.gracebio.com/)
Circle coverslip (25 mm, no.1 Menzel-Gläser, cat. no. CB00250RA1)
SuperFrost slide (Thermo Scientific SuperFrost Plus, Thermo Scientific)
Clear nail polish
Absorbent tissues
Dissection microscope
Confocal microscope (e.g., Zeiss LSM510 Meta with appropriate objectives)
Image analysis software (e.g., Zeiss Image Browser; available from Zeiss website)
REAGENT SETUP
Mice and embryo generation Adult mice are used in timed matings for embryo generation. The day of plug discovery is considered to be day 0. Embryos at the desired day of gestation (E8–E14) are dissected from the uterus18. Embryos must be handled with great care so as not to compromise tissue integrity.  Please note that all animals should be handled according to all relevant ethics regulations.
Please note that all animals should be handled according to all relevant ethics regulations.
Fixation solution Add 4 g of powdered PFA (4% (wt/vol), pH 7.4) in 100 ml of PBS and warm it in a microwave (stop before it starts boiling). Add 160 µl of 5 N NaOH solution to help the PFA dissolve. Adjust pH to 7.4 by adding HCl. Fixation solution can be aliquotted into 15-ml tubes (1–2 ml required per embryo) and stored at − 20 °C (up to ~6 months), and then diluted 1:1 in PBS at the time of dissection.  Wear gloves and a mask during preparation because PFA is toxic if swallowed, inhaled or absorbed through the skin.
Wear gloves and a mask during preparation because PFA is toxic if swallowed, inhaled or absorbed through the skin.
De/rehydration solutions Prepare 50% (vol/vol) methanol/PBS, 75% (vol/vol) methanol/PBS and 100% methanol. Prepare 40–50 ml per sample. Store the solutions at 4 °C until needed.
BSA blocking solution Prepare a 10% (wt/vol) stock in PBS, aliquot it into 1.5-ml Eppendorf tubes (2 ml required per sample) and store it in the freezer (−20 °C) for up to 2 years.
Antibody preparation We have used unlabeled primary antibodies with species/isotype-specific fluorochrome-conjugated secondary antibodies. Crucial to the success of imaging (signal to background) is the titration of the antibody (primary and secondary). Prepare isotype controls for negative controls.
PBS-MT solution Prepare PBS-MT solution using 1% (wt/vol) skim milk and 0.4% (vol/vol) Triton X-100. Dissolve 1 g of powdered skim milk and 0.4 ml of Triton X-100 in 100 ml of PBS. This solution can be stored at 4 °C for a few days.
PBS-T solution Prepare PBS-T solution using 0.4% (vol/vol) Triton X-100. Dissolve 0.4 ml of Triton X-100 in 100 ml of PBS. This solution can be stored at 4 °C for a few days.
BABB solution Mix one part benzyl alcohol with two parts benzyl benzoate. The solution should be freshly prepared for each use. Mix both in glass scintillation vials. For three samples, mix 1 ml of benzyl alcohol with 2 ml of benzylbenzoate. Use 1 ml of this solution to prepare the 1:1 BABB/methanol solution (50% BABB).  Wear gloves during preparation because BABB is toxic if absorbed through the skin.
Wear gloves during preparation because BABB is toxic if absorbed through the skin.
EQUIPMENT SETUP
Microscope We used a Zeiss confocal microscope (LSM510 Meta) equipped with three lasers of 488-, 543- and 633-nm wavelengths for fluorochrome excitation and emission. Other microscopes with similar equipment can be used. It is important to optimize the settings of the microscope before use. The preparations as dissected can be visualized with a ×20 objective (Plan-Neofluar ×20/NA 0.5). We also use a ×10 objective and ×40/×63 water-immersion objectives (EC Plan-Neofluar ×10/NA 0.3, HC PL APO CS ×10/NA 0.4, Achroplan ×40/NA 0.8 W, Achroplan ×63/NA 0.95 W). The ×40/×63 oil-immersion objectives cannot be used because their working distance is generally <0.5 mm and the samples are too thick.
Table 3 provides a summary of the lasers and filters that we have found to be optimal for c-Kit/CD31/SSEA-1 three-color staining. The pinhole diameter was set at 1 Airy unit and steps were set at 1.9–2.3 µm per z-section for a ×20 objective. To minimize the overlap when detecting fluorescent signals, we used the multitrack sequential mode. To make 3D reconstructed images, we took serial longitudinal pictures of the dorsal aorta region. As the width of the dorsal aorta is about 200 µm at 35 somite pairs, 100 z-stacks are required for 3D reconstruction. It will take about 45 min with scan line average number 8 and scan speed 8.
Table 3.
Lasers and filters for c-Kit/CD31/SSEA-1 three-color staining.
| Primary antibody |
Secondary antibody |
Excitation laser (nm) |
Emission filter (nm) |
|---|---|---|---|
| SSEA-1 | Alexa Fluor 488 | 488 | LP 475 |
| CD31 (biotinylated) | Streptavidin-Cy3 | 543 | BP 560-615 |
| c-Kit | Alexa Fluor 647 | 633 | LP 650 |
To correct the refractive index mismatch between the immersion medium and the embedding medium, LSM510 software has a compensative function, ‘Reff. Corr.’. The Reff. Corr. value for BABB is 1.559 (ref. 19). To quantify hematopoietic clusters in the E10.5 whole dorsal aorta, six or seven separate regions need to be scanned with ×20 objectives, and this will take 6–7 h (scan speed 8). However, if the default Reff. Corr. value of 1 is used, scanning will be completed in ~4 h and bleaching of samples can be reduced, although the final 3D picture is a little compressed along the z axis.
PROCEDURE
Dissection and fixation of embryos  2–4 h
2–4 h
-
1
Harvest embryos from a pregnant dam.
 All steps (until mounting at Step 24) are performed in 20-ml glass scintillation vials or 12-well plates.
All steps (until mounting at Step 24) are performed in 20-ml glass scintillation vials or 12-well plates. -
2
Dissect embryos, removing the placenta. The yolk sac can be left in place or can be removed; however, if you choose to remove it, then ensure that the arteries (vitelline and umbilical) are left intact (see Box 1 for preparation of younger mouse embryos and Supplementary Video 1 for a representative image).
-
3
Immediately place the embryos into PFA for fixation (2% (wt/vol) PFA in PBS, 20 min on ice).
 A single embryo needs about 1–2 ml of PFA. Fixation time depends on antibodies used for subsequent immunostaining.
A single embryo needs about 1–2 ml of PFA. Fixation time depends on antibodies used for subsequent immunostaining. To precisely stage embryos, count the somites after fixation.
To precisely stage embryos, count the somites after fixation. -
4
Wash embryos with PBS (three washes, 10 min per rinse, on ice).
-
5
Dehydrate embryos in 50% (vol/vol) methanol/PBS for 10 min, and then in 100% methanol, twice for 10 min, on ice.
 Embryos can be stored in 100% methanol at −20 °C for at least 6 months.
Embryos can be stored in 100% methanol at −20 °C for at least 6 months.
Box 1 | Preparation of younger mouse embryos for 3D imaging  1.5–2.5 h.
1.5–2.5 h.
Whole embryos, younger than E9, can be immunostained and imaged by using the following fixation procedure. An example of a final image is shown in Supplementary Video 1.
Trim the placenta but leave the yolk sac and chorionic disk intact.
Transfer embryos into a 35-mm Petri dish or a six-well plate.
Wash embryos two or three times with PBS.
Aspirate PBS slowly with a Pasteur pipette and allow the sagittal plane of the embryo to lie parallel to the surface of the Petri dish.
Add a small amount of 2% (wt/vol) PFA/PBS into the Petri dish, just enough to cover the embryo. Let it sit on ice for 10 min.
Add more 2% (wt/vol) PFA/PBS to continue the fixation for another 10–20 min.
Wash embryos with PBS, and continue from Step 5 of the main PROCEDURE. Embryos will be fixed in a position that allows for easy counting of somites and image acquisition.
Embryo trimming  15 min per embryo
15 min per embryo
-
6
For each embryo, cut off the head and leave the tail intact in order to help orient the sample later (Fig. 3).
-
7
Use insulin needles to make small cuts in the translucent area between the aorta and somites. Carefully push off the body wall (one side is enough). Leave the umbilical artery and fetal liver intact.
-
8
Cut all the way around the base of the hind limb buds to separate them from the umbilical artery.
-
9
Transfer embryos into scintillation vials containing 100% methanol. Use a nonsterile, 3-ml graduated plastic pipette to transfer embryos and to withdraw solutions during washes.
 The ends of the pipettes can be easily trimmed with scissors to make them wider.
The ends of the pipettes can be easily trimmed with scissors to make them wider. If the embryo sample gets stuck along the inside of the pipette, cut off the pipette tip at that point and ease it out with a pair of fine tweezers.
If the embryo sample gets stuck along the inside of the pipette, cut off the pipette tip at that point and ease it out with a pair of fine tweezers.
Rehydration  30–40 min
30–40 min
-
10
Rehydrate embryos on ice: 10 min in 75% (vol/vol) methanol/PBS (optional); 10 min in 50% (vol/vol) methanol/PBS; and twice for 10 min in PBS.
 Throughout the procedure, we transfer all the used wash solutions into an extra scintillation vial before discarding them, just in case an embryo is inadvertently sucked up into the pipette. The fluid is not discarded until all samples are accounted for.
Throughout the procedure, we transfer all the used wash solutions into an extra scintillation vial before discarding them, just in case an embryo is inadvertently sucked up into the pipette. The fluid is not discarded until all samples are accounted for.
Blocking  1–3 h
1–3 h
-
11If you use biotinylated antibody for primary antibody staining, use both option A for streptavidin/biotin blocking and option B for serum blocking. If not, use only option B.
- Streptavidin/biotin blocking
- Incubate embryos in streptavidin solution (streptavidin (three drops)/PBS (1 ml)) at room temperature (20–25 °C) for 15 min. Mix this gently with a pipette, as bubbles tend to be released as a reaction by-product.
- Wash embryos with PBS on ice for 10 min.
- Incubate embryos in biotin solution (three drops)/PBS (1 ml) for 15 min at room temperature.
- Wash embryos with PBS on ice for 10 min.
- Serum blocking
-
Incubate embryos in 2 ml of serum or BSA/PBS-MT solution on ice for 1 h.
 Serum—goat, donkey, rat or rabbit—should be the same as the host species of your secondary antibody (Table 2). Multiple sera can be used if you are using multiple antibodies. BSA can be used if you do not have appropriate serum. BSA is used at a final concentration of 0.2% (wt/vol) (40 µl of 10% (wt/vol) stock).
Serum—goat, donkey, rat or rabbit—should be the same as the host species of your secondary antibody (Table 2). Multiple sera can be used if you are using multiple antibodies. BSA can be used if you do not have appropriate serum. BSA is used at a final concentration of 0.2% (wt/vol) (40 µl of 10% (wt/vol) stock). At some point, the samples will be distributed into different vials. Do not do this until they are in PBS-MT. If you try to do it while samples are still in PBS, the samples will be sticky, and you will find them all over the inside (and even the outside) of your plastic pipette.
At some point, the samples will be distributed into different vials. Do not do this until they are in PBS-MT. If you try to do it while samples are still in PBS, the samples will be sticky, and you will find them all over the inside (and even the outside) of your plastic pipette.
-
Primary antibody staining  overnight
overnight
-
12
Add a single antibody or multiple antibodies diluted in PBS-MT to the samples (use separate vials if several combinations of antibody stainings are performed at the same time).
 Be sure to include a negative control (isotype control or no primary antibody), especially when testing the specificity of a new antibody.
Be sure to include a negative control (isotype control or no primary antibody), especially when testing the specificity of a new antibody. If background for your antibody is high, you can include BSA (2 mg ml−1).
If background for your antibody is high, you can include BSA (2 mg ml−1). -
13
Gently rock the samples in the cold room overnight. The rocker should be quite slow, taking about 15 s to rock back and forth once.
 Wrap vials in aluminum foil to shield them from light, but more importantly at this stage, to prevent the vials from rolling. Samples tend to get stuck to the sides of rolling vials.
Wrap vials in aluminum foil to shield them from light, but more importantly at this stage, to prevent the vials from rolling. Samples tend to get stuck to the sides of rolling vials.
Washing  3–8 h
3–8 h
-
14
Wash samples at least three times with PBS-MT, and rock them in a cold room (4 °C). Each wash is for a minimum duration of 1 h; the longer the better, but each wash should not exceed 24 h.
Secondary antibody staining  overnight
overnight
-
15
Add a single antibody or multiple secondary antibodies, and then rock samples in a cold room (4 °C) overnight. Antibody is diluted with PBS-MT.
Embryo washing, multiantibody staining  4–8 h (plus optional staining time)
4–8 h (plus optional staining time)
-
16
Wash embryos three times (1 h each) with PBS-MT and rock them in the cold room (4 °C).
-
17
(Optional) If you are using multiple primary antibodies from the same species, repeat Steps 12 through 16, depending on the number of primary antibodies that you are using.
 Each antibody will add 2 d to the procedure. Repeating is necessary only if you are using primary antibodies from the same species (Fig. 1). If primary antibodies are from different species, you can incubate embryos with multiple antibodies at the same time.
Each antibody will add 2 d to the procedure. Repeating is necessary only if you are using primary antibodies from the same species (Fig. 1). If primary antibodies are from different species, you can incubate embryos with multiple antibodies at the same time. -
18
Rinse three times (20 min each) with PBS-T and rock samples in a cold room (4 °C).
Clearing and mounting  2–3 h
2–3 h
-
19
Dehydrate embryos in methanol, rocking in a cold room (4 °C): 10 min in 50% (vol/vol) methanol/PBS and 10 min in 100% methanol.
 Samples can be stored in the refrigerator (4 °C) for a couple of days.
Samples can be stored in the refrigerator (4 °C) for a couple of days. -
20
Prepare 50 ml of 1% (wt/vol) low-melt agarose (melt it when you start the methanol/PBS dehydration (Step 19)), and allow it to cool to ~50 °C while you continue the dehydration steps.
 The agarose can be used for several experiments, but be aware that the agarose concentration may increase each time it is reheated.
The agarose can be used for several experiments, but be aware that the agarose concentration may increase each time it is reheated. -
21
Peel plastic off the FastWell to expose the adhesive, and then stick a round coverslip (0.13–0.16 mm thickness) on the FastWell.
 The FastWell thickness is 1 mm, and it will work well for embryos up to E11.5. Embryos that are late E11 or larger will require stacking two FastWells. Ensure that the coverslip is stuck down well; otherwise, the liquid will leak under the FastWell afterward. This is easiest to do (without breaking the coverslip) by placing the FastWell, coverslip up, on a flat surface and then pressing gently but firmly around the edges with your fingers.
The FastWell thickness is 1 mm, and it will work well for embryos up to E11.5. Embryos that are late E11 or larger will require stacking two FastWells. Ensure that the coverslip is stuck down well; otherwise, the liquid will leak under the FastWell afterward. This is easiest to do (without breaking the coverslip) by placing the FastWell, coverslip up, on a flat surface and then pressing gently but firmly around the edges with your fingers. -
22
Place the FastWell with the coverslip side down on the histology slide (the slide is just being used as a platform to support the coverslip).
-
23
Peel the plastic off the side of the FastWell facing you. Gently pick up the FastWell and look underneath to ensure that the coverslip is still firmly stuck to the FastWell. (Optional) You can put a small dot or circle on the spot where you plan to put the agarose. Make this mark on the side of the coverslip that is against the slide, so you can remove it later if it interferes with microscopy.
-
24
Remove one sample at a time from its vial after the last 100% methanol rinse (Step 19) and place it in a small Petri dish in 100% methanol.
-
25
Draw 200 ml of 100% methanol into a P200 pipette; set the pipette down and have it ready to use
-
26
Place an embryo on the coverslip and immediately add one drop of methanol from the P200 pipette onto the embryo to prevent it from drying up.
 Ensure that no dust is attached to the embryo before picking it up from the Petri dish.
Ensure that no dust is attached to the embryo before picking it up from the Petri dish. -
27
Draw 10 µl of agarose into a P20 pipette, and then dot a small amount (perhaps 2.5–5 µl) onto the coverslip in a dry area close to the sample.
-
28
By using fine bent tweezers, quickly move the embryo to the agarose before it solidifies (and before the methanol evaporates—the embryo should not dry up). The embryo will be adjacent to the agarose, with one end touching it, in order to immobilize it on the slide.
-
29
Immediately fill the well with 200 µl of 100% methanol.
 Try not to get the top of the FastWell wet.
Try not to get the top of the FastWell wet. -
30
Immediately after placing the embryo, remove the methanol and replace it with fresh 100% methanol. Do this several times, and let it sit for ~1 min in a covered slide box between rinses.
 The goal is to remove the water from the agarose.
The goal is to remove the water from the agarose. Do not remove all the methanol during these rinses, as this may cause an air bubble to appear in the embryo (e.g., within the dorsal aorta).
Do not remove all the methanol during these rinses, as this may cause an air bubble to appear in the embryo (e.g., within the dorsal aorta). -
31
Replace the methanol with 50% (vol/vol) BABB/ methanol; change two or three times, as described in Step 30.
-
32
Replace with 100% BABB two or three times until the sample is clear. You should just barely see an outline of the sample.
-
33
Place a second coverslip on the FastWell with tweezers.
 This is tricky—you need to remove enough BABB from the well so that when you place the coverslip the BABB does not overflow from the well and get between the coverslip and the FastWell. In contrast, too little BABB will result in a huge air bubble in the well. Some air is inevitable, but you need enough BABB in the well to cover the sample. This is the reason the sample is fastened down off-center, as the likelihood of having the BABB cover the sample is greater (you can tip the slide until it is covered).
This is tricky—you need to remove enough BABB from the well so that when you place the coverslip the BABB does not overflow from the well and get between the coverslip and the FastWell. In contrast, too little BABB will result in a huge air bubble in the well. Some air is inevitable, but you need enough BABB in the well to cover the sample. This is the reason the sample is fastened down off-center, as the likelihood of having the BABB cover the sample is greater (you can tip the slide until it is covered). -
34
Add four small drops (at four ‘corners’, i.e., 12:00, 3:00, 6:00, 9:00) of clear nail polish on the coverslip and place a SuperFrost slide on top.
-
35
Let the nail polish dry (~5 min). Thereafter, very carefully flip the slide upside down, and remove the lower-quality slide (carefully sliding it off) that was originally at the bottom (see Fig. 4 for a diagram of the completed slide).
 Sometimes, if the FastWell leaks, you can save the sample. Remove the FastWell from the coverslip to which the sample is attached. Clean carefully around the edges of the coverslip with a dry tissue (use 100% methanol to help remove the BABB). Once it is clean and dry, try mounting it again on a new FastWell. This is much easier to do when BABB is in the FastWell rather than methanol because BABB is slimy. We recommend remounting the sample beginning at the BABB step (Step 31). If the FastWell leaks at the methanol step (Steps 29–30), just go through to the 50% (vol/vol) BABB step, keeping in mind that the sample should not dry up. Replace 50% (vol/vol) BABB once or twice and then remount the sample to a new FastWell. In addition, if the embryo is not attached to the agarose while it is in methanol, you can remount it. However, if it is floating around in BABB, then leave it alone; it can still be used for imaging. Although we have not tried this, it might work to move a sample along with its attached agarose to a new FastWell/coverslip; continue with whatever step you are on, without trying to remount with agarose.
Sometimes, if the FastWell leaks, you can save the sample. Remove the FastWell from the coverslip to which the sample is attached. Clean carefully around the edges of the coverslip with a dry tissue (use 100% methanol to help remove the BABB). Once it is clean and dry, try mounting it again on a new FastWell. This is much easier to do when BABB is in the FastWell rather than methanol because BABB is slimy. We recommend remounting the sample beginning at the BABB step (Step 31). If the FastWell leaks at the methanol step (Steps 29–30), just go through to the 50% (vol/vol) BABB step, keeping in mind that the sample should not dry up. Replace 50% (vol/vol) BABB once or twice and then remount the sample to a new FastWell. In addition, if the embryo is not attached to the agarose while it is in methanol, you can remount it. However, if it is floating around in BABB, then leave it alone; it can still be used for imaging. Although we have not tried this, it might work to move a sample along with its attached agarose to a new FastWell/coverslip; continue with whatever step you are on, without trying to remount with agarose. Samples can be stored in the refrigerator (4 °C) for a couple of days.
Samples can be stored in the refrigerator (4 °C) for a couple of days.
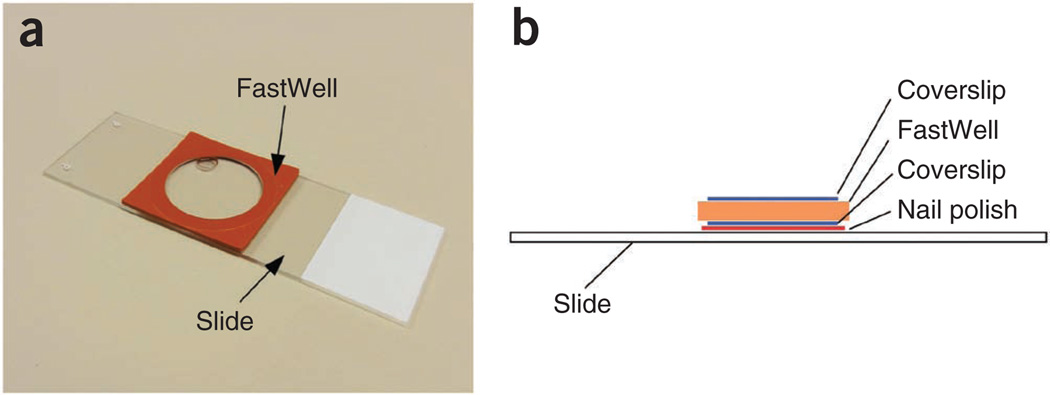
Figure 4.
Slide preparation. (a) A slide with a FastWell. (b) Diagram representing a side view of the completed slide. See Steps 20–35 for preparation.
Confocal microscopy  30–60 min to set up the microscope and ~45 min to scan one z-stack
30–60 min to set up the microscope and ~45 min to scan one z-stack
-
36
After allowing the mounted sample to return to room temperature (15–30 min), bring it to the confocal microscope.
 Embryos are best imaged immediately but can wait a couple of days when stored in the refrigerator (4 °C).
Embryos are best imaged immediately but can wait a couple of days when stored in the refrigerator (4 °C). -
37
Look for the embryo with fluorescent light (e.g., FITC filter) through the eyepieces.
-
38
Look for dorsal aorta by using continuous xy scanning of the CD31 staining channel (e.g., Cy3 for the 543-nm channel).
-
39
Adjust the image intensity by changing the laser intensity, detector gain and other parameters.
-
40
Place the center z plane at the middle of the dorsal aorta and set the top and bottom of the z axis to scan.
-
41
Collect the z-stacks (Fig. 5a).

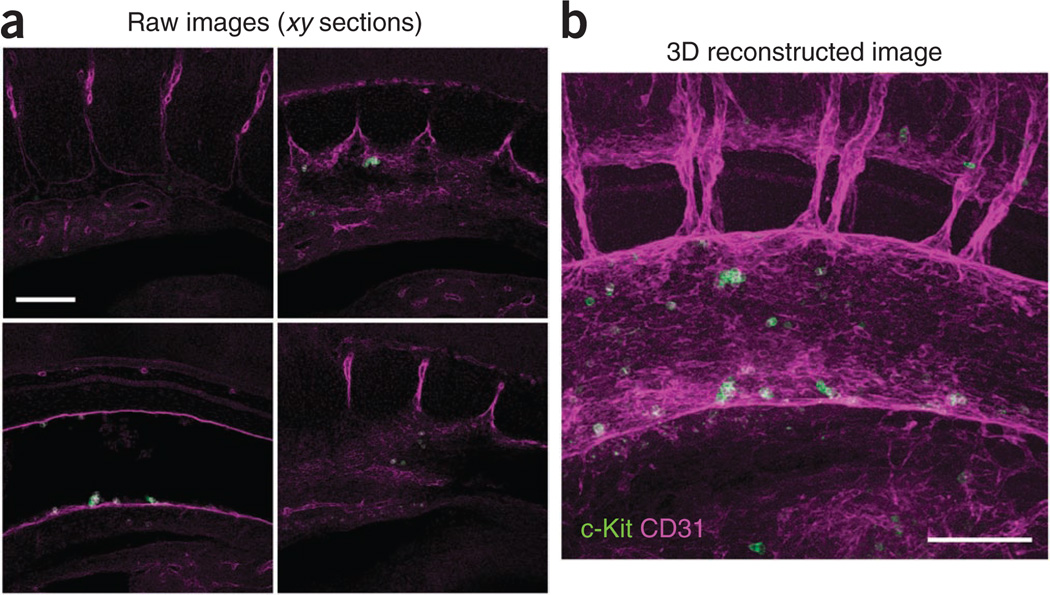
Figure 5.
Three-dimensional reconstruction of the dorsal aorta region stained with c-Kit (green)- and CD31 (magenta)-specific antibodies at E10.5 (35 somite pairs). (a) Representative raw images (xy confocal sections) of the sample. (b) 3D reconstructed image. Scale bars, 100 µm.
Higher-magnification images  up to 3–4 h
up to 3–4 h
-
42
(Optional) Change the objective from ×20 to ×40/×63 water-immersion type objective. Having confirmed the cells of interest by checking the lower-magnification picture (e.g., by making 3D reconstructed image, see Step 43), acquire images.

Image analysis  variable
variable
-
43Z-stacks obtained at Step 41 or 42 can be used for 3D reconstruction (A) and quantitative analysis (B).
- 3D reconstruction
- Create a 3D reconstructed image from z-stacks. By comparing them with background signals of isotype control samples, adjust the brightness of the image (Fig. 5b).
- Quantification of c-Kit+ hematopoietic cluster cells
-
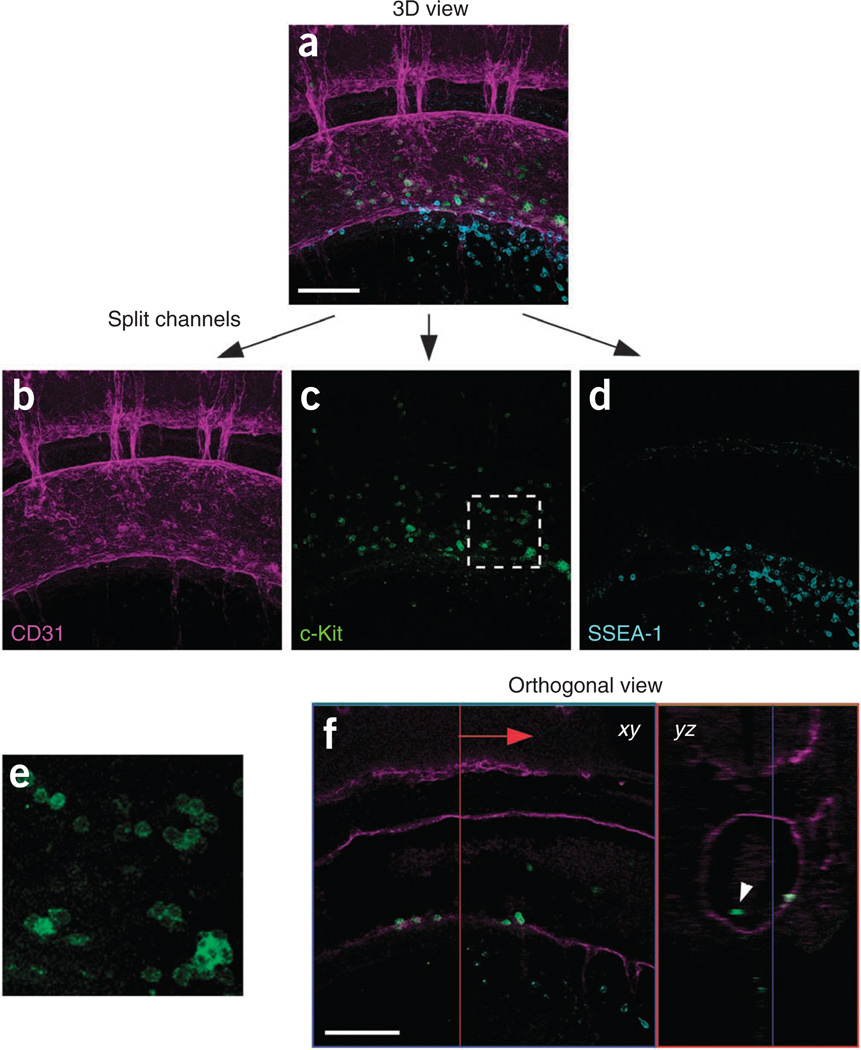
In the case of c-Kit/CD31 double staining, c-Kit+ hematopoietic cluster cells can be quantified. Split the 3D reconstruction image into each channel (Fig. 6a–d).
 If many c-Kit+ cells are observed in the reconstituted image, we advise that the z-stacks be separated into several parts in order to reduce the thickness of 3D images.
If many c-Kit+ cells are observed in the reconstituted image, we advise that the z-stacks be separated into several parts in order to reduce the thickness of 3D images. - Count c-Kit+ cells (Fig. 6c and e).
- Count circulating c-Kit+ cells (i.e., cells not attached to the wall of the dorsal aorta) by using the orthogonal view (Fig. 6f).
- To obtain the number of c-Kit+ hematopoietic cluster cells, subtract the number of circulating c-Kit+ cells obtained in 43B(iii) from the number of c-Kit+ cells obtained in 43B(ii).
-

Troubleshooting advice can be found in Table 4.
Figure 6.
Quantification procedure of c-Kit+ hematopoietic cluster cells in the dorsal aorta. (a) First, a 3D reconstructed image is made from serial xy section images. (b–d) After splitting into three channels (magenta, green and blue), the number of c-Kit+ cells (green) is counted. (e) Magnification of the boxed region in c. As c-Kit is a cell surface marker, the border of cells within clusters is clearly distinct. (f) The number of circulating c-Kit+ cells (i.e., cells that do not attach to the wall of dorsal aorta) is counted using an orthogonal view. Move the position of the yz plane (red line in xy data) manually, and identify circulating c-Kit+ cells in yz data (white arrowhead). To obtain the number of c-Kit+ hematopoietic cluster cells, the number of circulating c-Kit+ cells is subtracted from the number of c-Kit+ cells in image c. To cover the whole dorsal aorta, six or seven scans are required. Scale bars, 100 µm.
Table 4.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 41,42 | No or low signal detected (poor resolution) |
Incomplete or excess fixation | Optimize the fixation time for each antibody. In our experience, long fixation time (e.g., overnight) sometimes causes disruption of antigens |
| Confocal microscope is not properly set up |
Ensure that settings (e.g., filters, pinhole) are properly adjusted |
||
| Concentration of antibody is too low | Titrate the optimal concentration of antibody | ||
| Inappropriate choice of antibody | Check the host species and isotype | ||
| Embryo is too thick | Shave the embryo to reduce its thickness | ||
| High background signal | Concentration of antibody is too high | Titrate the optimal concentration of antibody | |
| Insufficient washing | Use longer washing time | ||
| Autofluorescence of embryonic tissues | We prefer to use far-red fluorochromes (e.g., Alexa Fluor 647), as there is little autofluorescence in the near-infrared range |
Steps 1–5, dissection and fixation of embryos: 2–4 h
Steps 6–9, embryo trimming: 15 min per embryo
Step 10, rehydration: 30–40 min
Step 11, blocking: 1–3 h
Steps 12–13, primary antibody staining: overnight
Step 14, washing: 3–8 h
Step 15, secondary antibody staining: overnight
Steps 16–18, embryo washing, multiantibody staining: Steps 16 and 18 (washing), 4–8 h; Step 17 (multiantibody staining, optional), 2 d per additional primary antibody
Steps 19–35, clearing and mounting: 2–3 h
Steps 36–41, confocal microscopy: 30–60 min to set up microscope and about 45 min to scan one z-stack
Box 1, preparation of younger mouse embryos: 1.5–2.5 h
ANTICIPATED RESULTS
c-Kit/CD31 staining
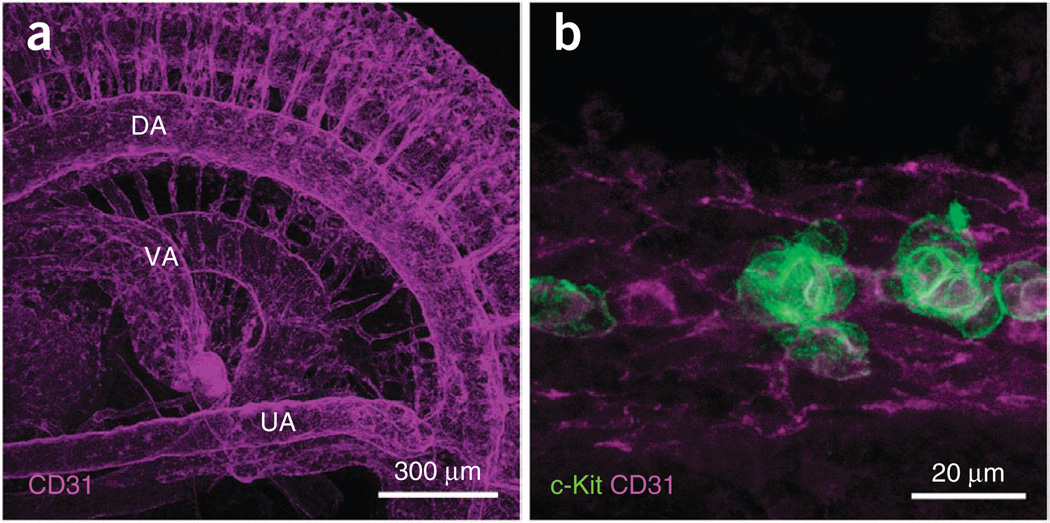
After 3D reconstruction, the distribution of c-Kit+ cells within the dorsal aorta can be seen through the semitransparent CD31+ endothelial layer. Representative raw images (xy images) and 3D pictures taken with a ×20 objective are shown in Figure 5. The brightness of raw images influences the final appearance of the 3D image. If the brightness of CD31 is too high, the CD31+ endothelial layer does not become semitransparent. This is not adjustable after collecting z-stacks. Therefore, before collecting z-stacks, adjust the intensity of CD31 by using the range indicator function (Zeiss Image Browser) to check the saturated pixels in the image. At E10.5, we detected 609 ± 84 c-Kit+ cells in the dorsal aorta7. In addition to a ×20 objective, ×10 and ×63 objectives can be used for low- and high-magnification images (Fig. 7).
Figure 7.
Three-dimensional reconstructed pictures at low and high magnifications. (a) E10.5 embryo stained with anti-CD31 antibody. The picture was taken with a ×10 objective (EC Plan-Neofluar ×10/NA 0.3). Blood vessels of various sizes throughout the embryo are observed. DA, dorsal aorta; UA, umbilical artery; VA, vitelline artery. (b) E10.5 embryo stained with anti–c-Kit and anti-CD31 antibodies. The picture was taken with a ×63 objective (Achroplan ×63/NA 0.95 W). c-Kit+ hematopoietic clusters attached to the CD31+ endothelial layer are observed.
Imaging nuclear proteins
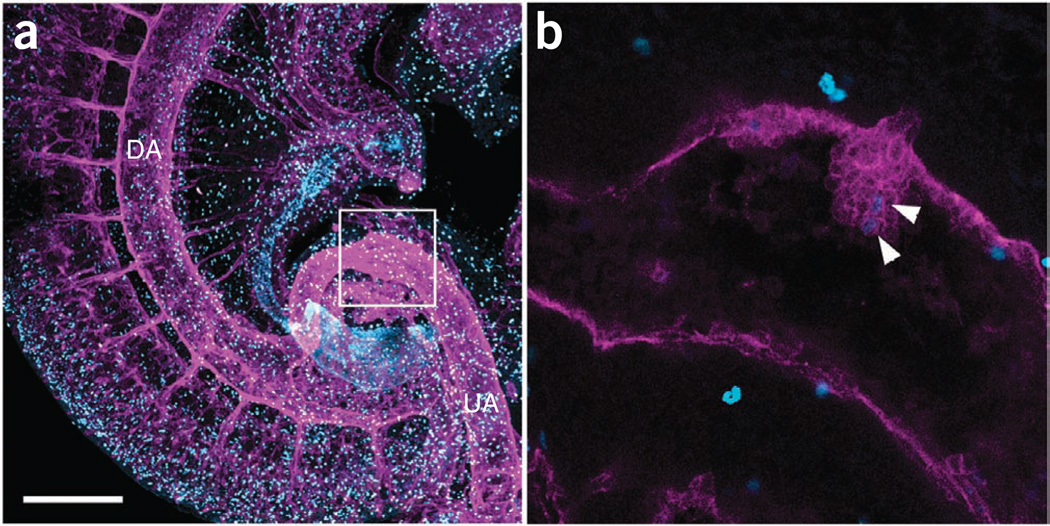
In addition to imaging cells immunostained with antibodies detecting cell surface proteins, the whole-mount immunostaining BABB method can be used to image nuclear proteins. For example, mitotic cells can be easily visualized after immunostaining with an anti-phosphohistone H3 antibody (Fig. 8).
Figure 8.
Three-dimensional reconstructed pictures showing nuclear expression of phosphohistone H3. (a) E10.5 embryo stained with anti-phosphohistone H3 antibody (blue) and anti-CD31 antibody (magenta). The picture was taken with a ×10 objective. Composite image of 83 slices (optical sections of 163.54 µm thickness) is shown. (b) Image of the umbilical artery (boxed region of panel a) showing a large hematopoietic cluster with two cells expressing phosphohistone H3 (arrowheads), taken with a ×20 objective (zoom 3.28). DA, dorsal aorta; UA, umbilical artery. Scale bar, 300 µm.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the laboratory for critical comments related to this work and also thank the following funding organizations: the US National Institutes of Health grants R37DK054077 (E.D.) and RO1HL091724 (N.A.S.), the Netherlands BSIK Innovation Program 03040 (E.D.) and the European Science Foundation Program EuroSTELLS 01-011 (E.D.), and the Netherlands Genomics Initiative—Cancer Genomic Distinguished Scientist Award (N.A.S.).
Footnotes
Note: Supplementary information is available via the HTML version of this article.
AUTHOR CONTRUBUTIONS T.Y. performed the experiments, optimized the protocol and wrote the paper; T.Y.-I., A.D.Y. and M.J.C. performed the experiments, N.A.S. performed the experiments and wrote the paper, and E.D. directed the research and wrote the paper.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Published online at http://www.natureprotocols.com/.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Dzierzak E, Speck NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat. Immunol. 2008;9:129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medvinsky A, Rybtsov S, Taoudi S. Embryonic origin of the adult hematopoietic system: advances and questions. Development. 2011;138:1017–1031. doi: 10.1242/dev.040998. [DOI] [PubMed] [Google Scholar]
- 3.Marshall CJ, Thrasher AJ. The embryonic origins of human haematopoiesis. Br. J. Haematol. 2001;112:838–850. doi: 10.1046/j.1365-2141.2001.02537.x. [DOI] [PubMed] [Google Scholar]
- 4.Tavian M, et al. Aorta-associated CD34+ hematopoietic cells in the early human embryo. Blood. 1996;87:67–72. [PubMed] [Google Scholar]
- 5.Gard DL. Confocal immunofluorescence microscopy of microtubules in amphibian oocytes and eggs. Methods Cell Biol. 1993;38:241–264. doi: 10.1016/s0091-679x(08)61006-7. [DOI] [PubMed] [Google Scholar]
- 6.Zucker RM, Hunter ES, Rogers JM, editors. Confocal Laser Scanning Microscopy of Morphology and Apoptosis in Organogenesis-Stage Mouse Embryos. Humana Press; 1999. pp. 191–202. [DOI] [PubMed] [Google Scholar]
- 7.Yokomizo T, Dzierzak E. Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development. 2010;137:3651–3661. doi: 10.1242/dev.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng CE, et al. A Runx1 intronic enhancer marks hemogenic endothelial cells and hematopoietic stem cells. Stem Cells. 2010;28:1869–1881. doi: 10.1002/stem.507. [DOI] [PubMed] [Google Scholar]
- 9.Yokomizo T, Ng CE, Osato M, Dzierzak E. Three-dimensional imaging of whole midgestation murine embryos show an intravascular localization for all hematopoietic clusters. Blood. 2011;117:6132–6134. doi: 10.1182/blood-2011-02-334037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Porrero JA, Godin IE, Dieterlen-Lievre F. Potential intraembryonic hemogenic sites at pre-liver stages in the mouse. Anat. Embryol. (Berl.) 1995;192:425–435. doi: 10.1007/BF00240375. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Porrero JA, et al. Antigenic profiles of endothelial and hemopoietic lineages in murine intraembryonic hemogenic sites. Dev. Comp. Immunol. 1998;22:303–319. doi: 10.1016/s0145-305x(98)00006-8. [DOI] [PubMed] [Google Scholar]
- 12.Ghiaur G, et al. Rac1 is essential for intraembryonic hematopoiesis and for the initial seeding of fetal liver with definitive hematopoietic progenitor cells. Blood. 2008;111:3313–3321. doi: 10.1182/blood-2007-08-110114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaffredo T, et al. Aortic remodelling during hemogenesis: is the chicken paradigm unique? Int. J. Dev. Biol. 2010;54:1045–1054. doi: 10.1387/ijdb.103062tj. [DOI] [PubMed] [Google Scholar]
- 14.Zovein AC, et al. Vascular remodeling of the vitelline artery initiates extravascular emergence of hematopoietic clusters. Blood. 2010;116:3435–3444. doi: 10.1182/blood-2010-04-279497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferkowicz MJ, Yoder MC, editors. Whole Embryo Imaging of Hematopoietic Cell Emergence and Migration. Springer Science-Business Media; 2008. pp. 143–155. [DOI] [PubMed] [Google Scholar]
- 16.Yokomizo T, et al. Requirement of Runx1/AML1/PEBP2αB for the generation of haematopoietic cells from endothelial cells. Genes Cells. 2001;6:13–23. doi: 10.1046/j.1365-2443.2001.00393.x. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida H, et al. Hematopoietic tissues, as a playground of receptor tyrosine kinases of the PDGF-receptor family. Dev. Comp. Immunol. 1998;22:321–332. doi: 10.1016/s0145-305x(98)00008-1. [DOI] [PubMed] [Google Scholar]
- 18.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 19.Becker K, Jahrling N, Kramer ER, Schnorrer F, Dodt HU. Ultramicroscopy: 3D reconstruction of large microscopical specimens. J. Biophotonics. 2008;1:36–42. doi: 10.1002/jbio.200710011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.