Abstract
Genetics in the post-genomic period is shifting from structural to functional genetics or genomics. Meanwhile, the use of twins is largely expanding from traditional heritability estimation for disease phenotypes to the study of both diseases and various molecular phenotypes, such as the regulatory phenotypes in functional genomics concerning gene expression and regulation, by engaging both classical twin design and marker-based gene mapping techniques in genetic epidemiology. New research designs have been proposed for making novel uses of twins in studying the molecular basis in the epigenetics of human diseases. Besides, twins not only serve as ideal samples for disease gene mapping using conventional genetic markers but also represent an excellent model for associating DNA copy number variations, a structural genetic marker, with human diseases. It is believed that, with the rapid development in biotechniques and new advances in bioinformatics, the unique samples of twins will make new contributions to our understanding of the nature and nurture in complex disease development and in human health. This paper aims at summarizing the new uses of twins in current genetic studies and suggesting novel proposes together with useful design and analytical strategies.
Keywords: Twins, Genetics, Genomics
The expanding role of twins
Twins are of special interest for genetic studies due to their genetic similarities and rearing-environment sharing. The last century witnessed successful uses of twins in exploring the genetic and environmental contributions to human diseases and complex traits. By comparing phenotype correlation patterns in monozygotic (MZ, identical) and dizygotic (DZ, fraternal) twin pairs, various genetic and environmental components can be assessed using the classical twin design for heritability estimation and correlation calculation. Based on variance decomposition that partitions the total phenotype variance (VP) into genetic variance with additive (VA) and dominant (VD) components and environmental variance with shared (VC) and non-shared (VE) components, the heritability is estimated as the proportion of total variance that is accounted for by the genetic components,
For example, based on data from the Danish Twin Registry, the heritability for type II diabetes was estimated as 26% (Poulsen et al. 1999) and the heritability for human lifespan about 25% (Herskind et al. 1996; Hjelmborg et al. 2006). These so-called classical twin studies make use of the unique genetic make-ups in MZ and DZ twins and infer the genetic importance in human diseases without individual genotypic or molecular marker information. Although with questionable assumptions, findings from twin studies have been of great importance in elucidating the genetic and environmental contributions to disease development and in directing the subsequent marker-based (linkage and association) analyses that aim at localizing or mapping specific genes involved.
With the completion of the Human Genome Project at the junction of twenty-first century, the study of human genetics has come to a post-genomic era. The availability of enormous amount of information of the human genome both poses challenges and creates opportunities for biomedical researchers. At the same time, there have been sweeping changes in genetic studies of human complex diseases. The international initiative of obtaining human genetic sequence variation and the use of single nucleotide polymorphisms (SNPs) as a source of markers for the variation have led to the high throughput genotyping techniques enabled by massive data acquisition systems. This facilitates, with ease, large-scale genome-wide scan of genes underlining human complex diseases where twins serve as perfectly matched samples for both linkage and association analyses (Boomsma et al. 2002). Perhaps the most significant change in the twenty-first century genetics will be the shift from structural genomics, where genes are regarded as a static concept, to functional genomics, where the dynamic patterns of the genes are analyzed jointly from gene interaction to gene regulation and to functional cluster or pathway analysis (Peltonen and McKusick 2001). Novel methodological and strategic innovations are called for in order to meet these emerging challenges. In this regard, strategic uses of twin samples can have an important impact on the study of human complex diseases in the functional genomics era. Table 1 provides a brief up-to-date overview for the various new uses of twins in genetic and genomic studies of human complex diseases.
Table 1.
Overview of new uses of twins in genetic and genomic studies of human complex phenotypes
| Applications | Phenotypes | Twin samples | Methods of analysis | Examples | Advantages over other designs |
|---|---|---|---|---|---|
| Genetic component in transcriptional regulation | Gene expression levels; DNA methylation measurement | MZ, DZ | Heritability estimate; ICC | York et al. (2005); Correa and Cheung (2004); Tan et al. (2005); Kaminsky et al. (2009) | Twins only design |
| Disease gene mapping | Genetic diseases | DZ pairs discordant for diseases | Non-parametric linkage analysis; S-TDT | De Moor et al. (2007); Livshits et al. (2007a, b); Perola et al. (2007); O’Connor et al. (2008) | Perfect match over age and family environment; and low probability of non-paternity as compared with sibling pairs |
| Genetical genomics | mRNA levels | DZ | Non-parametric linkage | Perfect match over age and family environment; and low probability of non-paternity as compared with sibling pairs | |
| Epigenetics of diseases | DNA methylation levels | MZ pairs discordant for diseases | Pair-matched analysis | Pietiläinen et al. (2008); Petronis et al. (2003); Kaminsky et al. (2008) | Identical twins only design |
| Structural genetic variations in diseases | DNA copy number variation | MZ pairs discordant for diseases | Pair-matched analysis | Bruder et al. (2008) | Perfect match for both genetic composition and family environmental factors as compared with population study design |
| Gene-environment interaction | Genetic diseases | MZ | Phenotype variability comparison | Spector et al. (2000); Martin (2000); Wray et al. (2008) | Identical twins only design |
Classical twin method in studying functional genomics
The transcriptomics or global (genome-wide) gene expression analysis has become an active field in functional genomics thanks to the microarray technology that enables global monitoring of massive gene expression profiles. Like other complex traits, the levels of gene expression are subject to both genetic and environmental influences. By treating the measured expression levels as quantitative phenotypes, twin methods can be used to assess the relative importance of genetic and environmental contributions to the regulation of transcriptional activities. For example, Correa and Cheung (2004) studied the genetic variation in radiation-induced gene expression using MZ twins. Intra-class correlation coefficient (ICC) was calculated for genes responsive to ionizing radiation before and after radiation exposure in MZ twin pairs. A notable increase in ICC was observed for most of the genes indicating a high genetic dissection in the transcriptional response to radiation exposure. By engaging both MZ and DZ twins, the typical twin design has been introduced in microarray-based global gene expression analysis to estimate the genetic component in transcriptional regulation. In a genome-wide gene expression study by York et al. (2005), broad sense heritability was estimated for 322 genes that showed high MZ to DZ ratio of ICC with most of the genes showing high heritability. A similar twin method was applied to gene expression data from elderly Danish twins by Tan et al. (2005). Their analysis using the classical twin method gave relatively high heritability estimates for the topmost active genes (from 0.23 to 0.44). It is worth to mention here that the ICC, a reliability measurement, has been frequently used in gene expression analysis of related individuals including twins because it is sensitive to both random and systematic variations. In the twin setting, an ICC can be calculated as
Here, MSB is between-pair mean square, and MSW is within-pair mean square. In contrast, the conventional correlation statistics such as the Pearson’s correlation coefficient can be sensitive to random variations only (Pellis 2003), the reason being that, in the ICC, data are centered and scaled using a pooled mean and standard deviation whereas in the correlation coefficient each variable is centered and scaled separately. A computer resampling method was introduced to assess statistical significance of an estimated ICC (Tan et al. 2005, 2008) by permuting the samples to form pseudo-twin pairs. Very recently, the twin method has been applied to studying global DNA methylation profiles in MZ and DZ twins (Kaminsky et al. 2009) with highly significant epigenetic differences reported in DZ twins as compared with that in MZ twins. Their results on twins suggested that the molecular mechanisms of heritability may not be limited to DNA sequences. It can be anticipated that heritable epigenetic changes will become a new dimension for the study of human heritable diseases or traits.
Twins for disease gene mapping
The fast development in high throughput SNP genotyping techniques using DNA microarray platforms, such as Illumina and Affymetrix, is revolutionizing the way we design and conduct genetic epidemiological studies of human complex traits or diseases. High-resolution genome-wide scan enabled by the high-density and informative SNP marker arrays makes it possible for researchers to fine map quantitative trait loci (QTL) that are responsible for human complex traits or diseases with even only minor effects.
The usefulness of twins in linkage mapping lays in the fact that DZ or fraternal twins are basically siblings. They can be used in non-parametric linkage analysis by comparing phenotype-dependent allele-sharing identical-by-decent (IBD) between members of a pair using regression or variance analysis. We emphasize that linkage scans in DZ twins are advantaged by the fact that DZ twins are matched for pre- and postnatal (shared rearing environmental) factors and especially perfectly matched for their ages. These factors could be potentially associated with the phenotypes of interest due to the nature of complex traits and thus need to be adjusted by adding extra covariates in the regression models for non-parametric linkage mapping, i.e.,
Here, y is a function of phenotype values for members of a pair; π is the estimated proportion of IBD sharing; x is the difference in covariate values; α, β, and γ are parameters to be estimated. When DZ twins instead of sib pairs are used, the covariates to the left of the regression model can be best matched so that the number of covariates in the model is reduced and thus an increase in power expected. Linkage mapping of late-onset diseases (missing parental genotypic information) using twins can be further advantaged by the low probability of non-paternity (MacGregor et al. 2000) that causes miscalculation of IBD probability and thus affects linkage results. Actually, the DZ twins have been frequently used in linkage mapping of QTL, especially in recent years (De Moor et al. 2007; Livshits et al. 2007a, b; Perola et al. 2007; O’Connor et al. 2008). Among the important findings, the GenomEUtwin project reported an evidence for common Caucasian loci that are linked with body stature in a consortium of 6,602 European twins (Perola et al. 2007). Likewise, association mapping of human disease or quantitative trait genes can be advantaged when applying sib-pair based methods such as S-TDT (Spielman and Ewens 1998) or QTDT (Abecasis et al. 2000; Ewens et al. 2008) to DZ twins.
It is unfortunate that the usefulness of MZ twins in gene mapping is largely limited due to their complete within-pair IBD sharing. However, efforts have been taken in making use of MZ twins for studying gene-by-environment interactions (Spector et al. 2000; MacGregor et al. 2000), an important issue in the study of complex traits. Martin (2000) proposed an MZ twin-based approach for detecting gene-environment interactions by assessing the within-pair difference for a measured phenotype in identical twins of a particular genotype. The rational is that a gene’s environmental sensitivity can be demonstrated by greater within-pair variance for the phenotype. Very recently, this approach has been applied to investigate the interaction between serotonin receptor (5HTTLPR) genotypes and environmental risk factors including depression and stressful life events (Wray et al. 2008). Note that such MZ twin-based association approach is characterized by assessing the genetic effect on the variability instead of the level of a phenotype.
Twins for genetical genomics
The term “genetical genomics” refers to the study of the genetic basis of gene expression (Jansen and Nap 2001). The existence of genetic control over mRNA levels as revealed by classical twin and family-based heritability studies means that the molecular phenotype of gene expression is amenable to genetic analysis for mapping the quantitative trait loci that are responsible for gene expression regulation referred to as expression QTL or eQTL (Li and Burmeister 2005).
By combining high throughput SNP genotyping and gene expression profiling, it is possible to map genomic regions (eQTL or eSNPs) that are associated with a change in the expression phenotype using linkage (Schadt et al. 2003; Morley et al. 2004; Monks et al. 2004) and association (Cheung et al. 2005) approaches. The eQTLs can be classified into cis (local) or trans (distant) acting eQTLs according to the location of the transcription site and that of the eQTL or eSNP significantly correlated with the transcriptional activity (Rockman and Kruglyak 2006). A cis-acting eQTL is positioned at the same location in the transcription site; otherwise, it is termed as a trans-acting eQTL. There have been two important gene mapping analyses on gene expression in humans (Morley et al. 2004; Monks et al. 2004) with both cis- and trans-acting eQTLs localized.
As a complex phenotype, the expression of a specific gene can be affected by genetic and non-genetic factors including multiple environmental influences. In this case, the advantages of deploying twins in disease gene mapping also applies in linkage analysis of the expression phenotypes. Among the advantages of twins in linkage studies mentioned above, we emphasize the importance of controlling for age effect because molecular phenotypes are subject to variation due to developmental stages or as a result of aging. Tan et al. (2008) reported significant differential gene expression patterns between grandparents and grandchildren. Their analysis revealed many differentially regulated genes with predominantly more genes turned off or down-regulated perhaps as a result of aging. The twin-based analysis provides a perfect match of age where age becomes a pair-specific covariate and can be easily included in the linkage models. With well-defined samples such as twins, the linkage analysis technique is yet to extend to other molecular phenotypes in studying genetical genomics, for example, the mapping of genomic sites that regulate the activities of microRNAs or control the status of DNA methylation.
Twins for studying epigenetics
Epigenetics studies the environmental impacts on traits acquired within our lifetime through chromatin modifications (involving methylations of cytosine at cytosine–guanine dinucleotides, CpG, and acetylations and methylations of DNA-bound histones) that alter regulation of gene expression (Rodenhiser and Mann 2006; Gibson 2008). Alterations in gene expression due to global epigenetic changes accumulating over time (Fraga et al. 2005) can have important influences on disease susceptibilities (Mathers 2008; Maier and Olek 2002) adding new evidence that nature and nurture are, in fact, inextricably linked. Elucidating the complex interplay between environmental exposure and gene expression that contributes to complex phenotypes poses a new challenge.
Identical twins are like clones and can serve as a good and unique model for studying the epigenetics of complex phenotypes because the genetic background is perfectly matched within each pair (Petronis 2006; Poulsen et al. 2007). A special design of “clonal control” has been introduced by which MZ twins discordant for a phenotype of interest are sampled from independent families (Fig. 1) and intra-pair epigenetic differences determined by high throughput global fingerprinting techniques for measuring epigenetic modifications. As shown in Fig. 1, the “clonal control” design identifies differential DNA methylation sites by taking advantage of the perfect match in within-pair genetic make-ups in identical twins. The differential epigenetic modification patterns can be further associated with a specific environmental exposure with rearing environments controlled by the design. Pietiläinen et al. (2008) recently studied the global transcript profiles of fat in MZ twins discordant for BMI and reported interesting findings suggesting the substantial roles of mitochondrial energy and amino acid metabolism behind acquired obesity and insulin resistance. The “clonal controls” design has been used in epigenetic studies of other complex traits as well including schizophrenia (Petronis et al. 2003), aging (Fraga et al. 2005), and behavior traits (Kaminsky et al. 2008). In one of our ongoing projects in the EU-funded Lifespan Network (http://www.lifespannetwork.nl), large samples of MZ twins extremely discordant on birth weight are being collected for epigenetic profiling and for linking epigenetic variations with aging phenotypes across ages.
Fig. 1.
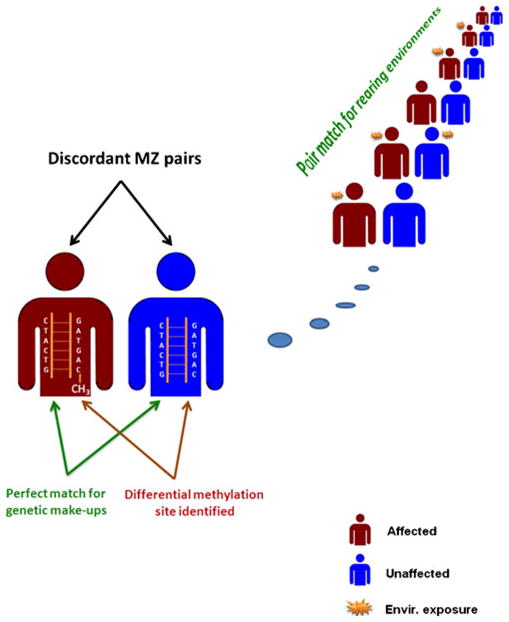
The “clonal control” design collects independent pairs of identical twins discordant for a phenotype (red: affected; blue: unaffected). The perfect match for within-pair genetic make-ups in identical twins offers a unique opportunity for localizing differential epigenetic modification sites. The identified differential epigenetic modification patterns can be further associated with a specific environmental exposure (yellow asterisk) after controlling for rearing environments
We emphasize that the use of identical twins discordant for a disease phenotype enables identification of genomic sites that are under differential epigenetic modification conditional on disease affection status. With this information, epidemiological and bioinformatics tools should help to further link the identified epigenetic changes with observed environmental exposures (Fig. 1). In contrast to the traditional epidemiological approaches, such a novel practice could help to set up a “causal” connection from macro-epidemiological variables to micro-molecular responses and to the final disease phenotypes (Fig. 2) thus providing better understanding of human disease etiology.
Fig. 2.
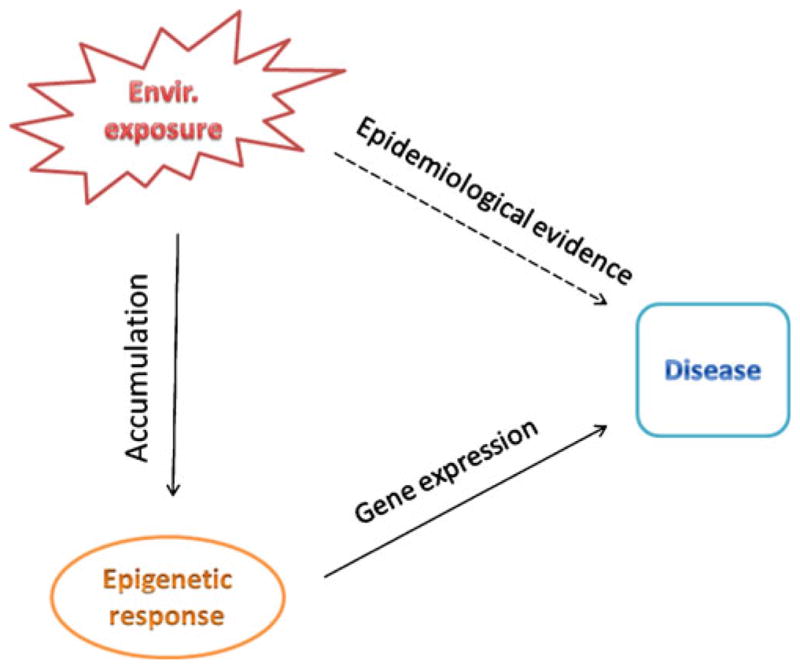
Different from the traditional epidemiological approach, the use of identical twins in epigenetic studies of human diseases links the macro-epidemiological variables such as environmental exposure with molecular epigenetic modification that leads to the final disease phenotype
Twins for studying structural genetic variations
As the most explored subtype of structural genetic variations, the DNA copy number variation (CNV) is a new genetic marker for whole genome association studies. The recent application of CNV in disease studies advantaged by the development of high-resolution CNV screening techniques such as the microarray-based comparative genome hybridization (array CGH; Pinkel et al. 1998) is likely to bring deeper insights into the contribution of CNV to common diseases (de Smith et al. 2008). Although the conventional case-control design popular for gene marker-based association analysis can be used in assessing the correlation between CNV and a certain disease, the use of twins in studying CNV-disease associations represents an excellent model because any intra-pair genetic difference in MZ twins can provide direct evidence for somatic mosaicism. In a recent study by Bruder et al. (2008), CNV analysis was performed on 19 MZ twin pairs either concordant or discordant on neurodegenerative phenotypes. Their results suggested that phenotypically discordant MZ twins could serve as a powerful tool for identifying genomic regions harboring disease-or trait-influencing CNV loci.
Moreover, the “normal” MZ twins also provide an ideal model for studying structural genetic variations in healthy individuals which is suspected to exist in a large number of genomic regions (de Smith et al. 2008). Because chromosomal structural variations are common in somatic development, it would be interesting to find out whether CNVs accumulate with age and whether these changes are associated with aging-related phenotypes. To this end, CNV profiling in age-stratified MZ twins could represent a unique model for the genetic study of human aging.
Concluding remarks
To summarize, the new use of twins in genetic studies in the functional genomics era is characterized by (1) the extended application of classical twin method from disease studies to the study of both diseases and emerging molecular phenotypes in functional genomics; (2) QTL mapping for complex disease phenotypes and for regulatory phenotypes; (3) “clonal control” using identical twins in epigenetic studies of complex diseases and traits; and (4) new uses of twins in studying structural genetic variations. The expanding use of the unique samples of twins triggered by new development in biomedical techniques, and bioinformatics is certain to bring new implications that could help improve our understanding of the nature and/or nurture of complex diseases and human health.
Acknowledgments
This work was jointly supported by Novo Nordisk Foundation and European Foundation for the Study of Diabetes. Part of this work was supported by and carried out within the EU-funded Network of Excellence LifeSpan (FP6 036894) and supported by grants NIH/NIA P01 AG08761 and from The National Program for Research Infrastructure 2007 from the Danish Agency for Science Technology and Innovation. The Danish Aging Research Center is supported by a grant from the VELUX foundation.
Contributor Information
Qihua Tan, Email: qtan@health.sdu.dk, The Danish Twin Registry and The Danish Aging Research Center, Institute of Public Health, University of Southern Denmark, J. B. Winsløws Vej 9B, DK-5000 Odense C, Denmark. Department of Clinical Genetics, Odense University Hospital, Sdr. Boulevard 29, DK-5000 Odense C, Denmark. Epidemiology, Institute of Public Health, University of Southern Denmark, Winsløws Vej 9B, DK-5000 Odense C, Denmark.
Kirsten Ohm Kyvik, Institute of Regional Health Services Research, University of Southern Denmark, Winsløwparken 19. 3, DK-5000 Odense C, Denmark.
Torben A. Kruse, Department of Clinical Genetics, Odense University Hospital, Sdr. Boulevard 29, DK-5000 Odense C, Denmark
Kaare Christensen, The Danish Twin Registry and The Danish Aging Research Center, Institute of Public Health, University of Southern Denmark, J. B. Winsløws Vej 9B, DK-5000 Odense C, Denmark.
References
- Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66(1):279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat Rev Genet. 2002;3(11):872–882. doi: 10.1038/nrg932. [DOI] [PubMed] [Google Scholar]
- Bruder CE, Piotrowski A, Gijsbers AA, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82 (3):763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437(7063):1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa CR, Cheung VG. Genetic variation in radiation-induced expression phenotypes. Am J Hum Genet. 2004;75(5):885–890. doi: 10.1086/425221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moor MH, Spector TD, Cherkas LF, Falchi M, Hottenga JJ, Boomsma DI, De Geus EJ. Genome-wide linkage scan for athlete status in 700 British female DZ twin pairs. Twin Res Hum Genet. 2007;10(6):812–820. doi: 10.1375/twin.10.6.812. [DOI] [PubMed] [Google Scholar]
- De Smith AJ, Walters RG, Froguel P, Blakemore AI. Human genes involved in copy number variation: mechanisms of origin, functional effects and implications for disease. Cytogenet Genome Res. 2008;123(1–4):17–26. doi: 10.1159/000184688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens WJ, Li M, Spielman RS. A review of family-based tests for linkage disequilibrium between a quantitative trait and a genetic marker. PLoS Genet. 2008;4(9):e1000180. doi: 10.1371/journal.pgen.1000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. The environmental contribution to gene expression profiles. Nat Rev Genet. 2008;9:575–581. doi: 10.1038/nrg2383. [DOI] [PubMed] [Google Scholar]
- Herskind AM, McGue M, Holm NV, Sorensen TI, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97:319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- Hjelmborg JB, Iachine I, Skytthe A, Vaupel JW, McGue M, Koskenvuo M, Kaprio J, Pedersen NL, Christensen K. Genetic influence on human lifespan and longevity. Hum Genet. 2006;119:312–321. doi: 10.1007/s00439-006-0144-y. [DOI] [PubMed] [Google Scholar]
- Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17(7):388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- Kaminsky Z, Petronis A, Wang SC, Levine B, Ghaffar O, Floden D, Feinstein A. Epigenetics of personality traits: an illustrative study of identical twins discordant for risk-taking behaviour. Twin Res Hum Genet. 2008;11(1):1–11. doi: 10.1375/twin.11.1.1. [DOI] [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, Wong AH, Feldcamp LA, Virtanen C, Halfvarson J, Tysk C, McRae AF, Visscher PM, Montgomery GW, Gottesman II, Martin NG, Petronis A. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41(2):240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- Li J, Burmeister M. Genetical genomics: combining genetics with gene expression analysis. Hum Mol Genet. 2005;14(2):R163–169. doi: 10.1093/hmg/ddi267. [DOI] [PubMed] [Google Scholar]
- Livshits G, Kato BS, Wilson SG, Spector TD. Linkage of genes to total lean body mass in normal women. J Clin Endocrinol Metab. 2007a;92(8):3171–3176. doi: 10.1210/jc.2007-0418. [DOI] [PubMed] [Google Scholar]
- Livshits G, Kato BS, Zhai G, Hart DJ, Hunter D, MacGregor AJ, Williams FM, Spector TD. Genomewide linkage scan of hand osteoarthritis in female twin pairs showing replication of quantitative trait loci on chromosomes 2 and 19. Ann Rheum Dis. 2007b;66(5):623–627. doi: 10.1136/ard.2006.060236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor AJ, Snieder H, Schork NJ, Spector TD. Twins. Novel uses to study complex traits and genetic diseases. Trends Genet. 2000;16(3):131–134. doi: 10.1016/s0168-9525(99)01946-0. [DOI] [PubMed] [Google Scholar]
- Maier S, Olek A. Diabetes: a candidate disease for efficient DNA methylation profiling. J Nutr. 2002;132(8 Suppl):2440S–2443S. doi: 10.1093/jn/132.8.2440S. [DOI] [PubMed] [Google Scholar]
- Martin N. Gene-environment interaction and twin studies. In: Spector TD, Snieder H, MacGregor AJ, editors. Advances in twin and sib-pair analysis. Greenwich Medical Media; London: 2000. pp. 143–150. [Google Scholar]
- Mathers JC. Session 2: personalised nutrition. Epigenomics: a basis for understanding individual differences? Proc Nutr Soc. 2008;67 (4):390–394. doi: 10.1017/S0029665108008744. [DOI] [PubMed] [Google Scholar]
- Monks SA, Leonardson A, Zhu H, Cundiff P, Pietrusiak P, Edwards S, Phillips JW, Sachs A, Schadt EE. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet. 2004;75 (6):1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430(7001):743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor DT, Zhu G, Rao F, Taupenot L, Fung MM, Das M, Mahata SK, Mahata M, Wang L, Zhang K, Greenwood TA, Shih PA, Cockburn MG, Ziegler MG, Stridsberg M, Martin NG, Whitfield JB. Heritability and genome-wide linkage in US and Australian twins identify novel genomic regions controlling chromogranin a: implications for secretion and blood pressure. Circulation. 2008;118(3):247–257. doi: 10.1161/CIRCULATIONAHA.107.709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellis L, Franssen-van Hal NL, Burema J, Keijer J. The intraclass correlation coefficient applied for evaluation of data correction, labeling methods, and rectal biopsy sampling in DNA microarray experiments. Physiol Genomics. 2003;16(1):99–106. doi: 10.1152/physiolgenomics.00111.2003. [DOI] [PubMed] [Google Scholar]
- Peltonen L, McKusick VA. Genomics and medicine. Dissecting human disease in the postgenomic era. Science. 2001;291(5507):1224–1229. doi: 10.1126/science.291.5507.1224. [DOI] [PubMed] [Google Scholar]
- Perola M, Sammalisto S, Hiekkalinna T, et al. Combined genome scans for body stature in 6,602 European twins: evidence for common Caucasian loci. PLoS Genet. 2007;3(6):e97. doi: 10.1371/journal.pgen.0030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A. Epigenetics and twins: three variations on the theme. Trends Genet. 2006;22(7):347–350. doi: 10.1016/j.tig.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Petronis A, Gottesman II, Kan P, Kennedy JL, Basile VS, Paterson AD, Popendikyte V. Monozygotic twins exhibit numerous epigenetic differences: clues to twin discordance? Schizophr Bull. 2003;29(1):169–178. doi: 10.1093/oxfordjournals.schbul.a006988. [DOI] [PubMed] [Google Scholar]
- Pietiläinen KH, Naukkarinen J, Rissanen A, Saharinen J, Ellonen P, Keränen H, Suomalainen A, Götz A, Suortti T, Yki-Järvinen H, Oresic M, Kaprio J, Peltonen L. Global transcript profiles of fat in monozygotic twins discordant for BMI: pathways behind acquired obesity. PLoS Med. 2008;5(3):e51. doi: 10.1371/journal.pmed.0050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20(2):207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H. Heritability of type 2 (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance—a population-based twin study. Diabetologia. 1999;42:139–145. doi: 10.1007/s001250051131. [DOI] [PubMed] [Google Scholar]
- Poulsen P, Esteller M, Vaag A, Fraga MF. The epigenetic basis of twin discordance in age-related diseases. Pediatr Res. 2007;61(5 Pt 2):38R–42R. doi: 10.1203/pdr.0b013e31803c7b98. [DOI] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7(11):862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- Rodenhiser D, Mann M. Epigenetics and human disease: translating basic biology into clinical applications. CMAJ. 2006;174 (3):341–348. doi: 10.1503/cmaj.050774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, Linsley PS, Mao M, Stoughton RB, Friend SH. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Spector TD, MacGregor AJ, Snieder H. Uses of twins in studying the genetics of complex traits. GeneScreen. 2000;1:93–95. [Google Scholar]
- Spielman RS, Ewens WJ. A sibship test for linkage in the presence of association: the sib transmission/disequilibrium test. Am J Hum Genet. 1998;62(2):450–458. doi: 10.1086/301714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Christensen K, Christiansen L, Frederiksen H, Bathum L, Dahlgaard J, Kruse TA. Genetic dissection of gene expression observed in whole blood samples of elderly Danish twins. Hum Genet. 2005;117:267–274. doi: 10.1007/s00439-005-1308-x. [DOI] [PubMed] [Google Scholar]
- Tan Q, Zhao J, Li S, Christiansen L, Kruse TA, Christensen K. Differential and correlation analyses of microarray gene expression data in the CEPH Utah families. Genomics. 2008;92 (2):94–100. doi: 10.1016/j.ygeno.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Coventry WL, James MR, Montgomery GW, Eaves LJ, Martin NG. Use of monozygotic twins to investigate the relationship between 5HTTLPR genotype, depression and stressful life events: an application of Item Response Theory. Novartis Found Symp. 2008;293:48–67. doi: 10.1002/9780470696781.ch4. [DOI] [PubMed] [Google Scholar]
- York TP, Miles MF, Kendler KS, Jackson-Cook C, Bowman ML, Eaves LJ. Epistatic and environmental control of genome-wide gene expression. Twin Res Hum Genet. 2005;8(1):5–15. doi: 10.1375/1832427053435418. [DOI] [PubMed] [Google Scholar]


