Abstract
An efficient uncoupling process is generally considered to have a protective effect on the aging muscle by slowing down its age-related decay. Genetic polymorphisms in the Uncoupling Protein 3 (UCP3) gene, whose product is mainly expressed in skeletal muscle, were suggested to be associated with hand grip (HG) performances in elderly populations. Considering the population specificity of the quality of aging, we aimed to add further support to this evidence by analyzing the association between four SNPs in the UCP3 gene and relative haplotypes in two large cohorts of middle aged (N = 708) and oldest old Danes (N = 908). We found that the variability at rs1685354 and rs11235972 was associated with HG levels both at single and haplotypic level in both cohorts. Furthermore, taking advantage of large cohort and period survival data of the oldest cohort, we tested the association of each SNP with survival at 10 years from the baseline visit. Interestingly, we found that allele A at rs11235972, associated in this cohort with lowest HG scores, influences also the survival patterns, with people carrying this allele showing higher mortality rates. On the whole, our work supports the role of UCP3 gene in functional status and survival at old age.
Keywords: Uncoupling proteins, Hand grip, Longevity
1. Introduction
The reduction of muscle mass in the elderly, known as sarcopenia, becomes so significant after the fifth decade of life that it is widely studied as one of the most important markers of human aging. Furthermore, sarcopenia is regarded as one of the major contributors to frailty (Morley et al., 2001), considering that the loss of muscle mass influences metabolic adaptation, immunological response to disease, capability to respond to environmental stress (Schrager et al., 2003) and so has a relevant impact on the overall homeostasis at old age. Operatively, sarcopenia is measured through the reduction of hand grip strength, which is considered a measure of the overall skeletal muscle mass, observed to gradually and significantly decrease as the individual ages (Doherty, 2003). Hand grip strength is generally considered a very reliable marker of the functional status, as well as one of the most effective predictors of disability and mortality in the elderly (Rantanen et al., 1999; Metter et al., 2002; Nybo et al., 2003; Newman et al., 2006). Furthermore, twin studies demonstrated that about 50% of the observed variation in hand grip strength can be explained by genetic effects (Frederiksen et al., 2002), thus pointing out this phenotype as a useful marker for the identification of genes affecting mid- and late-life physical functioning. Several associations have been reported between hand grip and the variability of genes involved in inflammatory response, myostatin signaling, neuron and bone metabolism (Roth et al., 2001; Arking et al., 2006; De Mars et al., 2007; Walsh et al., 2009; Dato et al., 2010; Windelinckx et al., 2011). Recently the Uncoupling Protein (UCP) genes have been put forward as candidate genes, because of their role in energy metabolism. Of particular interest is the Uncoupling Protein 3 (UCP3) gene, since this is expressed in skeletal muscle where it regulates fatty acid metabolism, oxidative status, and Reactive Oxygen Species (ROS) production. A previous report showed that a functional polymorphism, located in the promoter region of the UCP3 gene (rs1800849), significantly affects hand grip strength in an elderly population from southern Italy (Crocco et al., 2011), suggesting a correlation between the uncoupling process and the regulation of muscle metabolism/catabolism in the elderly.
In addition, recent evidences report as the genetic variability of UCP2, UCP3 and UCP4 affects the individual’s chances of surviving up to a very old age in the same population, thus confirming the relevance of the uncoupling pathway as candidate for human longevity (Rose et al., 2011a, 2011b).
In this study we aimed to add further support to the association of UCP3 variability with hand grip strength, by investigating four Single Nucleotide Polymorphisms (SNPs) and their relevant haplotypes in a different population, composed by two large cohorts of middle aged and oldest old Danes, well characterized by a phenotypic point of view. Moreover, taking advantage of the longitudinal study design of the oldest cohort, we investigated the association of UCP3 variations with survival at old ages.
2. Materials and methods
2.1. Samples
Two Danish samples were analyzed, for a total of 1616 (621 men and 995 women) subjects. The oldest old sample included 908 subjects 93 years old (265 males and 643 females), drawn from the Danish 1905 Cohort, a population-based nationwide survey of all Danish people born in Denmark in 1905 (Nybo et al., 2001, 2003; Christensen et al., 2008). Briefly, 2262 people were initially recruited in 1998, when they were 92-93 years of age. Vital status was ascertained every year, from the first visit to January 1st 2010 or until death whichever came first, resulting in a mean follow-up time for survivors of 11.4 years (range: 11.2-11.6). Information on death for all the cohort members was retrieved from the Danish Central Population Register, which since 1968 keeps a record of all those living in Denmark and which is continuously updated (Pedersen et al., 2006). The participation rate of the survey was 63% and comparisons of demographical characteristics of participants with non participants demonstrated that recruited people were a fairly non-selected group of the 1905 Cohort (Nybo et al., 2001).
The younger sample (mean age 50.5) included 708 subjects (356 males and 352 females) randomly selected from the Study of Middle-Aged Danish Twins (MADT) (Gaist et al., 2000). Recruitment was started in 1998, when 2640 intact twin pairs from 22 consecutive birth years (1931–1952) were chosen via the Danish Central Population Register. The sample analyzed in this study includes only one twin from each twin pair.
Both surveys included multidimensional face-to-face interviews, aimed at the collection of socio-demographic information, assessment of physical, cognitive, depressive status, sensory impairments, medications, self-reported health status and DNA sampling. Permission to collect blood samples and usage of register based information was granted by The Danish National Committee on Biomedical Research Ethics.
2.2. Measurement of hand grip strength
Hand grip (HG) strength was measured by a handheld dynamometer (SMEDLEY’s dynamometer TTM, Tokyo, Japan) while the subject was sitting with the arm close to his/her body. The test was repeated three times with the stronger hand; the maximum of these values was used in the analyses. When a test was not carried out, it was specified whether it was due to physical disabilities or because the subject refused to participate.
2.3. SNP selection and genotyping
For the analysis of UCP3 genetic variability, SNPs were chosen in the genomic region 73,387,985–73,402,778 of chromosome 11, which corresponds to the coding region of UCP3 gene plus 5000 base pairs (bp) upstream and 1000 bps down stream (NCBI B36 assembly), in order to take into consideration possible regulatory elements of the gene. Tagging SNPs covering as much as possible of the known common genetic variation in UCP3 were chosen by analyzing HapMap genotype SNP data in HaploView software (http://www.broadinstitute.org/haploview/haploview, Barrett et al., 2005). In this analysis, the following criteria were adopted: Minimum Allele Frequency (MAF) of at least 5%, ‘pair wise tagging only’, r2 ≥ 0.8, LOD = 3 and a minimum distance between SNPs = 60 bp. The same program has been used for obtaining the graphical overview of Linkage Disequilibrium (LD) (definition of blocks based on confidence interval algorithm, as reported in Gabriel et al., 2002). Pairwise measures of LD between the analyzed loci were quantified by the correlation coefficient r2.
DNA was isolated from blood spot samples using the QIAamp DNA Mini and Micro Kits (Qiagen). Genotyping of the 4 UCP3 SNPs (rs11235972, rs1685354, rs3781907 and rs647126, general characteristics reported in Table 1SM, Supplementary material) was performed as described in Soerensen et al. (2012), by using the Illumina GoldenGate platform (Illumina Inc) and data cleaning was carried out according to Illumina Inc’s recommendations.
2.4. Statistical analyses
For each UCP3 polymorphism, allele frequencies were estimated by gene counting from the observed genotypes. Hardy–Weinberg equilibrium was tested by a Monte-Carlo approach based on 10,000 random allele permutations (Weir, 1996). Standard errors for alleles were computed according to the hypothesis of the multinomial distribution.
2.4.1. Single-locus analysis
In order to assess phenotype–genotype associations we used the Fmax statistics proposed by Lettre et al., 2007. Briefly, this approach allows obtaining a robust association test for quantitative traits within a linear regression framework using the best F statistics (Fmax) observed for different genetic models (additive, dominant and recessive) taking into account also possible covariates’ effects.
In the present study, the Fmax test has been applied to investigate the relationship between UCP3 polymorphisms and the HG performances. The variables age at intake, gender and Body Mass Index (BMI) were used as adjunctive covariates in the regression analysis related to the MADT Cohort, while gender and BMI were included in the case of 1905 Cohort. A permutation approach (10,000 permutations) was used to evaluate the significance of the statistics obtained.
2.4.2. Haplotypic analysis
In order to model the effect of the UCP3 haplotypes on the HG performances, we used a haplotype-based association analysis within the Generalized Linear Model (GLM) framework, that allows to handle ambiguous haplotypes. The haplo.score function of haplo.stat package in R has been used to obtain the GLM-based score statistics for testing both global and individual haplotype effects on the HG performance (Schaid et al., 2002). In this model the effects of the different haplotypes were assessed by assuming an additive model. As in the previous case, the variables age at intake, gender and BMI were used as adjunctive covariates in the MADT Cohort, while gender and BMI were included in the case of 1905 Cohort. A permutation approach has been used to evaluate the significance of the scores obtained.
2.4.3. Survival analysis
In order to evaluate if the detected effects of the analyzed UCP3 polymorphisms on the HG performance might finally result in differential patterns of survival of the different relevant genotypes, we evaluated survival after 10 years from the baseline visit in the 1905 Cohort by using Accelerated Failure Time (AFT) models (Bradburn et al., 2003). Sex, BMI and the four UCP3 SNPs (a SNP each time) were considered as covariates. As reported in the paper by Dato et al. (2011), AFT models can be preferred over the more commonly used Cox regression model (Cox, 1972), particularly when, as in the case of the variable “sex” for the 1905 Cohort, proportional hazard assumptions are not respected (Schoenfeld, 1982, see Supplementary materials). Moreover, with respect to the less intuitive hazard function provided by Cox models, AFT models allow obtaining a clearer explanation of covariate effects, by directly linking the estimation of survival time with the values of covariates: a positive regression coefficient indicates that the covariate is associated with higher survival chances; conversely for negative coefficients. In addition, recent study demonstrated the usefulness of AFT models also in aging research (Swindell, 2009). Then, from the fitted AFT models, sex-adjusted survival curves were obtained, analyzing each SNP independently from each other. Fitting of AFT models was carried out by assuming a Weibull distribution. Subjects alive or immigrated after 10 years from the baseline visit were considered as censored.
Survival analysis of UCP3 haplotypes was carried out by using AFT models as well. In particular, first the probabilities of the individual haplotype combinations (diplotypes) were estimated by using an Expectation Maximization (EM) algorithm implemented in the haplo.stats package of R. Second, N dummy variables, corresponding to the number of different observed haplotypes, were obtained coding the reconstructed diplotype in an additive fashion (the count of a particular haplotype in the diplotype). Finally, AFT models were used for estimating the statistical significance of each haplotype, with the EM – estimated probabilities used as weights. Since this approach underestimates the standard errors of rare haplotypes, they were not included in the model when their frequency was lower than 5%. As in the previous case, sex and BMI were used as adjunctive covariates in the model.
All statistical analyses were carried out using R statistical environment (R Development Core Team, 2010. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria 2011, URL http://www.R-project.org). In particular, survival package was used for survival analyses; haplo.stats and Design packages for haplotypes association analyses (Sinnwell and Schaid, 2009; Harrell and Frank, 2009). A significance level of 0.05 was set for all the tests.
3. Results
Table 1 shows the distribution of the observed genotypes and allele frequencies of UCP3 SNPs in the MADT and 1905 Cohorts. For each SNP no significant departure from Hardy–Weinberg equilibrium was observed in both samples.
Table 1.
Absolute (Abs) and relative (Rel) genotypic and allelic frequencies ± standard errors (SE) of UCP3 gene polymorphisms in the MADT and 1905 Cohorts.
| dbSNP ID | Genotype/allele | MADT Cohort |
1905 Cohort |
||
|---|---|---|---|---|---|
| Abs (Rel ± SE) | p-value (HWE) | Abs (Rel ± SE) | p-value (HWE) | ||
| rs647126 | G/G | 267 (0.29 ± 0.015) | 0.141 | 217 (0.31 ± 0.017) | 0.447 |
| G/A | 471 (0.52 ± 0.017) | 359 (0.51 ± 0.019) | |||
| A/A | 170 (0.19 ± 0.013) | 132 (0.19 ± 0.015) | |||
| G | 1005 (0.55 ± 0.012) | 793 (0.56 ± 0.013) | |||
| A | 811 (0.45) | 623 (0.44) | |||
| rs1685354 | A/A | 494 (0.54 ± 0.017) | 0.270 | 381 (0.54 ± 0.019) | 0.244 |
| A/G | 343 (0.38 ± 0.016) | 285 (0.40 ± 0.018) | |||
| G/G | 71 (0.08 ± 0.009) | 42 (0.06 ± 0.009) | |||
| A | 1331 (0.73 ± 0.010v | 1047 (0.74 ± 0.012) | |||
| G | 485 (0.27) | 369 (0.26) | |||
| rs3781907 | A/A | 424 (0.47 ± 0.017) | 0.538 | 330 (0.47 ± 0.019) | 0.602 |
| A/G | 399 (0.44 ± 0.016) | 311 (0.44 ± 0.019) | |||
| G/G | 85 (0.09 ± 0.010) | 67 (0.09 ± 0.011) | |||
| A | 1247 (0.69 ± 0.011) | 971 (0.69 ± 0.012) | |||
| G | 569 (0.31) | 445 (0.31) | |||
| rs11235972 | G/G | 475 (0.52 ± 0.017) | 0.614 | 376 (0.53 ± 0.019) | 0.698 |
| G/A | 368 (0.41 ± 0.016) | 283 (0.40 ± 0.018) | |||
| A/A | 65 (0.07 ± 0.009) | 49 (0.07 ± 0.010) | |||
| G | 1318 (0.73 ± 0.010) | 1035 (0.73 ± 0.012) | |||
| A | 498 (0.27) | 381 (0.27) | |||
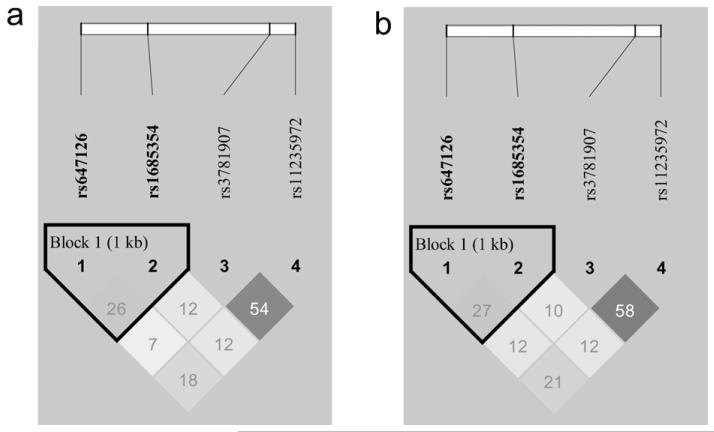
In order to model the effect of the UCP3 haplotypes on the HG performance, firstly we evaluated the degree of LD between pairs of loci. We found a modest degree of LD between the analyzed polymorphisms. In particular, in the 1905 Cohort, we found LD between rs647126 and rs1685354 (r2 = 0.271), which in turn was also linked to the rs3781907 (r2 = 0.102), which in turn was linked to the rs11235972 (r2 = 0.587). The same LD pattern was observed in the MADT Cohort. In fact, in this sample we found LD between rs647126 and rs1685354 (r2 = 0.263), which in turn was also linked to the rs3781907 (r2 = 0.121), which in turn was linked to the rs11235972 (r2 = 0.543). The LD structures of UCP3 SNPs in the two analyzed samples are shown in Fig. 1(a and b).
Fig. 1.
LD structures of UCP3 SNPs in the 1905 (a) and MADT (B) Cohort.
3.1. Single-locus analysis in the MADT Cohort
Table 2 reports for each UCP3 SNP the test statistics for the different genetic models (dominant, additive and recessive) obtained in the MADT Cohort with the relevant p-values.
Table 2.
Results of the Fmax association tests between the analyzed polymorphisms (risk allele in parenthesis) and the hand grip performance in the MADT Cohort.
| SNP | Additive model |
Dominant model |
Recessive model |
F max | p c | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| F a | β | p β b | F | β | pβ | F | β | pβ | |||
| rs647126 (A) | 488.75 | 0.740 | 0.035 | 488.78 | 1.116 | 0.034 | 486.01 | 0.773 | 0.215 | 488.78 | 0.075 |
| rs1685354 (G) | 484.85 | −0.223 | 0.578 | 484.78 | 0.238 | 0.626 | 490.28 | −2.528 | 0.014 | 490.28 | 0.029 |
| rs3781907 (G) | 486.78 | −0.577 | 0.124 | 485.87 | −0.579 | 0.237 | 486.39 | −1.159 | 0.163 | 486.78 | 0.230 |
| rs11235972 (A) | 485.37 | −0.364 | 0.354 | 484.87 | −0.281 | 0.566 | 485.79 | −1.099 | 0.253 | 485.79 | 0.451 |
F-statistics under the additive, dominant and recessive models.
p-values of the regression coefficients associated with the analyzed polymorphism under the three genetic models.
p-value adjusted for multiple testing of different genetic models (based on 10,000 random permutations), obtained using the approach proposed by Lettre et al., 2007 (see Section 2).
The table clearly shows that the variability of the rs1685354 is significantly associated to the HG performance (p = 0.029), with subjects carrying GG genotype (recessive model) showing lower HG scores than the allele A carriers. A marginal effect for rs647126 was also detected, for which carrying the A allele (dominant model) significantly increased the HG scores (p = 0.075).
3.2. Haplotypic analysis in the MADT Cohort
We evaluated the effect of the UCP3 haplotypes on the HG performance by applying a GLM based algorithm. Table 3 reports the estimation of the haplotype frequencies of the combined UCP3 SNPs and their association with the HG performance in the MADT Cohort.
Table 3.
Estimation of haplotype frequencies in the UCP3 SNPs and association with hand grip performance in MADT Cohort.
| Haplotype | rs647126 | rs1685354 | rs3781907 | rs11235972 | Freq.a | Score | p b |
|---|---|---|---|---|---|---|---|
| GAGA | G | A | G | A | 0.21491 | −1.33136 | 0.182 |
| AAGG | A | A | G | G | 0.05587 | −1.23035 | 0.208 |
| GAGG | G | A | G | G | 0.01512 | −0.60371 | 0.558 |
| GGAG | G | G | A | G | 0.25016 | −0.60117 | 0.549 |
| GAAG | G | A | A | G | 0.04281 | −0.58967 | 0.546 |
| GAAA | G | A | A | A | 0.02988 | −0.44337 | 0.655 |
| GGGG | G | G | G | G | 0.00715 | 0.08633 | 0.930 |
| AAGA | A | A | G | A | 0.01973 | 1.23184 | 0.218 |
| AAAG | A | A | A | G | 0.35878 | 2.33717 | 0.019 |
Estimated haplotype frequency.
Monte-Carlo p-value from 106 replications.
From this GLM model, considering age, gender and BMI as covariates, the global score statistic in the MADT Cohort was 9.54 (p = 0.410). The analysis of the scores associated with each haplotype indicated that the haplotype AAAG was significantly associated with higher HG scores (p = 0.019). Consistently with the single locus analysis, we found that SNP combinations including significant alleles resulted in significantly different haplotypic effects on the HG performances.
3.3. Single-locus analysis in the 1905 Cohort
Table 4 reports for each UCP3 SNP the test statistics for the different genetic models with the relevant p-values as obtained in the 1905 Cohort.
Table 4.
Results of the Fmax association tests with respect to the analyzed polymorphisms (risk allele in parenthesis) in the 1905 Cohort.
| SNP | Additive model |
Dominant model |
Recessive model |
F max | p c | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| F a | β | pβ b | F | β | pβ | F | β | pβ | |||
| rs647126 (A) | 201.54 | 0.153 | 0.542 | 201.44 | −0.161 | 0.670 | 202.71 | 0.692 | 0.116 | 202.71 | 0.225 |
| rs1685354 (G) | 204.54 | 0.648 | 0.016 | 203.62 | 0.699 | 0.043 | 203.41 | 1.236 | 0.054 | 204.55 | 0.037 |
| rs3781907 (G) | 202.98 | −0.454 | 0.086 | 203.28 | −0.643 | 0.062 | 201.55 | −0.370 | 0.531 | 203.28 | 0.120 |
| rs11235972 (A) | 206.60 | −0.844 | 0.002 | 207.47 | −1.139 | < 0.001 | 201.92 | −0.685 | 0.305 | 207.47 | 0.002 |
F-statistics under the additive, dominant and recessive models.
p-values of the regression coefficients associated with the analyzed polymorphism under the three .genetic models.
p-value adjusted for multiple testing of different genetic models (based on 10,000 random permutations), obtained using the approach proposed by Lettre et al. (2007) (see Section 2).
The table clearly shows that the rs11235972 and rs1685354 variations had significant effects on the HG performance. In fact, as it regards for the rs11235972 variation, subjects carrying the A allele (dominant model) showed lower HG scores than those homozygotes for the G allele (p < 0.001), whereas for the rs1685354 variation, the comparison of the F statistics makes the additive model the most plausible, indicating that carrying the G allele positively influenced the HG scores in an additive manner.
3.4. Haplotypic analysis in the 1905 Cohort
Table 5 reports the estimation of the haplotype frequencies of the combined UCP3 SNPs and their association with the HG performance in the 1905 Cohort.
Table 5.
Estimation of haplotype frequencies in the UCP3 SNPs and association with hand grip performance in 1905 Cohort.
| Haplotype | rs647126 | rs1685354 | rs3781907 | rs11235972 | Freq.a | Score | p b |
|---|---|---|---|---|---|---|---|
| GAGA | G | A | G | A | 0.225 | −2.385 | 0.008 |
| GAAA | G | A | A | A | 0.024 | −1.893 | 0.068 |
| AAAA | A | A | A | A | 0.006 | −0.780 | 0.455 |
| GAAG | G | A | A | G | 0.032 | −0.644 | 0.515 |
| AAGA | A | A | G | A | 0.016 | −0.255 | 0.794 |
| GGGA | G | G | G | A | 0.003 | 0.032 | 0.972 |
| GAGG | G | A | G | G | 0.012 | 0.252 | 0.803 |
| AAGG | A | A | G | G | 0.042 | 0.459 | 0.640 |
| AAAG | A | A | A | G | 0.377 | 0.567 | 0.575 |
| AGAG | A | G | A | G | 0.003 | 1.358 | 0.156 |
| GGAG | G | G | A | G | 0.246 | 1.974 | 0.051 |
| GGGG | G | G | G | G | 0.012 | 2.396 | 0.029 |
Estimated haplotype frequency.
Monte-Carlo p-value from 106 replications.
From this GLM model, considering gender and BMI as covariates, the global score statistic was 19.76 (p = 0.090). The data reported in Table 5 indicates that haplotypes GAGA, GAAA, GGAG and GGGG were significantly associated with the HG performance in this sample. In particular, haplotypes GAGA (p = 0.008) and GAAA (p = 0.068) points to be associated with lower HG scores, while haplotypes GGAG (p = 0.051) and GGGG (p = 0.029) were associated to higher scores. Also in this case we found that SNP combinations directly including alleles showing significant single-locus associations resulted in significantly different haplotype effects on the HG performances.
3.5. Survival analysis
Table 6 reports the estimated parameters of the fitted AFT models in the 1905 Cohort, including sex and the different SNPs as covariates.
Table 6.
Estimated coefficients of the fitted Accelerated Failure Time (AFT) model in 1905-Cohort including sex and one SNP (among rs11235972, rs1685354, rs3781907 and rs647126) as covariates. Standard errors (SE), Wald test statistics (z) and significance (p-value) are also reported.
| Coefficient | SE | z | p-value | |
|---|---|---|---|---|
| rs11235972 | ||||
| Intercept | 3.994 | 0.038 | 104.500 | <0.001 |
| Sex = Male | −0.171 | 0.057 | −3.020 | 0.002 |
| SNP | −0.090 | 0.041 | 2.190 | 0.029 |
| Log(scale) | −0.269 | 0.028 | −9.750 | <0.001 |
| rs1685354 | ||||
| Intercept | 3.954 | 0.039 | 102.640 | <0.001 |
| Sex = Male | −0.176 | 0.057 | −3.105 | 0.002 |
| SNP | −0.013 | 0.041 | −0.300 | 0.762 |
| Log(scale) | −0.267 | 0.028 | −9.680 | <0.001 |
| rs3781907 | ||||
| Intercept | 3.994 | 0.040 | 100.480 | <0.001 |
| Sex = Male | −0.169 | 0.057 | −2.980 | 0.003 |
| SNP | −0.080 | 0.041 | −1.960 | 0.050 |
| Log(scale) | −0.269 | 0.028 | −9.750 | <0.001 |
| rs647126 | ||||
| Intercept | 3.921 | 0.047 | 83.870 | <0.001 |
| Sex = Male | −0.173 | 0.057 | −3.040 | 0.002 |
| SNP | 0.028 | 0.037 | 0.760 | 0.447 |
| Log(scale) | −0.268 | 0.028 | −9.680 | <0.001 |
In Table 6 a positive coefficient indicates that an increment of the respective factor is associated with higher survival chances; conversely for negative coefficients. After 10 years, a significant effect on survival chances was observed for rs11235972 and rs3781907 (p = 0.029 and p = 0.050, respectively). In particular, since in these models each SNP was coded as 0,1 or 2 according to the count of the minor (less frequent) allele in the genotype, carriers of the A allele at rs11235972 variation and carriers of the G allele at rs3781907 (that are the less frequent alleles for these SNPs) showed a reduced survival chance (coefficient = −0.090 and −0.080, respectively). In addition, male sex was associated with a reduced survival probability in all four models (p < 0.01). Interestingly, the rs11235972-A variation was also associated with a decreased HG performance in the previous analysis in this cohort. As for the rs3781907, despite the borderline association with HG performance (p = 0.062, see Table 4), it significantly influenced survival in this cohort.
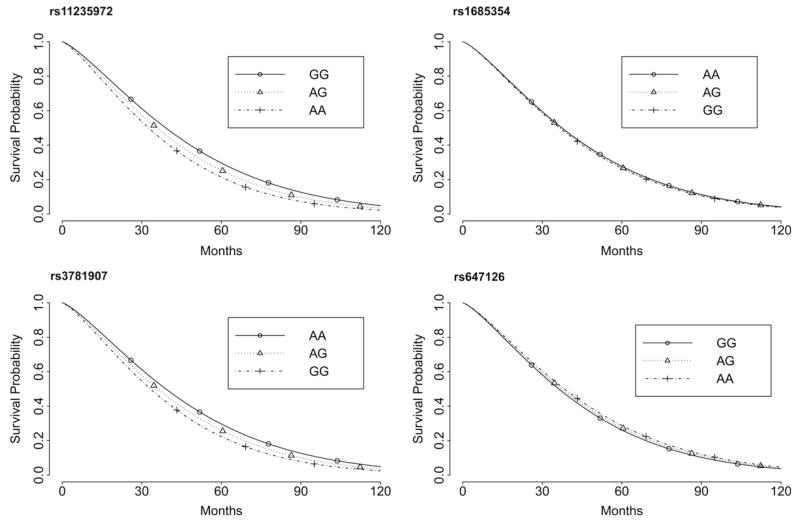
For each polymorphism, the survival functions obtained from the fitted AFT models are shown in Fig. 2.
Fig. 2.
Panel showing the survival functions of the different genotypes at the four analyzed UCP3 SNPS in the 1905 Cohort sample, as obtained from the fitted AFT models.
Considering the previously demonstrated LD between SNPs and the haplotypic effect on the HG performances, we then carried out a haplotype-based survival analysis, as previously described in Section 2. No significant association was found between any UCP3 haplotype and survival in the Danish 1905 sample (data not shown).
4. Discussion
Several studies have hypothesized a correlation between the uncoupling process and the regulation of muscle metabolism in the elderly (Cortright et al., 1999; Nabben and Hoeks, 2008). Considering the association demonstrated between UCP3 genetic variability and HG strength in a southern Italian population (Crocco et al., 2011), and taking into account the different quality of aging demonstrated between southern and northern Europeans (Jeune et al., 2006; Andersen-Ranberg et al., 2009), in this paper we wanted to further test this association in a larger sample of 1616 individuals (908 nonagenarians and 708 younger subjects), belonging to a northern European population, the Danes. To this aim, we carried out an association analysis between four common variants in the UCP3 gene and HG performance, both at single and haplotypic levels. We found that UCP3 markers previously associated with age-related phenotypes (diabetes, obesity, serum lipid levels and BMI) (van Abeelen et al., 2008; Salopuro et al., 2009) significantly influenced also the HG performance in the analyzed cohorts. In particular, the minor allele of rs1685354 had significant effects on the HG performance both in the younger and in the older sample, although in opposite sense. In nonagenarians, the less frequent allele of rs11235972 was negatively associated with HG, while a marginal positive effect was observed for rs647126 in the younger sample.
With the exception of rs11235972 in the 1905 Cohort, no other association hold a Bonferroni’s correction (p < 0.0125). However, this correction seems more suitable when searching for associations without a priori hypotheses but too conservative when assessing a specific research question, as in our case, with a candidate gene chosen for its relevance with respect to the analyzed phenotype (Perneger, 1998). Furthermore, because this work confirmed a previous association between UCP3 variability and hand grip levels at old age, we believe that also borderline and trend results should be taken into account.
Although the candidate SNPs analyzed in this study are not functional, data from the HapMap consortium report a complete LD between the functional variant rs1800849, influencing the HG strength in a southern Italian elderly population (Crocco et al., 2011) and rs11235972 (r2 = 1). A less strong LD exists between rs1800849 and the other SNPs tested (r2 = 0.5 with rs3781907, r2 = 0.23 with rs647126, r2 = 0.22 with rs1685354). Consequently, we may argue that the association observed in this work can be due to a functional linkage of our polymorphisms with the rs1800849, or they could both be in LD with the actual causal variant.
The observed association of rs1685354 with a lower HG in middle-aged individuals and a higher HG in nonagenarians could suggest a possible antagonistic pleiotropic effect of this polymorphism (or the associated causal variant) on the phenotype. In younger individuals, a more efficient uncoupling process, promoting the dissipation of proton gradient, reduces ATP level and so energy availability, which may be associated with a lower muscular functioning in an energy-demanding organism; on the contrary, in a different metabolic scenario, possibly characterized by a loss of muscle mass, a higher expression of uncoupling proteins may replace dysfunctional mitochondria by stimulating the mitochondrial biogenesis, as suggested for UCP2 by Wu et al. (1999), and so represent an adaptation of the aging organism to the new physiological status. In support of this hypothesis is the evidence coming from studies on the regulation of UCP proteins, where it emerges that their activity is modified when facing different physiological and nutritional stimuli (Ricquier et al., 2000). Furthermore, the observation of a correlation between genetic variants harboring higher gene expression of UCP genes with a better functional performance in the elderly, may be due to the fact that a more efficient uncoupling reduce the ROS production, thus preventing or reducing the age related decay of the aging muscle (Nabben et al., 2008; Mookerjee et al., 2010).
When investigating the association of UCP3 haplotypes (consisting of the four SNPs) with HG, in line with the single locus analysis, we found that some haplotypes show different effects on HG performances in both cohorts. In particular, the haplotype AAAG was significantly associated with higher HG scores in MADT Cohort, while in the 1905 sample haplotypes GAGA and GAAA were associated with lower, and GGAG and GGGG were associated with higher scores. Both the single and haplotypic associations remain significant after correction for multiple testing, therefore they can be considered statistically robust. Accordingly, genetic variants being associated with lower HG at the single-SNP level also show lower HG at the haplotypic level.
Considering the importance of sarcopenia and loss of muscle mass in the aging process, we tested the possible association of UCP3 single variants with survival in the 1905 Cohort by using AFT models. Taking advantage of the large cohort and follow-up period available in our dataset, we found that UCP3 SNPs were also associated with survival at 10 years in this cohort. Consistently with the previous association results, rs11235972, which negatively influenced HG scores, was also associated with a lower survival chance, with rs11235972-A carriers showing higher mortality than non carriers. A borderline significant association with survival was also observed for rs3781907, even if the variant did not show any significant association with HG levels. On the contrary, despite rs1685354 had significant effects on the HG performance, no significant association was detected in terms of survival. A comparison with the classical Cox regression proportional hazard models, carried out by using each SNP and sex as covariates, confirmed these results (Table 2SM).
Finally, taking into consideration the modest LD found between SNPs analyzed in this study, we tested the effect of their interaction with respect to survival. Despite the association previously found with HG phenotypes, we could not demonstrate any significant association with survival for UCP3 haplotypes, probably because of the level of statistical correction needed, and the scarce frequency of some haplotypes in our sample. These considerations, however, did not affect the previous associations found between single polymorphisms with survival in this cohort, rather strengthen the hypothesis of an association with another functional variant in the UCP3 gene region.
4.1. Conclusions
On the whole, our work confirms the association between UCP3 variability and muscular strength, measured through HG, also in the Danish population. Despite the differences in HG performances found in nonagenarians and 50+ individuals across Europe (Jeune et al., 2006; Andersen-Ranberg et al., 2009), UCP3 genetic polymorphisms could be considered a common and informative target for better evaluating the muscular functionality and then the quality of aging in both northern and southern European populations. Furthermore, the different associations found between UCP3 polymorphism in individuals belonging to different age range may be due to the fact that UCP proteins act in a complex pathway, where genetic variability may also interact with several transcription/replication factors directly regulating mitochondrial metabolism involved in muscular functioning: the coordination of these factors into a program responsive to the environment is complex and may represent one of the mechanism used by the aging organism to adapt itself to the new physiological scenario (Franceschi et al., 2000).
In line with this hypothesis are the recent data emphasizing the role of UCP variations on the metabolic efficiency (Mookerjee et al., 2010), which underline as the impact of UCPs’ gene variation on human longevity can be due to the coordination of their different functions rather than the sole well known effect on oxidative stress.
In any case, our work adds a piece of evidence to the hypothesis that the variability of the UCP3 gene, involved in the control of energetic metabolism, can influence the survival at old age. In general, the association of UCPs genetic variability with survival, if confirmed in different population by future replication studies, suggests a crucial role of their metabolic pathway in human longevity.
Supplementary Material
Acknowledgments
This study was supported by the Max-Planck Institute for Demographic Research (Rostock, Germany), the National Institute on Aging (P01 AG08761), The Novo Nordisk Foundation, the Aase and Ejnar Danielsen Foundation, the Augustinus Foundation, the Brødrene Hartmann Foundation, the King Christian the 10th Foundation and the Einer Willumsens Mindelegat Foundation. The Danish Aging Research Center is supported by a grant from the VELUX Foundation. Researchers from University of Calabria were supported by the European Union Seventh Framework Programme (FP7/2007-2011) under grant agreement no. 259679 and from “Fondi di Ateneo”.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.mad.2012.06.004.
References
- Andersen-Ranberg K, Petersen I, Frederiksen H, Mackenbach J, Christensen K. Cross-national differences in grip strength among 50+ year-old Europeans: results from the SHARE study. European Journal of Ageing. 2009;6(3):227–236. doi: 10.1007/s10433-009-0128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arking DE, Fallin DM, Fried LP, Li T, Beamer BA, Xue QL, Chakravarti A, Walston J. Variation in the ciliary neurotrophic factor gene and muscle strength in older Caucasian women. Journal of the American Geriatrics Society. 2006;54(5):823–826. doi: 10.1111/j.1532-5415.2006.00693.x. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bradburn MJ, Clark TG, Love SB, Altman DG. Survival analysis part II: multivariate data analysis – an introduction to concepts and methods. British Journal of Cancer. 2003;89(3):431–436. doi: 10.1038/sj.bjc.6601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, McGue M, Petersen I, Jeune B, Vaupel JW. Exceptional longevity does not result in excessive levels of disability. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(36):13274–13279. doi: 10.1073/pnas.0804931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortright RN, Zheng D, Jones JP, Fluckey JD, Di Carlo SE, Grujic D, Lowell BB, Dohm GL. Regulation of skeletal muscle UCP-2 and UCP-3 gene expression by exercise and denervation. American Journal of Physiology. 1999;276(1 Pt 1):E217–E221. doi: 10.1152/ajpendo.1999.276.1.E217. [DOI] [PubMed] [Google Scholar]
- Cox DR. Regression models and life-tables. Journal of the Royal Statistical Society. 1972;34:187–220. [Google Scholar]
- Crocco P, Montesanto A, Passarino G, Rose G. A common polymorphism in the UCP3 promoter influences hand grip strength in elderly people. Biogerontology. 2011;12(3):265–271. doi: 10.1007/s10522-011-9321-z. [DOI] [PubMed] [Google Scholar]
- Dato S, Krabbe KS, Thinggaard M, Pedersen BK, Christensen K, Bruunsgaard H, Christiansen L. Commonly studied polymorphisms in inflammatory cytokine genes show only minor effects on mortality and related risk factors in zonagenarians. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences. 2010;65(3):225–235. doi: 10.1093/gerona/glp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dato S, Montesanto A, Lagani V, Jeune B, Christensen K, Passarino G. Frailty phenotypes in the elderly based on cluster analysis: a longitudinal study of two Danish cohorts Evidence for a genetic influence on frailty. Age (Dordr) 2011 May; doi: 10.1007/s11357-011-9257-x. http://dx.doi.org/10.1007/s11357-011-9257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mars G, Windelinckx A, Beunen G, Delecluse C, Lefevre J, Thomis MA. Polymorphisms in the CNTF and CNTF receptor genes are associated with muscle strength in men and women. Journal of Applied Physiology. 2007;102(5):1824–1831. doi: 10.1152/japplphysiol.00692.2006. [DOI] [PubMed] [Google Scholar]
- Doherty TJ. Aging and sarcopenia. Journal of Applied Physiology. 2003;95(4):1717–1727. doi: 10.1152/japplphysiol.00347.2003. [DOI] [PubMed] [Google Scholar]
- Franceschi C, Valensin S, Bonafè M, Paolisso G, Yashin AI, Monti D, De Benedictis G. The network and the remodeling theories of aging: historical background and new perspectives. Experimental Gerontology. 2000;35:879–896. doi: 10.1016/s0531-5565(00)00172-8. [DOI] [PubMed] [Google Scholar]
- Frederiksen H, Gaist D, Petersen HC, Hjelmborg J, McGue M, Vaupel JW, Christensen K. Hand grip strength: a phenotype suitable for identifying genetic variants affecting mid- and late-life physical functioning. Genetic Epidemiology. 2002;23(2):110–122. doi: 10.1002/gepi.1127. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, De Felice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gaist D, Bathum L, Skytthe A, Jensen TK, McGue M, Vaupel JW, Christensen K. Strength and anthropometric measures in identical and fraternal twins: no evidence of masculinization of females with male co-twins. Epidemiology. 2000;11(3):340–343. doi: 10.1097/00001648-200005000-00020. [DOI] [PubMed] [Google Scholar]
- Harrell FE., Jr. Design: Design Package. R Package Version 2.3-0. 2009 [Google Scholar]
- Jeune B, Skytthe A, Cournil A, Greco V, Gampe J, Berardelli M, Andersen-Ranberg K, Passarino G, Debenedictis G, Robine JM. Handgrip strength among nonagenarians and centenarians in three European regions. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences. 2006;61(7):707–712. doi: 10.1093/gerona/61.7.707. [DOI] [PubMed] [Google Scholar]
- Lettre G, Lange C, Hirschhorn JN. Genetic model testing and statistical power in population-based association studies of quantitative traits. Genetic Epidemiology. 2007;31(4):358–362. doi: 10.1002/gepi.20217. [DOI] [PubMed] [Google Scholar]
- Metter EJ, Talbot LA, Schrager M, Conwit R. Skeletal muscle strength as a predictor of all-cause mortality in healthy men. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences. 2002;57(10):B359–B365. doi: 10.1093/gerona/57.10.b359. [DOI] [PubMed] [Google Scholar]
- Mookerjee SA, Divakaruni AS, Jastroch M, Brand MD. Mitochondrial uncoupling and lifespan. Mechanisms of Ageing and Development. 2010;131(7-8):463–472. doi: 10.1016/j.mad.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JE, Baumgartner RN, Roubenoff R, Mayer J, Nair KS. Sarcopenia. Journal of Laboratory and Clinical Medicine. 2001;137(4):231–243. doi: 10.1067/mlc.2001.113504. [DOI] [PubMed] [Google Scholar]
- Nabben M, Hoeks J, Briedé JJ, Glatz JF, Moonen-Kornips E, Hesselink MK, Schrauwen P. The effect of UCP3 overexpression on mitochondrial ROS production in skeletal muscle of young versus aged mice. FEBS Letters. 2008;582(30):4147–4152. doi: 10.1016/j.febslet.2008.11.016. [DOI] [PubMed] [Google Scholar]
- Nabben M, Hoeks J. Mitochondrial uncoupling protein 3 and its role in cardiac- and skeletal muscle metabolism. Physiology and Behavior. 2008;94(2):259–269. doi: 10.1016/j.physbeh.2007.11.039. [DOI] [PubMed] [Google Scholar]
- Newman AB, Kupelian V, Visser M, Simonsick EM, Goodpaster BH, Kritchevsky SB, Tylavsky FA, Rubin SM, Harris TB. Strength, but not muscle mass, is associated with mortality in the health, aging and body composition study cohort. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences. 2006;61(1):72–77. doi: 10.1093/gerona/61.1.72. [DOI] [PubMed] [Google Scholar]
- Nybo H, Gaist D, Jeune B, Bathum L, McGue M, Vaupel JW, Christensen K. The Danish 1905 Cohort: a genetic-epidemiological nationwide survey. Journal of Aging and Health. 2001;13(1):32–46. doi: 10.1177/089826430101300102. [DOI] [PubMed] [Google Scholar]
- Nybo H, Petersen HC, Gaist D, Jeune B, Andersen K, McGue M, Vaupel JW, Christensen K. Predictors of mortality in 2,249 nonagenarians – the Danish 1905-Cohort Survey. Journal of the American Geriatrics Society. 2003;51(10):1365–1373. doi: 10.1046/j.1532-5415.2003.51453.x. [DOI] [PubMed] [Google Scholar]
- Pedersen CB, Gøtzsche H, Møller JO, Mortensen PB. The Danish Civil Registration System. A cohort of eight million persons. Danish Medical Bulletin. 2006;53(4):441–449. [PubMed] [Google Scholar]
- Perneger TV. What’s wrong with Bonferroni adjustments. British Medical Journal. 1998;316(7139):1236–1238. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantanen T, Guralnik JM, Foley D, Masaki K, Leveille S, Curb JD, White L. Midlife hand grip strength as a predictor of old age disability. Journal of the American Medical Association. 1999;281(6):558–560. doi: 10.1001/jama.281.6.558. [DOI] [PubMed] [Google Scholar]
- Ricquier D, Miroux B, Larose M, Cassard-Doulcier AM, Bouillaud F. Endocrine regulation of uncoupling proteins and energy expenditure. International Journal of Obesity and Related Metabolic Disorders. 2000;24:86–88. doi: 10.1038/sj.ijo.0801286. [DOI] [PubMed] [Google Scholar]
- Rose G, Crocco P, De Rango F, Montesanto A, Passarino G. Further support to the uncoupling-to-survive theory: the genetic variation of human UCP genes is associated with longevity. PLoS One. 2011a;6(12):e29650. doi: 10.1371/journal.pone.0029650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G, Crocco P, D’Aquila P, Montesanto A, Bellizzi D, Passarino G. Two variants located in the upstream enhancer region of human UCP1 gene affect gene expression and are correlated with human longevity. Experimental Gerontology. 2011b;46(11):897–904. doi: 10.1016/j.exger.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Roth SM, Schrager MA, Ferrell RE, Riechman SE, Metter EJ, Lynch NA, Lindle RS, Hurley BF. CNTF genotype is associated with muscular strength and quality in humans across the adult age span. Journal of Applied Physiology. 2001;90(4):1205–1210. doi: 10.1152/jappl.2001.90.4.1205. [DOI] [PubMed] [Google Scholar]
- Salopuro T, Pulkkinen L, Lindström J, Kolehmainen M, Tolppanen AM, Eriksson JG, Valle TT, Aunola S, Ilanne-Parikka P, Keinänen-Kiukaanniemi S, Tuomilehto J, Laakso M, Uusitupa M. Variation in the UCP2 and UCP3 genes associates with abdominal obesity and serum lipids: the Finnish Diabetes Prevention Study. BMC Medical Genetics. 2009;10:94. doi: 10.1186/1471-2350-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. American Journal of Human Genetics. 2002;70(2):425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld D. Partial residuals for the proportional hazards regression model. Biometrika. 1982;69:239–241. [Google Scholar]
- Schrager M, Bandinelli S, Maggi S, Ferrucci L. Sarcopenia: twenty open questions for a research agenda. Basic and Applied Myology. 2003;13(4):203–208. [Google Scholar]
- Sinnwell JP, Schaid DJ. haplo.stats: Statistical Analysis of Haplotypes with Traits and Covariates when Linkage Phase is Ambiguous. R Package Version 1.4.4. 2009. [Google Scholar]
- Soerensen M, Dato S, Tan Q, Thinggaard M, Kleindorp R, Beekman M, Suchiman HED, Jacobsen R, McGue M, Stevnsner T, Bohr VA, de Craen AJ, Westendorp RGJ, Schreiber S, Slagboom E, Nebel A, Christensen K, Christiansen L. Common genetic variation in the GH-IGF-1-insulin signalling DNA damage signalling and repair and pro-antioxidant pathways is associated with human longevity. Experimental Gerontology. 2012;47(5):379–387. doi: 10.1016/j.exger.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR. Accelerated failure time models provide a useful statistical framework for aging research. Experimental Gerontology. 2009;44:190–200. doi: 10.1016/j.exger.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Abeelen AF, de Krom M, Hendriks J, Grobbee DE, Adan RA, van der Schouw YT. Variations in the uncoupling protein-3 gene are associated with specific obesity phenotypes. European Journal of Endocrinology. 2008;158(5):669–676. doi: 10.1530/EJE-07-0834. Erratum in: European Journal of Endocrinology 2008, 159(1), 159. [DOI] [PubMed] [Google Scholar]
- Walsh S, Kelsey BK, Angelopoulos TJ, Clarkson PM, Gordon PM, Moyna NM, Visich PS, Zoeller RF, Seip RL, Bilbie S, Thompson PD, Hoffman EP, Price TB, Devaney JM, Pescatello LS. CNTF 1357 G -> A polymorphism and the muscle strength response to resistance training. Journal of Applied Physiology. 2009;107(4):1235–1240. doi: 10.1152/japplphysiol.90835.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sinauer Associates; Sunderland: 1996. [Google Scholar]
- Windelinckx A, De Mars G, Huygens W, Peeters MW, Vincent B, Wijmenga C, Lambrechts D, Delecluse C, Roth SM, Metter EJ, Ferrucci L, Aerssens J, Vlietinck R, Beunen GP, Thomis MA. Comprehensive fine mapping of chr12q12-14 and follow-up replication identify activin receptor 1B (ACVR1B) as a muscle strength gene. European Journal of Human Genetics. 2011;19(2):208–215. doi: 10.1038/ejhg.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.