Abstract
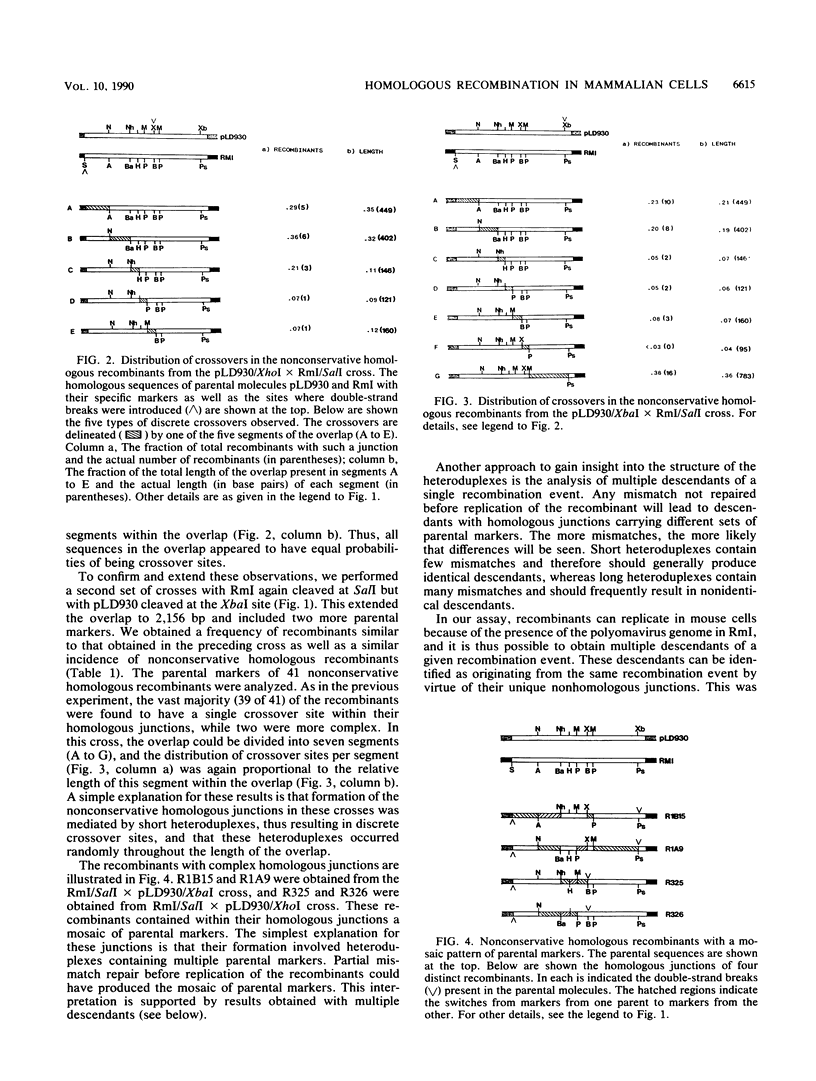
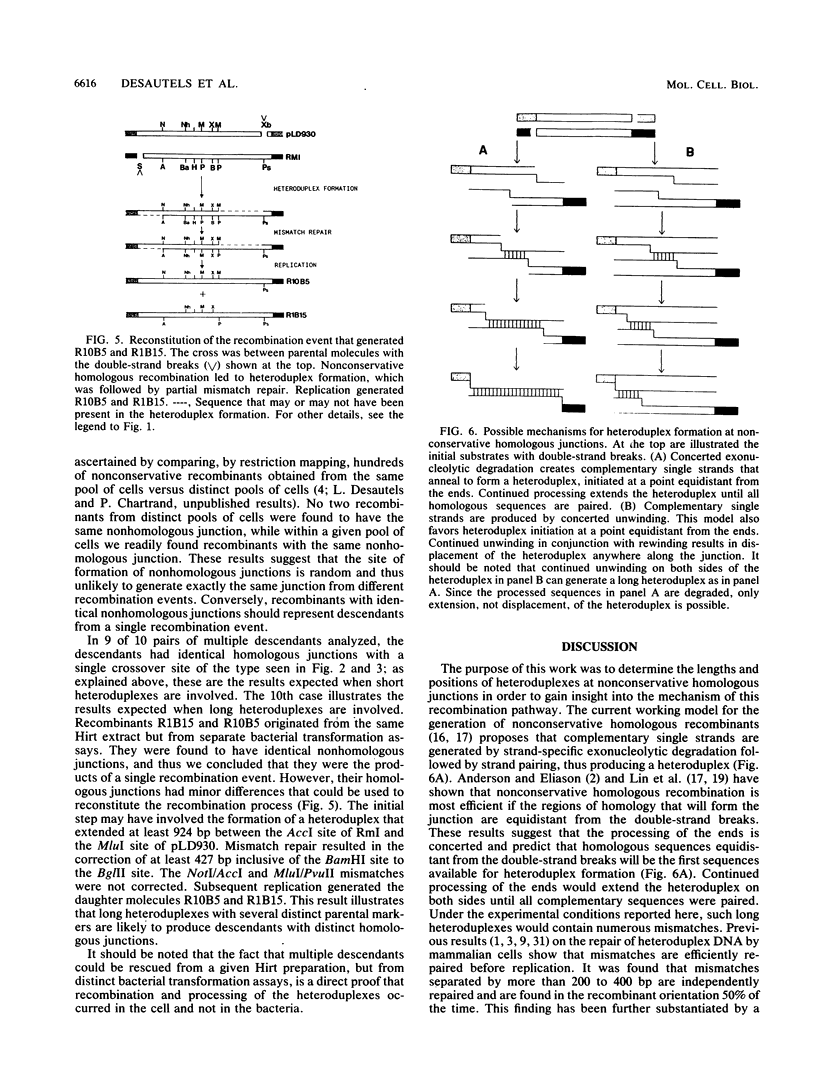
Homologous recombination in mammalian cells between extrachromosomal molecules, as well as between episomes and chromosomes, can be mediated by a nonconservative mechanism. It has been proposed that the key steps in this process are the generation (by double-strand cleavage) of overlapping homologous ends, the creation of complementary single-strand ends (either by strand-specific exonuclease degradation or by unwinding of the DNA helix), and finally the creation of heteroduplex DNA by the annealing of the single-strand ends. We have analyzed in detail the structure of nonconservative homologous junctions and determined the contribution of each end to the formation of the junction. We have also analyzed multiple descendants from single recombination events. Two types of junctions were found. The majority (90%) of the junctions were characterized by a single crossover site. These crossover sites were distributed randomly throughout the junction. The remaining 10% of the junctions had mosaic patterns of parental markers. Furthermore, in 9 of 10 cases, multiple descendants from a single recombination event were identical. Thus, it appears that in most cases few parental markers were involved in junction formation. This finding suggests that nonconservative homologous junctions are mediated mainly by short heteroduplexes of a few hundred base pairs or less. These results are discussed in terms of the current models of nonconservative homologous recombination.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abastado J. P., Cami B., Dinh T. H., Igolen J., Kourilsky P. Processing of complex heteroduplexes in Escherichia coli and Cos-1 monkey cells. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5792–5796. doi: 10.1073/pnas.81.18.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. A., Eliason S. L. Recombination of homologous DNA fragments transfected into mammalian cells occurs predominantly by terminal pairing. Mol Cell Biol. 1986 Sep;6(9):3246–3252. doi: 10.1128/mcb.6.9.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayares D., Ganea D., Chekuri L., Campbell C. R., Kucherlapati R. Repair of single-stranded DNA nicks, gaps, and loops in mammalian cells. Mol Cell Biol. 1987 May;7(5):1656–1662. doi: 10.1128/mcb.7.5.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillette S., Chartrand P. Intermolecular recombination assay for mammalian cells that produces recombinants carrying both homologous and nonhomologous junctions. Mol Cell Biol. 1987 Jun;7(6):2248–2255. doi: 10.1128/mcb.7.6.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassuto E., Lightfoot L. A., Howard-Flanders P. Partial purification of an activity from human cells that promotes homologous pairing and the formation of heteroduplex DNA in the presence of ATP. Mol Gen Genet. 1987 Jun;208(1-2):10–14. doi: 10.1007/BF00330415. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S., Seidman M. M. Intramolecular recombination between transfected repeated sequences in mammalian cells is nonconservative. Mol Cell Biol. 1986 Jul;6(7):2520–2526. doi: 10.1128/mcb.6.7.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby V., Blattner F. Homologous recombination catalyzed by mammalian cell extracts in vitro. Science. 1984 Dec 7;226(4679):1213–1215. doi: 10.1126/science.6334360. [DOI] [PubMed] [Google Scholar]
- Fishel R. A., Detmer K., Rich A. Identification of homologous pairing and strand-exchange activity from a human tumor cell line based on Z-DNA affinity chromatography. Proc Natl Acad Sci U S A. 1988 Jan;85(1):36–40. doi: 10.1073/pnas.85.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folger K. R., Thomas K., Capecchi M. R. Efficient correction of mismatched bases in plasmid heteroduplexes injected into cultured mammalian cell nuclei. Mol Cell Biol. 1985 Jan;5(1):70–74. doi: 10.1128/mcb.5.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganea D., Moore P., Chekuri L., Kucherlapati R. Characterization of an ATP-dependent DNA strand transferase from human cells. Mol Cell Biol. 1987 Sep;7(9):3124–3130. doi: 10.1128/mcb.7.9.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hsieh P., Meyn M. S., Camerini-Otero R. D. Partial purification and characterization of a recombinase from human cells. Cell. 1986 Mar 28;44(6):885–894. doi: 10.1016/0092-8674(86)90011-5. [DOI] [PubMed] [Google Scholar]
- Kenne K., Ljungquist S. A DNA-recombinogenic activity in human cells. Nucleic Acids Res. 1984 Apr 11;12(7):3057–3068. doi: 10.1093/nar/12.7.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucherlapati R. S., Spencer J., Moore P. D. Homologous recombination catalyzed by human cell extracts. Mol Cell Biol. 1985 Apr;5(4):714–720. doi: 10.1128/mcb.5.4.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K. M., Sternberg N. L. Extrachromosomal recombination in mammalian cells as studied with single- and double-stranded DNA substrates. Mol Cell Biol. 1987 Jan;7(1):129–140. doi: 10.1128/mcb.7.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Intermolecular recombination between DNAs introduced into mouse L cells is mediated by a nonconservative pathway that leads to crossover products. Mol Cell Biol. 1990 Jan;10(1):103–112. doi: 10.1128/mcb.10.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Repair of double-stranded DNA breaks by homologous DNA fragments during transfer of DNA into mouse L cells. Mol Cell Biol. 1990 Jan;10(1):113–119. doi: 10.1128/mcb.10.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez B., Rousset S., Coppey J. Homologous recombination intermediates between two duplex DNA catalysed by human cell extracts. Nucleic Acids Res. 1987 Jul 24;15(14):5643–5655. doi: 10.1093/nar/15.14.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryon E., Carroll D. Degradation of linear DNA by a strand-specific exonuclease activity in Xenopus laevis oocytes. Mol Cell Biol. 1989 Nov;9(11):4862–4871. doi: 10.1128/mcb.9.11.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S. P., Rich A., Fishel R. The human recombination strand exchange process. Genome. 1989;31(1):45–52. doi: 10.1139/g89-012. [DOI] [PubMed] [Google Scholar]
- Mudgett J. S., Taylor W. D. Recombination between irradiated shuttle vector DNA and chromosomal DNA in African green monkey kidney cells. Mol Cell Biol. 1990 Jan;10(1):37–46. doi: 10.1128/mcb.10.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman M. M. Intermolecular homologous recombination between transfected sequences in mammalian cells is primarily nonconservative. Mol Cell Biol. 1987 Oct;7(10):3561–3565. doi: 10.1128/mcb.7.10.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg C. M., Ellis J., Bernstein A. Introduction of specific point mutations into RNA polymerase II by gene targeting in mouse embryonic stem cells: evidence for a DNA mismatch repair mechanism. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4680–4684. doi: 10.1073/pnas.87.12.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman D. J., Milman G. Short-term, high-efficiency expression of transfected DNA. Mol Cell Biol. 1984 Aug;4(8):1641–1643. doi: 10.1128/mcb.4.8.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Wake C. T., Vernaleone F., Wilson J. H. Topological requirements for homologous recombination among DNA molecules transfected into mammalian cells. Mol Cell Biol. 1985 Aug;5(8):2080–2089. doi: 10.1128/mcb.5.8.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S., Liskay R. M. Differential effects of base-pair mismatch on intrachromosomal versus extrachromosomal recombination in mouse cells. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5340–5344. doi: 10.1073/pnas.84.15.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss U., Wilson J. H. Repair of single-stranded loops in heteroduplex DNA transfected into mammalian cells. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1619–1623. doi: 10.1073/pnas.84.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]



