Abstract
In the title compound, {[Cu(C24H15N2O4)2(H2O)2]·4C3H7NO}n, the CuII ion, lying on an inversion center, is six-coordinated by two N atoms from two 4-[6-(4-carboxyphenyl)-4,4′-bipyridin-2-yl]benzoate (L) ligands, two deprotonated carboxylate O atoms from two other symmetry-related L ligands and two water molecules in a slightly distorted octahedral geometry. The CuII atoms are linked by the bridging ligands into a layer parallel to (101). The presence of intralayer O—H⋯O hydrogen bonds and π–π interactions between the pyridine and benzene rings [centroid–centroid distances = 3.808 (2) and 3.927 (2) Å] stabilizes the layer. Further O—H⋯O hydrogen bonds link the layers and the dimethylformamide solvent molecules.
Related literature
For the design of metal-organic coordination polymers, see: Ge & Song (2012 ▶); Herm et al. (2011 ▶); Liu et al. (2010 ▶); Wang et al. (2010 ▶). For a related structure, see: Xia et al. (2012 ▶). 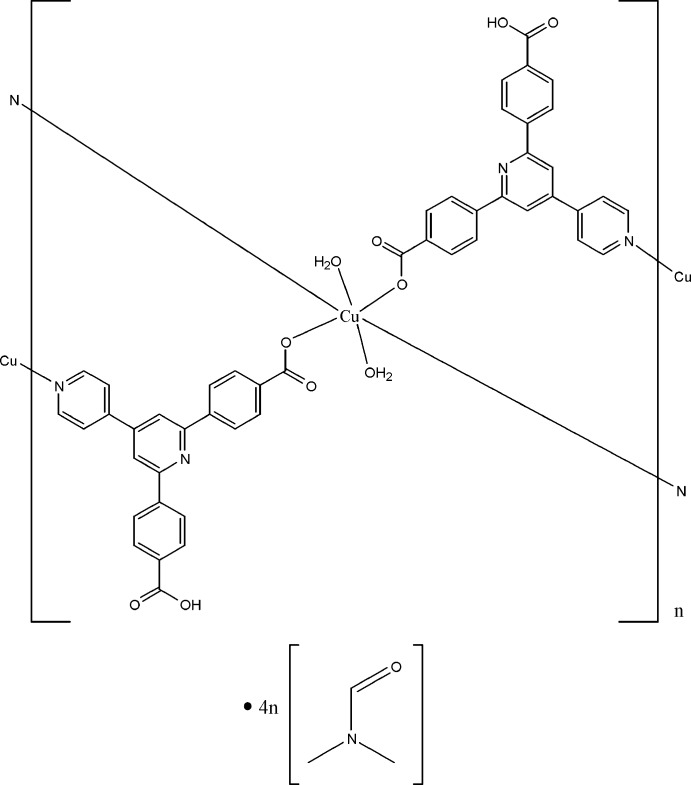
Experimental
Crystal data
[Cu(C24H15N2O4)2(H2O)2]·4C3H7NO
M r = 1182.73
Monoclinic,
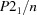
a = 7.7161 (17) Å
b = 17.550 (4) Å
c = 20.947 (4) Å
β = 96.800 (4)°
V = 2816.6 (10) Å3
Z = 2
Mo Kα radiation
μ = 0.46 mm−1
T = 293 K
0.27 × 0.25 × 0.20 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2001 ▶) T min = 0.885, T max = 0.913
14622 measured reflections
5226 independent reflections
3371 reflections with I > 2σ(I)
R int = 0.058
Refinement
R[F 2 > 2σ(F 2)] = 0.061
wR(F 2) = 0.185
S = 1.04
5226 reflections
376 parameters
H-atom parameters constrained
Δρmax = 0.93 e Å−3
Δρmin = −0.39 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: XP in SHELXTL and DIAMOND (Brandenburg, 1999 ▶); software used to prepare material for publication: SHELXTL (Sheldrick, 2008 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813006430/hy2619sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813006430/hy2619Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O4—H4A⋯O2i | 0.82 | 1.86 | 2.584 (4) | 146 |
| O1W—H1A⋯O5ii | 0.85 | 1.98 | 2.808 (5) | 165 |
| O1W—H1B⋯O2iii | 0.85 | 1.95 | 2.758 (4) | 159 |
Symmetry codes: (i) 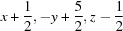 ; (ii)
; (ii) 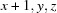 ; (iii)
; (iii) 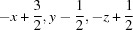 .
.
Acknowledgments
The authors are grateful for financial aid from The First Hospital of Jilin University.
supplementary crystallographic information
Comment
Metal-organic coordination polymers (MOCPs) with infinite one-, two- or three-dimensional structures are assembled with metal ions or polynuclear clusters as nodes and organic ligands as linkers (Herm et al., 2011; Liu et al., 2010). Recently, the chemists have devoted themselves to design and synthesize MOCPs, not only due to their potential applications in the realm of gas adsorption and separation, catalysis, magnetism, luminescence, host–guest chemistry and etc, but also for their aesthetic and often complicated architectures and topologies (Ge & Song, 2012; Wang et al., 2010). In order to extend the investigations in this field, we used a multifunctional ligand, 4,4'-(4,4'-bipyridine-2,6-diyl)dibenzoic acid (bpydbH2) to design and synthesize the title copper(II) complex and report its structure here.
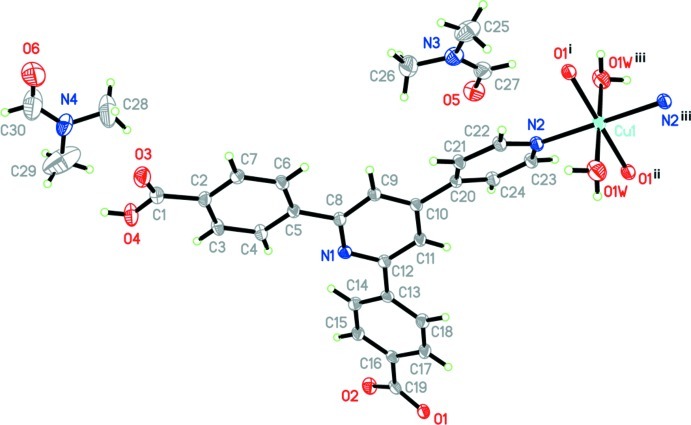
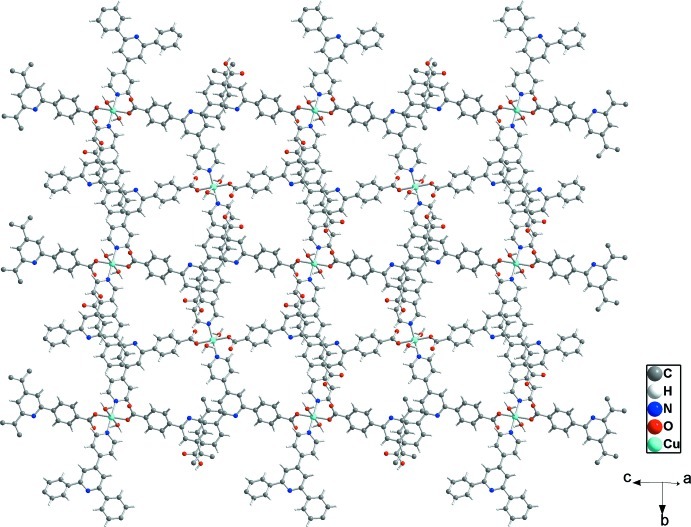
The asymmetric unit of the title compound contains one CuII ion lying on an inversion center, one anionic bpydbH ligand, one aqua ligand and two lattice DMF molecules. As shown in Fig. 1, the CuII ion is six-coordinated by two N atoms from two bpydbH ligands, two deprotonated carboxylate O atoms from two other symmetry-related bpydbH ligands and two aqua ligands, furnishing a slightly distorted octahedral geometry. The bond distances and angles are in a normal range (Xia et al., 2012). The Cu nodes are extended by the bridging bpydbH linkers into a layer parallel to (101) (Fig. 2). The presence of intralayer O—H···O hydrogen bonds and π–π interactions between the pyridine and benzene rings [centroid–centroid diatances = 3.808 (2) and 3.927 (2) Å] stabilizes the single layer.
Experimental
Cu(NO3)2.3H2O (0.0063 g, 0.025 mmol) and bpydbH2 (0.0099 g, 0.025 mmol) were suspended in a mixed solvent of dimethylformamide (DMF) (4 ml) and H2O (0.5 ml), and heated in a 15 ml Teflon-lined stainless-steel autoclave at 80°C for 3 days. After the autoclave was cooled to room temperature slowly, green crystals were collected by filtration and washed with DMF, and dried in air (yield: 65% based on Cu).
Refinement
H atoms on C and carboxyl O atoms were positioned geometrically and refined as riding atoms, with C—H = 0.93, 0.96 and O—H = 0.82 Å and with Uiso(H) = 1.2(1.5 for methyl and carboxyl)Ueq(C,O). H atoms of water molecules were located in a difference Fourier map and refined as riding atoms, with Uiso(H) = 1.5Ueq(O).
Figures
Fig. 1.
The asymmetric unit of the title compound. Displacement ellipsoids are drawn at the 30% probability level. [Symmetry codes: (i) 1/2 + x, 3/2 - y, -1/2 + z; (ii) 3/2 - x, -1/2 + y, 1/2 - z; (iii) 2 - x, 1 - y, -z.]
Fig. 2.
View of the layer structure of the title compound.
Crystal data
| [Cu(C24H15N2O4)2(H2O)2]·4C3H7NO | F(000) = 1238 |
| Mr = 1182.73 | Dx = 1.395 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 5226 reflections |
| a = 7.7161 (17) Å | θ = 1.0–26.0° |
| b = 17.550 (4) Å | µ = 0.46 mm−1 |
| c = 20.947 (4) Å | T = 293 K |
| β = 96.800 (4)° | Block, green |
| V = 2816.6 (10) Å3 | 0.27 × 0.25 × 0.20 mm |
| Z = 2 |
Data collection
| Bruker APEXII CCD diffractometer | 5226 independent reflections |
| Radiation source: fine-focus sealed tube | 3371 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.058 |
| φ and ω scans | θmax = 25.5°, θmin = 2.0° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −9→7 |
| Tmin = 0.885, Tmax = 0.913 | k = −21→21 |
| 14622 measured reflections | l = −20→25 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.061 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.185 | H-atom parameters constrained |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0891P)2 + 2.0095P] where P = (Fo2 + 2Fc2)/3 |
| 5226 reflections | (Δ/σ)max < 0.001 |
| 376 parameters | Δρmax = 0.93 e Å−3 |
| 0 restraints | Δρmin = −0.39 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cu1 | 1.0000 | 0.5000 | 0.0000 | 0.0362 (2) | |
| C1 | 0.8452 (6) | 1.2336 (2) | −0.1386 (2) | 0.0432 (10) | |
| C2 | 0.8060 (5) | 1.1653 (2) | −0.10151 (19) | 0.0347 (9) | |
| C3 | 0.7251 (5) | 1.1715 (2) | −0.0455 (2) | 0.0369 (10) | |
| H3 | 0.6854 | 1.2186 | −0.0330 | 0.044* | |
| C4 | 0.7040 (5) | 1.1079 (2) | −0.0088 (2) | 0.0350 (9) | |
| H4 | 0.6517 | 1.1128 | 0.0288 | 0.042* | |
| C5 | 0.7592 (5) | 1.0363 (2) | −0.02669 (18) | 0.0286 (8) | |
| C6 | 0.8380 (5) | 1.0305 (2) | −0.08253 (19) | 0.0352 (9) | |
| H6 | 0.8762 | 0.9832 | −0.0953 | 0.042* | |
| C7 | 0.8607 (5) | 1.0943 (2) | −0.11976 (19) | 0.0376 (10) | |
| H7 | 0.9132 | 1.0894 | −0.1573 | 0.045* | |
| C8 | 0.7389 (5) | 0.9695 (2) | 0.01565 (19) | 0.0302 (8) | |
| C9 | 0.7683 (5) | 0.8950 (2) | −0.00365 (18) | 0.0304 (9) | |
| H9 | 0.7972 | 0.8853 | −0.0447 | 0.036* | |
| C10 | 0.7540 (5) | 0.83558 (19) | 0.03927 (18) | 0.0280 (8) | |
| C11 | 0.7072 (5) | 0.8537 (2) | 0.09959 (18) | 0.0314 (9) | |
| H11 | 0.6958 | 0.8154 | 0.1295 | 0.038* | |
| C12 | 0.6774 (5) | 0.9291 (2) | 0.11507 (18) | 0.0299 (8) | |
| C13 | 0.6253 (5) | 0.9511 (2) | 0.17864 (18) | 0.0301 (9) | |
| C14 | 0.5242 (5) | 1.0156 (2) | 0.18392 (19) | 0.0358 (9) | |
| H14 | 0.4924 | 1.0455 | 0.1478 | 0.043* | |
| C15 | 0.4702 (5) | 1.0358 (2) | 0.24194 (19) | 0.0372 (10) | |
| H15 | 0.4004 | 1.0786 | 0.2445 | 0.045* | |
| C16 | 0.5196 (5) | 0.9925 (2) | 0.29668 (18) | 0.0312 (9) | |
| C17 | 0.6242 (5) | 0.9290 (2) | 0.29215 (19) | 0.0378 (10) | |
| H17 | 0.6586 | 0.8998 | 0.3285 | 0.045* | |
| C18 | 0.6777 (5) | 0.9089 (2) | 0.23352 (19) | 0.0366 (9) | |
| H18 | 0.7494 | 0.8667 | 0.2310 | 0.044* | |
| C19 | 0.4600 (6) | 1.0145 (2) | 0.3597 (2) | 0.0361 (10) | |
| C20 | 0.7944 (5) | 0.75539 (19) | 0.02344 (17) | 0.0288 (8) | |
| C21 | 0.9191 (5) | 0.73870 (19) | −0.01677 (18) | 0.0307 (9) | |
| H21 | 0.9689 | 0.7778 | −0.0384 | 0.037* | |
| C22 | 0.9695 (5) | 0.6647 (2) | −0.02478 (19) | 0.0339 (9) | |
| H22 | 1.0543 | 0.6549 | −0.0517 | 0.041* | |
| C23 | 0.7732 (5) | 0.6214 (2) | 0.04100 (19) | 0.0360 (9) | |
| H23 | 0.7197 | 0.5811 | 0.0598 | 0.043* | |
| C24 | 0.7177 (5) | 0.6942 (2) | 0.05158 (18) | 0.0342 (9) | |
| H24 | 0.6294 | 0.7025 | 0.0774 | 0.041* | |
| C25 | 0.6692 (8) | 0.6406 (4) | −0.1720 (3) | 0.0829 (19) | |
| H25A | 0.7059 | 0.6773 | −0.2016 | 0.124* | |
| H25B | 0.6144 | 0.5984 | −0.1955 | 0.124* | |
| H25C | 0.7689 | 0.6229 | −0.1442 | 0.124* | |
| C26 | 0.5118 (8) | 0.7558 (3) | −0.1443 (3) | 0.0756 (16) | |
| H26A | 0.5739 | 0.7741 | −0.1783 | 0.113* | |
| H26B | 0.5493 | 0.7833 | −0.1055 | 0.113* | |
| H26C | 0.3889 | 0.7633 | −0.1558 | 0.113* | |
| C27 | 0.4770 (6) | 0.6364 (3) | −0.0897 (2) | 0.0543 (12) | |
| H27 | 0.5082 | 0.5854 | −0.0843 | 0.065* | |
| C28 | 0.4517 (16) | 1.2769 (5) | −0.2557 (5) | 0.203 (6) | |
| H28A | 0.4153 | 1.2642 | −0.2148 | 0.304* | |
| H28B | 0.5656 | 1.2561 | −0.2586 | 0.304* | |
| H28C | 0.3703 | 1.2562 | −0.2895 | 0.304* | |
| C29 | 0.4063 (11) | 1.3970 (6) | −0.2090 (4) | 0.157 (4) | |
| H29A | 0.3787 | 1.3613 | −0.1770 | 0.235* | |
| H29B | 0.3054 | 1.4274 | −0.2228 | 0.235* | |
| H29C | 0.4999 | 1.4294 | −0.1912 | 0.235* | |
| C30 | 0.5028 (11) | 1.3864 (4) | −0.3123 (4) | 0.119 (3) | |
| H30 | 0.4860 | 1.4387 | −0.3166 | 0.142* | |
| N1 | 0.6925 (4) | 0.98644 (16) | 0.07362 (15) | 0.0305 (7) | |
| N2 | 0.9010 (4) | 0.60580 (17) | 0.00477 (15) | 0.0331 (8) | |
| N3 | 0.5471 (5) | 0.6755 (2) | −0.13415 (19) | 0.0533 (10) | |
| N4 | 0.4576 (6) | 1.3570 (2) | −0.2622 (2) | 0.0619 (11) | |
| O1 | 0.5313 (4) | 0.98064 (14) | 0.40897 (13) | 0.0395 (7) | |
| O2 | 0.3418 (4) | 1.06386 (16) | 0.35908 (14) | 0.0488 (8) | |
| O3 | 0.9397 (5) | 1.23445 (17) | −0.18056 (16) | 0.0582 (9) | |
| O4 | 0.7673 (4) | 1.29571 (17) | −0.11906 (16) | 0.0607 (9) | |
| H4A | 0.7916 | 1.3324 | −0.1405 | 0.091* | |
| O5 | 0.3740 (4) | 0.6624 (2) | −0.05498 (17) | 0.0651 (10) | |
| O6 | 0.5670 (7) | 1.3545 (2) | −0.3567 (2) | 0.1082 (17) | |
| O1W | 1.3019 (4) | 0.54357 (17) | 0.02770 (15) | 0.0552 (8) | |
| H1A | 1.3236 | 0.5847 | 0.0087 | 0.083* | |
| H1B | 1.2828 | 0.5552 | 0.0656 | 0.083* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu1 | 0.0628 (5) | 0.0222 (3) | 0.0263 (4) | 0.0050 (3) | 0.0164 (3) | 0.0015 (3) |
| C1 | 0.054 (3) | 0.033 (2) | 0.042 (3) | −0.001 (2) | 0.005 (2) | 0.0034 (19) |
| C2 | 0.036 (2) | 0.031 (2) | 0.037 (2) | −0.0049 (17) | 0.0019 (18) | 0.0061 (17) |
| C3 | 0.040 (2) | 0.0251 (19) | 0.046 (3) | 0.0037 (16) | 0.010 (2) | 0.0004 (17) |
| C4 | 0.039 (2) | 0.0273 (19) | 0.041 (2) | 0.0008 (17) | 0.0146 (19) | 0.0013 (17) |
| C5 | 0.032 (2) | 0.0272 (19) | 0.027 (2) | −0.0010 (16) | 0.0060 (16) | 0.0009 (15) |
| C6 | 0.048 (2) | 0.0254 (18) | 0.033 (2) | 0.0009 (17) | 0.0094 (19) | −0.0029 (16) |
| C7 | 0.052 (3) | 0.035 (2) | 0.028 (2) | −0.0037 (18) | 0.0104 (19) | 0.0002 (17) |
| C8 | 0.034 (2) | 0.0256 (18) | 0.032 (2) | 0.0004 (16) | 0.0066 (17) | 0.0011 (16) |
| C9 | 0.037 (2) | 0.0284 (19) | 0.027 (2) | 0.0006 (16) | 0.0088 (17) | −0.0017 (16) |
| C10 | 0.033 (2) | 0.0240 (18) | 0.028 (2) | −0.0001 (15) | 0.0067 (16) | −0.0014 (15) |
| C11 | 0.042 (2) | 0.0257 (19) | 0.027 (2) | 0.0008 (16) | 0.0090 (17) | 0.0019 (16) |
| C12 | 0.036 (2) | 0.0275 (19) | 0.027 (2) | −0.0004 (16) | 0.0085 (17) | 0.0000 (16) |
| C13 | 0.038 (2) | 0.0284 (19) | 0.025 (2) | −0.0028 (16) | 0.0074 (17) | −0.0047 (15) |
| C14 | 0.048 (2) | 0.033 (2) | 0.027 (2) | 0.0025 (17) | 0.0097 (19) | 0.0007 (16) |
| C15 | 0.050 (2) | 0.030 (2) | 0.034 (2) | 0.0050 (18) | 0.0104 (19) | −0.0020 (17) |
| C16 | 0.041 (2) | 0.0259 (19) | 0.029 (2) | −0.0039 (16) | 0.0126 (17) | −0.0045 (16) |
| C17 | 0.051 (3) | 0.032 (2) | 0.032 (2) | 0.0009 (18) | 0.0092 (19) | 0.0052 (17) |
| C18 | 0.047 (2) | 0.031 (2) | 0.033 (2) | 0.0078 (18) | 0.0118 (19) | −0.0013 (17) |
| C19 | 0.050 (2) | 0.026 (2) | 0.035 (2) | −0.0054 (18) | 0.014 (2) | −0.0043 (17) |
| C20 | 0.039 (2) | 0.0239 (18) | 0.024 (2) | 0.0012 (16) | 0.0051 (16) | −0.0003 (15) |
| C21 | 0.043 (2) | 0.0225 (18) | 0.029 (2) | −0.0019 (16) | 0.0141 (17) | 0.0005 (15) |
| C22 | 0.047 (2) | 0.0275 (19) | 0.030 (2) | −0.0006 (17) | 0.0146 (18) | −0.0004 (16) |
| C23 | 0.052 (3) | 0.028 (2) | 0.030 (2) | −0.0039 (18) | 0.0140 (19) | 0.0014 (16) |
| C24 | 0.045 (2) | 0.031 (2) | 0.029 (2) | 0.0012 (17) | 0.0158 (18) | −0.0017 (16) |
| C25 | 0.081 (4) | 0.119 (5) | 0.052 (4) | 0.025 (4) | 0.022 (3) | 0.004 (3) |
| C26 | 0.097 (4) | 0.062 (3) | 0.069 (4) | −0.003 (3) | 0.017 (3) | 0.009 (3) |
| C27 | 0.056 (3) | 0.053 (3) | 0.054 (3) | −0.005 (2) | 0.005 (3) | −0.002 (2) |
| C28 | 0.303 (15) | 0.088 (6) | 0.196 (11) | −0.064 (8) | −0.057 (10) | 0.066 (7) |
| C29 | 0.132 (7) | 0.259 (12) | 0.082 (6) | 0.104 (8) | 0.025 (5) | 0.000 (6) |
| C30 | 0.167 (8) | 0.079 (5) | 0.123 (7) | 0.022 (5) | 0.067 (6) | 0.031 (5) |
| N1 | 0.0353 (17) | 0.0303 (17) | 0.0272 (18) | −0.0014 (13) | 0.0090 (14) | −0.0030 (13) |
| N2 | 0.049 (2) | 0.0260 (16) | 0.0265 (18) | 0.0009 (14) | 0.0128 (15) | 0.0000 (13) |
| N3 | 0.054 (2) | 0.062 (3) | 0.046 (2) | 0.000 (2) | 0.0151 (19) | 0.0015 (19) |
| N4 | 0.071 (3) | 0.059 (3) | 0.061 (3) | 0.012 (2) | 0.027 (2) | 0.019 (2) |
| O1 | 0.0655 (19) | 0.0274 (14) | 0.0276 (16) | −0.0003 (13) | 0.0135 (14) | −0.0001 (11) |
| O2 | 0.071 (2) | 0.0377 (16) | 0.0413 (19) | 0.0105 (15) | 0.0232 (15) | −0.0045 (13) |
| O3 | 0.082 (2) | 0.0458 (19) | 0.051 (2) | −0.0038 (17) | 0.0217 (19) | 0.0064 (15) |
| O4 | 0.081 (2) | 0.0349 (17) | 0.070 (2) | 0.0016 (16) | 0.0251 (19) | 0.0134 (16) |
| O5 | 0.060 (2) | 0.075 (2) | 0.064 (2) | −0.0093 (18) | 0.0235 (19) | −0.0012 (19) |
| O6 | 0.179 (5) | 0.077 (3) | 0.083 (3) | −0.005 (3) | 0.075 (3) | −0.021 (2) |
| O1W | 0.072 (2) | 0.0441 (18) | 0.053 (2) | −0.0041 (16) | 0.0239 (17) | −0.0006 (15) |
Geometric parameters (Å, º)
| Cu1—O1i | 1.980 (3) | C19—O2 | 1.256 (5) |
| Cu1—O1ii | 1.980 (3) | C19—O1 | 1.260 (5) |
| Cu1—N2 | 2.015 (3) | C20—C21 | 1.383 (5) |
| Cu1—N2iii | 2.015 (3) | C20—C24 | 1.390 (5) |
| C1—O3 | 1.207 (5) | C21—C22 | 1.371 (5) |
| C1—O4 | 1.332 (5) | C21—H21 | 0.9300 |
| C1—C2 | 1.480 (5) | C22—N2 | 1.345 (5) |
| C2—C7 | 1.383 (5) | C22—H22 | 0.9300 |
| C2—C3 | 1.398 (5) | C23—N2 | 1.342 (5) |
| C3—C4 | 1.375 (5) | C23—C24 | 1.374 (5) |
| C3—H3 | 0.9300 | C23—H23 | 0.9300 |
| C4—C5 | 1.393 (5) | C24—H24 | 0.9300 |
| C4—H4 | 0.9300 | C25—N3 | 1.439 (6) |
| C5—C6 | 1.385 (5) | C25—H25A | 0.9600 |
| C5—C8 | 1.490 (5) | C25—H25B | 0.9600 |
| C6—C7 | 1.388 (5) | C25—H25C | 0.9600 |
| C6—H6 | 0.9300 | C26—N3 | 1.447 (6) |
| C7—H7 | 0.9300 | C26—H26A | 0.9600 |
| C8—N1 | 1.340 (5) | C26—H26B | 0.9600 |
| C8—C9 | 1.394 (5) | C26—H26C | 0.9600 |
| C9—C10 | 1.390 (5) | C27—O5 | 1.226 (5) |
| C9—H9 | 0.9300 | C27—N3 | 1.323 (6) |
| C10—C11 | 1.392 (5) | C27—H27 | 0.9300 |
| C10—C20 | 1.487 (5) | C28—N4 | 1.413 (9) |
| C11—C12 | 1.388 (5) | C28—H28A | 0.9600 |
| C11—H11 | 0.9300 | C28—H28B | 0.9600 |
| C12—N1 | 1.343 (5) | C28—H28C | 0.9600 |
| C12—C13 | 1.487 (5) | C29—N4 | 1.411 (8) |
| C13—C14 | 1.385 (5) | C29—H29A | 0.9600 |
| C13—C18 | 1.387 (5) | C29—H29B | 0.9600 |
| C14—C15 | 1.377 (5) | C29—H29C | 0.9600 |
| C14—H14 | 0.9300 | C30—O6 | 1.238 (8) |
| C15—C16 | 1.391 (5) | C30—N4 | 1.255 (7) |
| C15—H15 | 0.9300 | C30—H30 | 0.9300 |
| C16—C17 | 1.385 (5) | O1—Cu1iv | 1.980 (3) |
| C16—C19 | 1.499 (5) | O4—H4A | 0.8200 |
| C17—C18 | 1.387 (5) | O1W—H1A | 0.8501 |
| C17—H17 | 0.9300 | O1W—H1B | 0.8489 |
| C18—H18 | 0.9300 | ||
| O1i—Cu1—O1ii | 180.0 | O2—C19—C16 | 117.9 (4) |
| O1i—Cu1—N2 | 91.16 (11) | O1—C19—C16 | 116.7 (4) |
| O1ii—Cu1—N2 | 88.84 (11) | C21—C20—C24 | 117.2 (3) |
| O1i—Cu1—N2iii | 88.84 (11) | C21—C20—C10 | 121.0 (3) |
| O1ii—Cu1—N2iii | 91.16 (11) | C24—C20—C10 | 121.7 (3) |
| N2—Cu1—N2iii | 180.0 | C22—C21—C20 | 120.2 (3) |
| O3—C1—O4 | 123.2 (4) | C22—C21—H21 | 119.9 |
| O3—C1—C2 | 124.8 (4) | C20—C21—H21 | 119.9 |
| O4—C1—C2 | 112.0 (4) | N2—C22—C21 | 122.6 (4) |
| C7—C2—C3 | 118.9 (3) | N2—C22—H22 | 118.7 |
| C7—C2—C1 | 119.8 (4) | C21—C22—H22 | 118.7 |
| C3—C2—C1 | 121.1 (4) | N2—C23—C24 | 123.0 (3) |
| C4—C3—C2 | 120.0 (4) | N2—C23—H23 | 118.5 |
| C4—C3—H3 | 120.0 | C24—C23—H23 | 118.5 |
| C2—C3—H3 | 120.0 | C23—C24—C20 | 119.6 (4) |
| C3—C4—C5 | 121.4 (4) | C23—C24—H24 | 120.2 |
| C3—C4—H4 | 119.3 | C20—C24—H24 | 120.2 |
| C5—C4—H4 | 119.3 | N3—C25—H25A | 109.5 |
| C6—C5—C4 | 118.3 (3) | N3—C25—H25B | 109.5 |
| C6—C5—C8 | 122.2 (3) | H25A—C25—H25B | 109.5 |
| C4—C5—C8 | 119.5 (3) | N3—C25—H25C | 109.5 |
| C5—C6—C7 | 120.8 (4) | H25A—C25—H25C | 109.5 |
| C5—C6—H6 | 119.6 | H25B—C25—H25C | 109.5 |
| C7—C6—H6 | 119.6 | N3—C26—H26A | 109.5 |
| C2—C7—C6 | 120.6 (4) | N3—C26—H26B | 109.5 |
| C2—C7—H7 | 119.7 | H26A—C26—H26B | 109.5 |
| C6—C7—H7 | 119.7 | N3—C26—H26C | 109.5 |
| N1—C8—C9 | 122.8 (3) | H26A—C26—H26C | 109.5 |
| N1—C8—C5 | 115.0 (3) | H26B—C26—H26C | 109.5 |
| C9—C8—C5 | 122.2 (3) | O5—C27—N3 | 124.9 (5) |
| C10—C9—C8 | 119.2 (3) | O5—C27—H27 | 117.6 |
| C10—C9—H9 | 120.4 | N3—C27—H27 | 117.6 |
| C8—C9—H9 | 120.4 | N4—C28—H28A | 109.5 |
| C9—C10—C11 | 117.7 (3) | N4—C28—H28B | 109.5 |
| C9—C10—C20 | 122.1 (3) | H28A—C28—H28B | 109.5 |
| C11—C10—C20 | 120.2 (3) | N4—C28—H28C | 109.5 |
| C12—C11—C10 | 119.9 (3) | H28A—C28—H28C | 109.5 |
| C12—C11—H11 | 120.1 | H28B—C28—H28C | 109.5 |
| C10—C11—H11 | 120.1 | N4—C29—H29A | 109.5 |
| N1—C12—C11 | 122.3 (3) | N4—C29—H29B | 109.5 |
| N1—C12—C13 | 115.9 (3) | H29A—C29—H29B | 109.5 |
| C11—C12—C13 | 121.8 (3) | N4—C29—H29C | 109.5 |
| C14—C13—C18 | 118.8 (3) | H29A—C29—H29C | 109.5 |
| C14—C13—C12 | 119.9 (3) | H29B—C29—H29C | 109.5 |
| C18—C13—C12 | 121.3 (3) | O6—C30—N4 | 128.2 (7) |
| C15—C14—C13 | 120.9 (4) | O6—C30—H30 | 115.9 |
| C15—C14—H14 | 119.6 | N4—C30—H30 | 115.9 |
| C13—C14—H14 | 119.6 | C8—N1—C12 | 118.2 (3) |
| C14—C15—C16 | 120.3 (4) | C23—N2—C22 | 117.3 (3) |
| C14—C15—H15 | 119.9 | C23—N2—Cu1 | 121.6 (2) |
| C16—C15—H15 | 119.9 | C22—N2—Cu1 | 120.9 (3) |
| C17—C16—C15 | 119.2 (4) | C27—N3—C25 | 121.1 (5) |
| C17—C16—C19 | 120.7 (4) | C27—N3—C26 | 121.6 (4) |
| C15—C16—C19 | 120.1 (3) | C25—N3—C26 | 117.3 (4) |
| C16—C17—C18 | 120.2 (4) | C30—N4—C29 | 125.9 (7) |
| C16—C17—H17 | 119.9 | C30—N4—C28 | 120.2 (7) |
| C18—C17—H17 | 119.9 | C29—N4—C28 | 113.8 (7) |
| C17—C18—C13 | 120.6 (4) | C19—O1—Cu1iv | 128.1 (3) |
| C17—C18—H18 | 119.7 | C1—O4—H4A | 109.5 |
| C13—C18—H18 | 119.7 | H1A—O1W—H1B | 107.4 |
| O2—C19—O1 | 125.4 (4) | ||
| O3—C1—C2—C7 | −8.6 (7) | C19—C16—C17—C18 | 179.6 (4) |
| O4—C1—C2—C7 | 173.1 (4) | C16—C17—C18—C13 | −1.0 (6) |
| O3—C1—C2—C3 | 166.8 (4) | C14—C13—C18—C17 | 2.5 (6) |
| O4—C1—C2—C3 | −11.4 (6) | C12—C13—C18—C17 | −178.2 (3) |
| C7—C2—C3—C4 | 1.3 (6) | C17—C16—C19—O2 | −167.7 (4) |
| C1—C2—C3—C4 | −174.2 (4) | C15—C16—C19—O2 | 12.3 (5) |
| C2—C3—C4—C5 | −1.1 (6) | C17—C16—C19—O1 | 11.0 (5) |
| C3—C4—C5—C6 | 0.6 (6) | C15—C16—C19—O1 | −168.9 (4) |
| C3—C4—C5—C8 | 178.0 (4) | C9—C10—C20—C21 | 30.9 (6) |
| C4—C5—C6—C7 | −0.2 (6) | C11—C10—C20—C21 | −146.3 (4) |
| C8—C5—C6—C7 | −177.6 (4) | C9—C10—C20—C24 | −153.5 (4) |
| C3—C2—C7—C6 | −1.0 (6) | C11—C10—C20—C24 | 29.2 (5) |
| C1—C2—C7—C6 | 174.6 (4) | C24—C20—C21—C22 | −3.5 (6) |
| C5—C6—C7—C2 | 0.5 (6) | C10—C20—C21—C22 | 172.2 (4) |
| C6—C5—C8—N1 | 167.3 (3) | C20—C21—C22—N2 | 0.6 (6) |
| C4—C5—C8—N1 | −10.0 (5) | N2—C23—C24—C20 | 0.7 (6) |
| C6—C5—C8—C9 | −11.9 (6) | C21—C20—C24—C23 | 2.9 (6) |
| C4—C5—C8—C9 | 170.7 (4) | C10—C20—C24—C23 | −172.8 (4) |
| N1—C8—C9—C10 | −1.8 (6) | C9—C8—N1—C12 | 1.4 (5) |
| C5—C8—C9—C10 | 177.3 (3) | C5—C8—N1—C12 | −177.9 (3) |
| C8—C9—C10—C11 | 1.2 (5) | C11—C12—N1—C8 | −0.4 (5) |
| C8—C9—C10—C20 | −176.1 (3) | C13—C12—N1—C8 | −179.9 (3) |
| C9—C10—C11—C12 | −0.3 (5) | C24—C23—N2—C22 | −3.6 (6) |
| C20—C10—C11—C12 | 177.1 (3) | C24—C23—N2—Cu1 | 171.8 (3) |
| C10—C11—C12—N1 | −0.1 (6) | C21—C22—N2—C23 | 3.0 (6) |
| C10—C11—C12—C13 | 179.4 (3) | C21—C22—N2—Cu1 | −172.4 (3) |
| N1—C12—C13—C14 | 28.6 (5) | O1i—Cu1—N2—C23 | −34.5 (3) |
| C11—C12—C13—C14 | −151.0 (4) | O1ii—Cu1—N2—C23 | 145.5 (3) |
| N1—C12—C13—C18 | −150.7 (4) | O1i—Cu1—N2—C22 | 140.8 (3) |
| C11—C12—C13—C18 | 29.8 (6) | O1ii—Cu1—N2—C22 | −39.2 (3) |
| C18—C13—C14—C15 | −2.7 (6) | O5—C27—N3—C25 | 179.0 (5) |
| C12—C13—C14—C15 | 178.1 (4) | O5—C27—N3—C26 | 2.5 (8) |
| C13—C14—C15—C16 | 1.3 (6) | O6—C30—N4—C29 | 171.2 (8) |
| C14—C15—C16—C17 | 0.3 (6) | O6—C30—N4—C28 | −10.0 (14) |
| C14—C15—C16—C19 | −179.8 (4) | O2—C19—O1—Cu1iv | −6.5 (6) |
| C15—C16—C17—C18 | −0.4 (6) | C16—C19—O1—Cu1iv | 174.9 (2) |
Symmetry codes: (i) −x+3/2, y−1/2, −z+1/2; (ii) x+1/2, −y+3/2, z−1/2; (iii) −x+2, −y+1, −z; (iv) −x+3/2, y+1/2, −z+1/2.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O4—H4A···O2v | 0.82 | 1.86 | 2.584 (4) | 146 |
| O1W—H1A···O5vi | 0.85 | 1.98 | 2.808 (5) | 165 |
| O1W—H1B···O2i | 0.85 | 1.95 | 2.758 (4) | 159 |
Symmetry codes: (i) −x+3/2, y−1/2, −z+1/2; (v) x+1/2, −y+5/2, z−1/2; (vi) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HY2619).
References
- Brandenburg, K. (1999). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2001). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Ge, X. & Song, S. (2012). Acta Cryst. E68, m1413. [DOI] [PMC free article] [PubMed]
- Herm, Z. R., Swisher, J. A., Smit, B., Krishna, R. & Long, J. R. (2011). J. Am. Chem. Soc. 133, 5664–5667. [DOI] [PubMed]
- Liu, Y., Xuan, W. & Cui, Y. (2010). Adv. Mater. 22, 4112–4135. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wang, G.-H., Lei, Y.-Q., Wang, N., He, R.-L., Jia, H.-Q., Hu, N.-H. & Xu, J.-W. (2010). Cryst. Growth Des. 10, 534–540.
- Xia, Q.-H., Guo, Z.-F., Liu, L., Wang, Z. & Li, B. (2012). Acta Cryst. E68, m1395. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813006430/hy2619sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813006430/hy2619Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report