Abstract
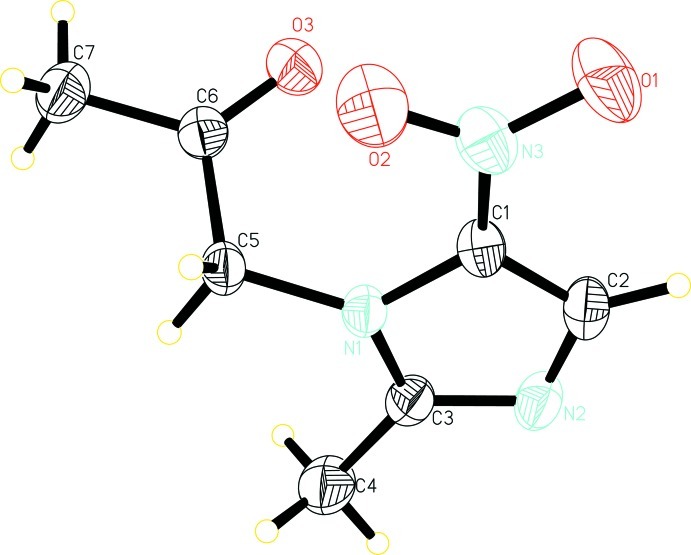
In the molecule of the title compound, C7H9N3O3, the nitro and carbonyl groups are tilted with respect to the imidazole ring by 9.16 (6) and 65.47 (7)°, respectively. Neighbouring chains are linked via C—H⋯N and C—H⋯O hydrogen bonds forming two-dimensional slab-like networks lying parallel to (01-1).
Related literature
For the antibiotic properties of metronidazole and mecnidazole, see: Lin et al. (2012 ▶); Almirall et al. (2011 ▶); Zhang et al. (2011 ▶). For the crystal structure of related imidazoles, see: Yousuf et al. (2012 ▶); Zeb et al. (2012 ▶).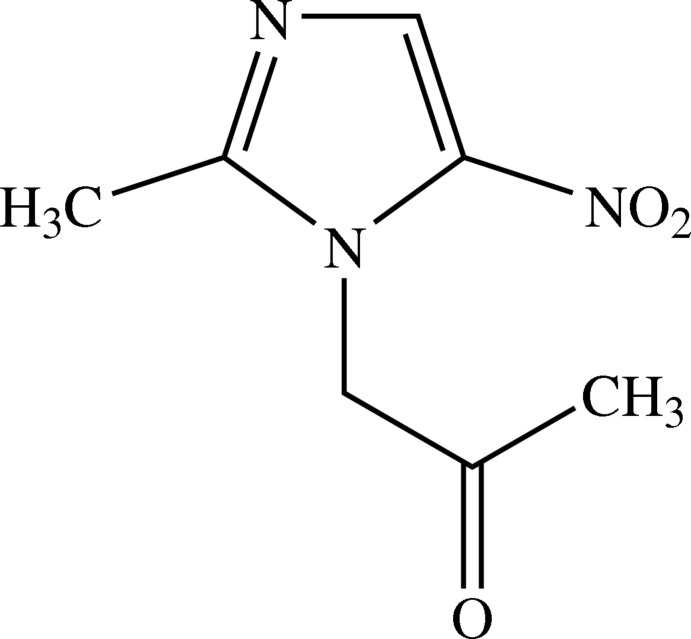
Experimental
Crystal data
C7H9N3O3
M r = 183.17
Monoclinic,
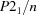
a = 4.7548 (4) Å
b = 12.3971 (9) Å
c = 14.8580 (11) Å
β = 97.350 (2)°
V = 868.62 (12) Å3
Z = 4
Mo Kα radiation
μ = 0.11 mm−1
T = 273 K
0.52 × 0.33 × 0.24 mm
Data collection
Bruker SMART APEX CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2000 ▶) T min = 0.944, T max = 0.974
5030 measured reflections
1614 independent reflections
1328 reflections with I > 2σ(I)
R int = 0.019
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.122
S = 1.06
1614 reflections
120 parameters
H-atom parameters constrained
Δρmax = 0.19 e Å−3
Δρmin = −0.15 e Å−3
Data collection: SMART (Bruker, 2000 ▶); cell refinement: SAINT (Bruker, 2000 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL, PARST (Nardelli, 1995 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813006569/rz5048sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813006569/rz5048Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813006569/rz5048Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2B⋯N2i | 0.93 | 2.56 | 3.361 (2) | 144 |
| C5—H5B⋯O2ii | 0.97 | 2.57 | 3.527 (2) | 167 |
| C7—H7B⋯O3iii | 0.96 | 2.49 | 3.340 (2) | 147 |
Symmetry codes: (i) 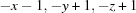 ; (ii)
; (ii) 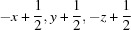 ; (iii)
; (iii) 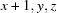 .
.
Acknowledgments
The authors gratefully acknowledge the Pakistan Academy of Sciences for funding project No. 5-9/PAS/8418 entitled ‘Biology-oriented Parallel Synthesis on Nitroimidazoles in Search of Better Therapeutic Agents’.
supplementary crystallographic information
Comment
Imidazole nuclei containing metronidazole and mecnidazole are widely used antibiotics, known to be effective against anaerobic microorganisms. These drugs employed to cure amoebiasis (Almirall et al., 2011) and protozoal infections (Lin et al., 2012). Secnidazoles is also reported to have anti-inflammatory and urease inhibiton activites (Zhang et al., 2011). The title compound is a derivative of secnidazole obtained during our attempts to make more effective structure analogues of this important antibacterial drug.
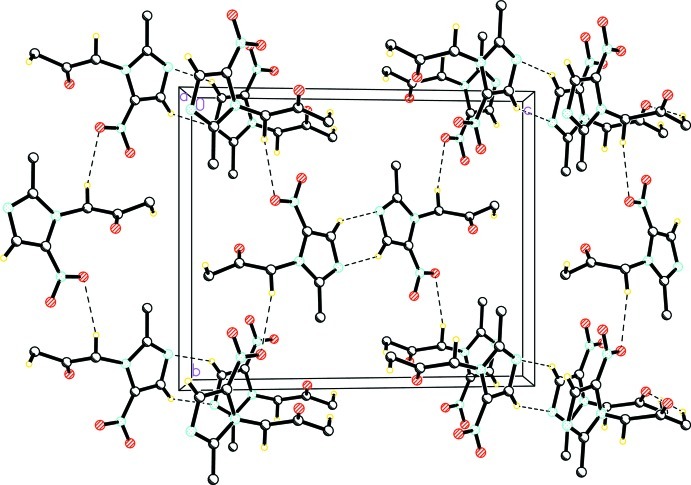
The structure of the title compound (Fig. 1) is similar to that of our previously published compound 2-(2-methyl-5-nitro-1H-imidazol-1-yl)-ethyl methanesulfonate (Zeb et al., 2012) with the difference that the ethyl methanesulfonate attached to the imidazole ring is replaced by an acetone (O3/C5—C7) group. Bond length and angles were found to be similar to those reported for related structures (Yousuf et al., 2012). In the crystal, molecules are linked by C2—H2B···N2, C5—H5B···O2 and C7—H7B···O3 intermolecular interactions (Table 1) to form a three-dimensional network (Fig. 2).
Experimental
Periodic acid (2.8 mmol, 0.64 g), pyridinium chlorochromate (PCC, 4 mol%) were suspended in acetonitrile (20 ml) and stirred vigorously for five minutes. The mixture was allowed to cool on an ice-salt bath followed by the addition of secnidazole (2.7 mmol, 0.50 g) and allowed to stir for 36 h at ambient temperature. After the completion of the reaction [TLC analysis], the reaction mixture was washed with brine/water (1:1 v/v), saturated aqueous Na2SO3 solution, dried (Na2SO4) and filtered. The filtrate was evaporated in vacuum to afford off-white crystals which were washed and recrystalized by dissolving in petroleum ether to obtained colorless crystals of the title compound (0.32 g, 64% yield) found suitable for single-crystal X-ray diffraction analysis.
Refinement
H atoms of methyl, methylene and methine carbon atoms were positioned geometrically with C—H = 0.93–0.96 Å and constrained to ride on their parent atoms with Uiso(H)= 1.2Ueq(C) or 1.5Ueq(C) for methyl H atoms. A rotating group model was applied to the methyl group.
Figures
Fig. 1.
The molecular structure of the title compound with displacement ellipsoids drawn at 30% probability level.
Fig. 2.
The crystal packing of the title compound. Intermolecular hydrogen bonds are shown as dashed lines.
Crystal data
| C7H9N3O3 | F(000) = 384 |
| Mr = 183.17 | Dx = 1.401 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 1790 reflections |
| a = 4.7548 (4) Å | θ = 2.8–26.7° |
| b = 12.3971 (9) Å | µ = 0.11 mm−1 |
| c = 14.8580 (11) Å | T = 273 K |
| β = 97.350 (2)° | Block, colorless |
| V = 868.62 (12) Å3 | 0.52 × 0.33 × 0.24 mm |
| Z = 4 |
Data collection
| Bruker SMART APEX CCD area-detector diffractometer | 1614 independent reflections |
| Radiation source: fine-focus sealed tube | 1328 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.019 |
| ω scan | θmax = 25.5°, θmin = 2.2° |
| Absorption correction: multi-scan (SADABS; Bruker, 2000) | h = −5→5 |
| Tmin = 0.944, Tmax = 0.974 | k = −14→15 |
| 5030 measured reflections | l = −14→17 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.122 | H-atom parameters constrained |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0591P)2 + 0.2124P] where P = (Fo2 + 2Fc2)/3 |
| 1614 reflections | (Δ/σ)max < 0.001 |
| 120 parameters | Δρmax = 0.19 e Å−3 |
| 0 restraints | Δρmin = −0.15 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | −0.1621 (4) | 0.29596 (12) | 0.33611 (13) | 0.0887 (5) | |
| O2 | 0.1562 (3) | 0.37418 (11) | 0.26792 (12) | 0.0791 (5) | |
| O3 | −0.2465 (3) | 0.53369 (11) | 0.15962 (9) | 0.0622 (4) | |
| N1 | −0.0008 (3) | 0.57463 (11) | 0.33181 (9) | 0.0424 (4) | |
| N2 | −0.2805 (3) | 0.59824 (13) | 0.43940 (10) | 0.0574 (4) | |
| N3 | −0.0337 (3) | 0.37691 (12) | 0.31638 (12) | 0.0589 (4) | |
| C1 | −0.1098 (3) | 0.47575 (13) | 0.35252 (12) | 0.0464 (4) | |
| C2 | −0.2784 (4) | 0.49243 (16) | 0.41793 (12) | 0.0554 (5) | |
| H2B | −0.3781 | 0.4389 | 0.4442 | 0.067* | |
| C3 | −0.1137 (4) | 0.64611 (14) | 0.38648 (11) | 0.0485 (4) | |
| C4 | −0.0467 (5) | 0.76284 (16) | 0.38973 (15) | 0.0728 (6) | |
| H4A | −0.1214 | 0.7951 | 0.4404 | 0.109* | |
| H4B | −0.1305 | 0.7965 | 0.3346 | 0.109* | |
| H4C | 0.1552 | 0.7725 | 0.3963 | 0.109* | |
| C5 | 0.1509 (3) | 0.60187 (13) | 0.25566 (11) | 0.0444 (4) | |
| H5A | 0.3240 | 0.5598 | 0.2597 | 0.053* | |
| H5B | 0.2034 | 0.6775 | 0.2595 | 0.053* | |
| C6 | −0.0235 (3) | 0.58087 (13) | 0.16507 (12) | 0.0451 (4) | |
| C7 | 0.1011 (4) | 0.62131 (17) | 0.08469 (13) | 0.0653 (5) | |
| H7A | −0.0270 | 0.6069 | 0.0307 | 0.098* | |
| H7B | 0.2783 | 0.5855 | 0.0810 | 0.098* | |
| H7C | 0.1326 | 0.6976 | 0.0905 | 0.098* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.1009 (12) | 0.0431 (8) | 0.1246 (15) | −0.0117 (8) | 0.0248 (10) | 0.0121 (8) |
| O2 | 0.0790 (10) | 0.0538 (9) | 0.1115 (13) | 0.0122 (7) | 0.0387 (9) | −0.0036 (8) |
| O3 | 0.0539 (8) | 0.0678 (9) | 0.0641 (9) | −0.0109 (6) | 0.0049 (6) | −0.0016 (6) |
| N1 | 0.0423 (7) | 0.0430 (8) | 0.0437 (8) | −0.0021 (6) | 0.0123 (6) | 0.0018 (6) |
| N2 | 0.0622 (9) | 0.0651 (10) | 0.0487 (9) | −0.0007 (7) | 0.0216 (7) | 0.0031 (7) |
| N3 | 0.0590 (9) | 0.0430 (9) | 0.0751 (11) | 0.0029 (7) | 0.0098 (8) | 0.0065 (7) |
| C1 | 0.0465 (9) | 0.0426 (9) | 0.0509 (10) | −0.0010 (7) | 0.0098 (7) | 0.0075 (7) |
| C2 | 0.0541 (10) | 0.0610 (12) | 0.0530 (11) | −0.0045 (9) | 0.0138 (8) | 0.0145 (9) |
| C3 | 0.0529 (9) | 0.0508 (10) | 0.0432 (9) | −0.0007 (8) | 0.0112 (8) | −0.0011 (7) |
| C4 | 0.0993 (16) | 0.0555 (12) | 0.0684 (14) | −0.0068 (11) | 0.0296 (12) | −0.0125 (10) |
| C5 | 0.0436 (8) | 0.0444 (9) | 0.0479 (9) | −0.0045 (7) | 0.0159 (7) | 0.0003 (7) |
| C6 | 0.0457 (9) | 0.0395 (9) | 0.0518 (10) | 0.0047 (7) | 0.0124 (7) | −0.0012 (7) |
| C7 | 0.0693 (12) | 0.0784 (14) | 0.0502 (11) | −0.0027 (10) | 0.0151 (9) | 0.0038 (10) |
Geometric parameters (Å, º)
| O1—N3 | 1.230 (2) | C3—C4 | 1.481 (3) |
| O2—N3 | 1.225 (2) | C4—H4A | 0.9600 |
| O3—C6 | 1.205 (2) | C4—H4B | 0.9600 |
| N1—C3 | 1.358 (2) | C4—H4C | 0.9600 |
| N1—C1 | 1.381 (2) | C5—C6 | 1.510 (2) |
| N1—C5 | 1.457 (2) | C5—H5A | 0.9700 |
| N2—C3 | 1.326 (2) | C5—H5B | 0.9700 |
| N2—C2 | 1.350 (3) | C6—C7 | 1.486 (3) |
| N3—C1 | 1.404 (2) | C7—H7A | 0.9600 |
| C1—C2 | 1.352 (2) | C7—H7B | 0.9600 |
| C2—H2B | 0.9300 | C7—H7C | 0.9600 |
| C3—N1—C1 | 104.93 (14) | C3—C4—H4C | 109.5 |
| C3—N1—C5 | 125.87 (14) | H4A—C4—H4C | 109.5 |
| C1—N1—C5 | 128.02 (14) | H4B—C4—H4C | 109.5 |
| C3—N2—C2 | 105.74 (15) | N1—C5—C6 | 112.47 (13) |
| O2—N3—O1 | 122.92 (17) | N1—C5—H5A | 109.1 |
| O2—N3—C1 | 119.63 (15) | C6—C5—H5A | 109.1 |
| O1—N3—C1 | 117.45 (17) | N1—C5—H5B | 109.1 |
| C2—C1—N1 | 107.35 (15) | C6—C5—H5B | 109.1 |
| C2—C1—N3 | 127.87 (16) | H5A—C5—H5B | 107.8 |
| N1—C1—N3 | 124.56 (15) | O3—C6—C7 | 123.21 (16) |
| N2—C2—C1 | 109.97 (15) | O3—C6—C5 | 121.44 (15) |
| N2—C2—H2B | 125.0 | C7—C6—C5 | 115.35 (14) |
| C1—C2—H2B | 125.0 | C6—C7—H7A | 109.5 |
| N2—C3—N1 | 112.01 (16) | C6—C7—H7B | 109.5 |
| N2—C3—C4 | 124.07 (16) | H7A—C7—H7B | 109.5 |
| N1—C3—C4 | 123.86 (16) | C6—C7—H7C | 109.5 |
| C3—C4—H4A | 109.5 | H7A—C7—H7C | 109.5 |
| C3—C4—H4B | 109.5 | H7B—C7—H7C | 109.5 |
| H4A—C4—H4B | 109.5 | ||
| C3—N1—C1—C2 | −0.39 (18) | C2—N2—C3—N1 | −0.6 (2) |
| C5—N1—C1—C2 | −168.41 (15) | C2—N2—C3—C4 | −177.73 (19) |
| C3—N1—C1—N3 | −175.31 (16) | C1—N1—C3—N2 | 0.60 (18) |
| C5—N1—C1—N3 | 16.7 (3) | C5—N1—C3—N2 | 168.96 (14) |
| O2—N3—C1—C2 | −168.38 (18) | C1—N1—C3—C4 | 177.77 (18) |
| O1—N3—C1—C2 | 11.0 (3) | C5—N1—C3—C4 | −13.9 (3) |
| O2—N3—C1—N1 | 5.5 (3) | C3—N1—C5—C6 | −106.10 (18) |
| O1—N3—C1—N1 | −175.14 (16) | C1—N1—C5—C6 | 59.6 (2) |
| C3—N2—C2—C1 | 0.3 (2) | N1—C5—C6—O3 | −9.0 (2) |
| N1—C1—C2—N2 | 0.1 (2) | N1—C5—C6—C7 | 171.59 (15) |
| N3—C1—C2—N2 | 174.76 (17) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2B···N2i | 0.93 | 2.56 | 3.361 (2) | 144 |
| C5—H5B···O2ii | 0.97 | 2.57 | 3.527 (2) | 167 |
| C7—H7B···O3iii | 0.96 | 2.49 | 3.340 (2) | 147 |
Symmetry codes: (i) −x−1, −y+1, −z+1; (ii) −x+1/2, y+1/2, −z+1/2; (iii) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: RZ5048).
References
- Almirall, P., Escobedo, A. A., Ayala, I., Alfonso, M., Salazar, Y., Cañete, R., Cimerman, S., Galloso, M., Olivero, I., Robaina, M. & Tornés, K. (2011). J. Parasitol. Res., Article ID 636857, doi:10.1155/2011/636857. [DOI] [PMC free article] [PubMed]
- Bruker (2000). SADABS, SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Lin, Y., Su, Y., Liao, X., Yang, N., Yang, X. & Choi, M. M. F. (2012). Talanta, 88, 646–652. [DOI] [PubMed]
- Nardelli, M. (1995). J. Appl. Cryst. 28, 659.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Yousuf, S., Zeb, A., Batool, F. & Basha, F. Z. (2012). Acta Cryst. E68, o2781. [DOI] [PMC free article] [PubMed]
- Zeb, A., Yousuf, S. & Basha, F. Z. (2012). Acta Cryst. E68, o1218. [DOI] [PMC free article] [PubMed]
- Zhang, H.-J., Zhu, D.-D., Li, Z.-L., Sun, J. & Zhu, H. L. (2011). Bioorg. Med. Chem. 19, 4513–4519. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813006569/rz5048sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813006569/rz5048Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813006569/rz5048Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report