Introduction
The last few years have seen an explosion of interest in the new field of cellular reprogramming. Given the extensive clinical experience with stem cells in bone marrow transplant, hematologists are particularly excited about future clinical opportunities for reprogrammed cell therapies. Here, we compare the benefits of induced and embryonic pluripotent stem cells, discuss current approaches to convert pluripotent stem cells into hematopoietic cells, and describe the utility of disease models for learning about and treating disease. These comments will provide an update for clinicians and scientists interested in the progress of stem cell research towards the clinic.
History of reprogramming
The first derivation of human embryonic stem (ES) cells in 1998 captured the imaginations of clinicians and scientists alike (1). These pluripotent cells had the ability to differentiate into any cell in the human body, suggesting revolutionary approaches to learning about and treating human disease. But research into this promising avenue of investigation flagged as opposition sprang up from opponents of embryo research. New policies severely limited federal funding of ES cell studies, impeding US scientists’ ability to work on this versatile new cell type.
With American research hobbled, scientists from around the globe asserted strong leadership positions in the field. Major stem cell research initiatives grew in the United Kingdom, Israel, Singapore, and Japan, fueling the excitement of stem cell communities located in these countries (2). One flagship center at Kyoto University, the Institute for Frontier Medical Sciences, was founded in 1998 with the goal of advancing the field of regenerative medicine by characterizing ES cells. At this institute, Shinya Yamanaka began tinkering with ES cells and trying to recreate their hitherto unmatched pluripotency. Through an ingenious series of experiments, Yamanaka and colleagues developed a new technology that can convert fibroblasts and other somatic cells into induced pluripotent stem (iPS) cells (3).
Yamanaka’s breakthrough research built upon previous demonstrations that one cell could be turned into another by expressing transcription factors specific to the target cell type: for example, expression of the muscle-specific transcription factor MyoD is sufficient to convert fibroblasts into muscle progenitor cells (4). Yamanaka and a graduate student named Kazutoshi Takahashi hypothesized that they could convert fibroblasts into pluripotent stem cells by forcing them to express embryonic transcription factors. To observe what they anticipated would be a very rare event, they used cells from a strain of mice that carried an antibiotic resistance gene under the control of an embryonic gene promoter (3). Adult cells from these mice would thus become resistant to antibiotics only if they adopted embryonic-like gene expression. By infecting these cells with retroviruses containing candidate genes, Takahashi and Yamanaka discovered combinations of transcription factors that conferred antibiotic resistance by activating an embryonic gene expression program. With this tool, they were able to establish that specific transcription factors could convert differentiated tissues into pluripotent stem cells.
Twenty-four genes involved in pluripotent cell identity were chosen as candidates for induction of pluripotency. No single factor was able to induce antibiotic resistance, but when all 24 were expressed at the same time, some rare cells successfully activated embryonic expression patterns and acquired resistance to the antibiotic. When these cells were grown in culture, about half of them demonstrated characteristics of pluripotent stem cells including morphology, growth rate, and expression of key embryonic genes. These cells were dubbed induced pluripotent stem (iPS) cells (3).
After this successful initial reprogramming of fibroblasts into pluripotent stem cells, the investigators began to narrow down the field of responsible genes. They infected cells with viruses containing all possible combination of 23 genes, leaving 1 gene out each time; those experiments that failed thus identified the genes that were required for reprogramming. This led to the identification of 4 genes as indispensable for efficient reprogramming: Oct3/4, Sox2, Klf4, and c-Myc (OSKM) (3). These genes are now colloquially referred to as the “Yamanaka factors” and comprise the 4 genes most commonly used to induce pluripotency.
The initial mouse iPS cells were evaluated for pluripotency by multiple assays. First, cell surface markers were investigated, which demonstrated the similarities between iPS and ES cells. Then microarrays comparing gene expression profiles between iPS and ES cells demonstrated that although the cell types were distinguishable, they shared virtually all characteristic expression patterns. Next, teratoma assays showed that the iPS cells were capable of differentiating into cell types of all three germ layers, a crucial assay to prove their pluripotency. Finally, the authors established that when iPS cells were injected into blastocysts, they contributed to all three germ layers in developing embryos (3). Since the initial report, murine iPS cells’ pluripotency has been further confirmed by the birth of live chimeras, germline transmission, and the most stringent test for pluripotency, tetraploid complementation, which entails injecting pluripotent cells into engineered tetraploid embryos, allowing the iPS cells to grow into a complete mouse (5-8).
Within 18 months of publication of the seminal paper describing mouse reprogramming, 3 independent laboratories reported successful derivation of human iPS cells (9-11), and shortly thereafter 2 groups produced iPS cells from patients with a multitude of diseases (12, 13). By this point in mid-2008, it was clear that Takahashi and Yamanaka’s new reprogramming technology was destined to revolutionize the field of medicine. The power to create patient-specific cells promised to provide invaluable models of human disease for in vitro research, and offered the prospect of autologous, rejection-proof cell transplantation therapies.
Relative merits of iPS and ES cells
Clinical
The chief safety concern of any iPS or ES cell-derived therapy is the risk of cancer. Pluripotent cells are characterized by the potential to form teratomas, typically benign encapsulated masses of disorganized tissues from all three embryonic germ layers— ectoderm, mesoderm, and endoderm. Should cell therapies from pluripotent sources be contaminated by any residual undifferentiated cells, then the risk of teratoma formation, or various growths due to partially or incompletely differentiated cells, remains. These risks have been recently reviewed in depth (14). From this perspective, iPS cells derived using the traditional viral approach are more worrisome than ES cells, because reprogramming genes function as oncogenes in pathologic contexts. Not only can viruses lead to insertional mutagenesis, but any cell with transgenic oncogenes in its genome stands the risk of future reactivation (15). Because of this concern, much effort has been put into creative approaches for deriving iPS cells without genomic modifications. The resulting transgene-free iPS cells are less likely to provoke safety concerns and are a better resource for therapeutics.
iPS cells have an important clinical advantage over ES cells in one important respect: the genome of an iPS cell matches the genome of the patient from whom it was derived. Thus, if transplantable tissues can be derived from iPS cells, they will be less apt to face immune rejection than tissues derived from allogeneic ES cells (16, 17). This advantage may be maintained even in cases where a patient’s disease is genetic. Because pluripotent cells expand indefinitely, scientists are working on developing gene targeting techniques to repair known genetic mutations (see Sidebar). The resulting iPS cells are isogenic to the patient but lack the disease-causing mutation.
The effect of reprogramming on genomic integrity is an active area of research. A few papers have demonstrated that early-passage iPS cells are likely to have more point mutations in protein-coding genes as well as more copy number variations than ES cell lines (18, 19). This suggests that before iPS cells can be used therapeutically, either the reprogramming process must be improved or each individual iPS line must undergo comprehensive testing for genomic changes induced by reprogramming.
Scientific
From a scientific perspective, iPS technology has greatly expanded the field of human genetic disease modeling. Before 2007, the only way to obtain human pluripotent stem cells carrying a particular genetic disease was to recruit parents undergoing pre-implantation genetic diagnosis and generate ES cells from their discarded blastocysts (20, 21). Now using a reprogramming approach, researchers can generate iPS cells from patients bearing virtually any remarkable genotype. This change is reflected in the large and growing number of disease-specific human pluripotent stem cell lines that have to date been described (22). These patient-specific cell lines are an invaluable tool for scientists to probe the pathology of disorders and identify novel therapeutic approaches.
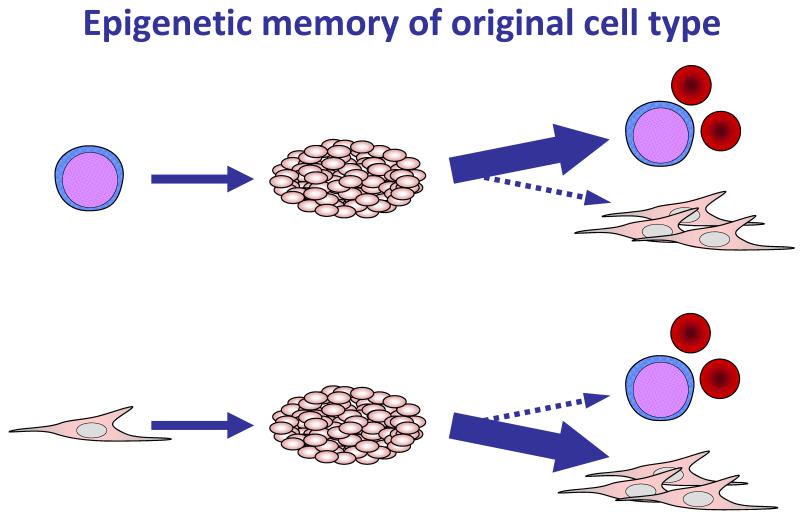
One challenge iPS cells pose to researchers is their sometimes partially reprogrammed state, which manifests as a sort of “epigenetic memory”, whereby iPS lines retain epigenetic signatures of their tissue of origin, and are most effective at differentiating into the tissue type from which they were originally derived (Figure 1). For example, some iPS lines derived from blood cells are better at differentiating into blood than are iPS lines derived from skin cells (23, 24). This feature suggests that some strains of iPS cells have not reached a true pluripotent state, and that further research into the epigenetics underlying reprogramming must be done before iPS cells can truly be as unbiased as ES cells.
Figure 1. Epigenetic memory of original cell type.
iPS cell lines differ in their ability to differentiate towards various cell lineages. In many cases, iPS cell lines differentiate more robustly towards cell types related to the lineage from which they were reprogrammed than towards unrelated cell types (23, 24). This phenomenon is referred to as epigenetic memory.
Ethical
Unlike ES cell isolation, derivation of iPS cells does not involve human blastocysts (1, 9). As some find research on human embryos unethical, it has been suggested that reprogrammed cells should replace all embryonic-derived cells in laboratories. However, research into the similarities and differences of iPS and ES cells is ongoing, and it has become clear that the two cell types are similar but not identical (25-27). As scientists learn more about human ES cells, they will be able to improve reprogramming and generate iPS cells that resemble them more closely. This goal, however, requires continued research on ES cells.
Logistical
From a regulatory, logistical, and financial standpoint, iPS cells are less cumbersome to employ in research than are ES cells. Derivation of ES cells is subject to close regulation by Embryonic Stem Cell Research Oversight Committees, and in the United States, research funding from the federal government is restricted to a limited subset of ES cell lines that have passed scrutiny by an ethical review committee of the National Institutes of Health. Thus, in addition to the considerable advantages of iPS cells for disease research applications, they likewise provide significant practical advantages relative to ES cells.
The blood system as a starting point
Currently, only one stem cell-based therapy is widely accepted and utilized worldwide: hematopoietic stem cell transplantation. This practice was pioneered by Dr. E. Donnall Thomas, who in 1959 demonstrated that the bone marrow from one identical twin could reconstitute the blood system of her sister, who suffered from leukemia (28). Initially, treatment success was limited to those rare patients who had identical twins, as transplantation of bone marrow from non-identical donors resulted in severe, systemic attack of the new immune system against the recipient’s organs, a condition termed graft-versus-host disease (GvHD). After ten years of exhaustive research into HLA-matching and immunosuppression, Dr. Thomas’ team successfully performed a bone marrow transplant (BMT) from one non-twin sibling to another (29). This extraordinary feat earned Dr. Thomas a Nobel Prize in 1990 for his contributions to cellular transplantation.
Since 1959, the practice of hematopoietic stem cell transplantation has become widespread. Advanced and inexpensive HLA-typing has allowed for the collection of millions of would-be bone marrow donors in national registries, and an increased understanding of hematology and immunology has lessened the risks associated with transplant. However, even with high-quality HLA matches, BMT still carries a treatment-related mortality of 5 – 20% in the first few months (30, 31). The dangers haunting this period include extreme neutropenia and consequent infections, failure of the transplant to engraft, and severe GvHD. Various features of the transplant determine the likelihood of these complications, including the number of hematopoietic stem cells that are transplanted and the degree of mismatches at both major and minor histocompatability loci. Most patients who survive the transplant itself have a good long-term prognosis, with overall 5-year survival rates around 70 - 80% for non-cancer patients (32-35).
The remarkable ability of hematopoietic stem cells to repopulate the entire blood system allows for a few hundred milliliters of donor marrow to reconstitute an entire organ in the recipient. Following intravenous infusion, the transplanted hematopoietic stem cells home to the bone marrow and engraft into the niches already present there. The blood system is thus an attractive starting place for working towards iPS cell-derived cellular therapy because no cellular organization is required for transplant to occur. Combined with bone marrow transplant’s long history and established clinical efficacy, the hematopoietic stem cell is arguably more feasible for clinical application than stem cells from any other organ system.
Conversion of pluripotent stem cells to transplantable hematopoietic stem cells
The quest to convert human pluripotent stem cells into functional, engraftable hematopoietic stem cells (HSCs) has been challenging and has not yet proven successful. This complex task requires a thorough understanding of the developmental events that lead to formation of HSCs in vivo and approaches to mimic these events in the laboratory.
Mouse models guide thinking
Because hematopoietic development in humans and mice bears striking similarities, researchers have turned to the mouse to learn about mammalian blood development. It is hoped that an exhaustive understanding of the signaling, migration, and molecular events in the developing mouse hematopoietic system will inform our ability to direct similar development in vitro from human pluripotent stem cells.
In both mouse and human, the first hematopoietic cells are found in the extra-embryonic yolk sac. These are referred to as primitive hematopoietic progenitors because the blood cells they produce have more embryonic characteristics than those that arise later in development. This is especially true of early red blood cells, which express embryonic forms of hemoglobin with greater affinity for oxygen, as appropriate for the hypoxic fetal environment. Around day 10.5 of mouse development, the first HSCs arise in the aortagonad-mesonephros region of the developing mouse embryo (36, 37). Hemogenic endothelium forms on the ventral wall of the aorta, squeezing the first true HSCs into the growing circulatory system (38-40). These definitive HSCs, possibly along with some primitive progenitors from the yolk sack, colonize the fetal liver (41). The main function of the liver appears to be to provide a permissive environment for the HSCs to grow and expand, as their numbers increase greatly at this stage of development (42). Finally, HSCs colonize the bone marrow, where they will be responsible for maintaining the immune and blood systems for the rest of the animal’s life (43).
Current in vitro approaches fall short of creating human HSCs
Two culture systems are currently employed when attempting to coax mammalian pluripotent stem cells down the path of hematopoietic differentiation: two-dimensional culture on supportive stromal cells, and aggregation of stem cells into three-dimensional balls of cells called embryoid bodies (EBs) (44, 45). In both approaches, exogenous cytokines are applied at strict intervals and concentrations to mimic the evolution of cellular identity that occurs during development. If these mouse cells are infected with viruses expressing homeodomain-containing transcriptional regulators like Cdx4 and/or HoxB4, these systems create cells that fulfill the definition of HSCs: long-term, multilineage engraftment (46-48). However, the activity of these factors does not appear to be as effective in human lines as it is in the mouse background (49, 50). Both EB creation and co-culture with stromal cells lead human pluripotent stem cells to differentiate into the hematopoietic lineage, as measured by robust expression of CD34, CD45, and other hematopoietic markers (50, 51). However, when these cells are transplanted into immunodeficient mice, they fail to generate stable, high level, long-term engraftment (50). The reasons for the observed differences between mouse and human pluripotent-to-hematopoietic stem cell differentiation are active areas of inquiry. Achieving stable engraftment of human HSCs derived from pluripotent stem cells remains a major unmet goal in hematology research.
iPS cells in treatment and research
Treatment
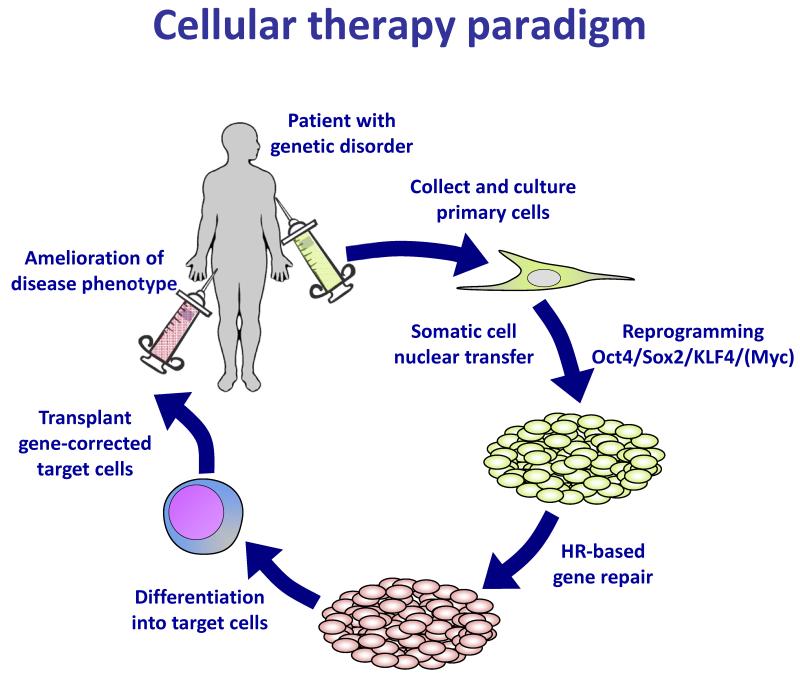
Reprogramming holds tremendous potential for clinical application. The inherent pluripotency of iPS cells, along with their genetic identity to specific patients, stokes the idea of providing cells or cellular products to patients suffering from a myriad of disorders. The theoretical applications of iPS cell technology extend to any disorder characterized by loss of a key cellular function, including the loss of dopaminergic neurons in Parkinson’s disease, the loss of beta cells in type I diabetes, and the loss of hematopoietic stem cells in aplastic anemias. In these disorders of cellular deficiency, the therapeutic paradigm is to create iPS cells from a patient, differentiate these cells into the relevant cell or tissue that has been destroyed by disease, and transplant the rejuvenated cells back into the patient to replace the missing tissue (Figure 2).
Figure 2. Cellular therapy paradigm.
This figure outlines the goals of stem cell-based regenerative medicine, first demonstrated in mouse by somatic cell nuclear transfer in 2002 (52) and iPS cell derivation in 2007 (53). The paradigm combines gene therapy and cellular therapy to provide a gene-corrected, autologous cell transplant to a patient.
This transplantation approach is unlikely to treat disorders that relate to complex cellular disorganization, such as developmental disorders of the brain, or disorders with a gain of activity, such as cancer, as these latter diseases have no obvious target for a replacement-based cure. Instead, these types of disorders are likely to benefit from iPS cell-based research models, which can recreate characteristics of the disease in easily studied in vitro systems, hopefully allowing for improvement of current therapies. Below, we describe how iPS cell transplants are anticipated to be used therapeutically by focusing on an early paper in the mouse hematopoietic system, and describe the current state of research into each of the hurdles faced before the technology can enter the clinic.
Research
Scientists can now create a pluripotent stem cell that bears the genotype of any person. The only necessary starting material is a small cell sample, allowing for creation of iPS cells from patients with any disease. As the field advances, the bar for publication of iPS cell-based disease research is being raised closer and closer to the ultimate goal of true disease modeling.
When the reprogramming field was in its infancy, the first papers simply demonstrated the successful creation of an iPS line from a patient with a genetic disorder. Now with reprogramming technology more widely available, numerous examples exist in which investigators have both generated patient-specific iPS cells and differentiated them into the cell type affected by the disease. In the future, we will see an exciting increase in the number of reports that identify a disease-specific phenotype in the patient-derived cells. These models, in turn, will lead to revelations about disease mechanisms and provide a basis for chemical screens to identify novel drug compounds. Below, we describe the essential features of a successful disease model and provide current examples in the hematopoietic system.
Treating diseases using stem cell-based therapies: an overview
Many clinicians and scientists hope that iPS cells will one day be used to provide patient-specific cellular therapy by generating autologous cells through reprogramming, correcting gene defects, differentiating the repaired cells into the disease relevant tissue, and returning healthy cells to the patient. The conceptual foundations for this approach were laid in 2002 with the first publication to progress “full circle” through the paradigm diagrammed in Figure 2 (52).
This paper focused on a murine genetic immunodeficiency caused by homozygous deletion of the Rag2 gene. Cell nuclei from an affected mouse were transplanted into enucleated oocytes by micromanipulation by a method called somatic cell nuclear transfer, the same technique that led to the cloning of Dolly the sheep. The goal of using nuclear transfer to treat disease, on the other hand, was to generate an embryo autologous to the donor, which then gave rise to a pluripotent stem cell line in vitro. The genetic defect was corrected using homologous recombination-based gene targeting, and the corrected cells were differentiated into hematopoietic progenitors. When these progenitors were transplanted into the mouse, they engrafted and began to produce immune cells, thereby establishing the proof of principle for treating a genetic blood disease by combined gene and cell therapy (52).
Within a year of the advent of reprogramming technology, this paradigm was adapted to iPS cells. Dr. Rudolf Jaenisch’s group started with a mouse with the genetic blood disease sickle cell anemia, and cured it using iPS-derived cells (53). Below, we will describe each step accomplished in the mouse and summarize the current state of research for applying these techniques to human patients.
The authors began by collecting fibroblasts from mice engineered to express the human sickle hemoglobin genes, then reprogramming them to iPS cells using retroviruses. As mentioned above, this viral approach carries risks of insertional mutagenesis and oncogene reactivation, and thus would not be suited to human clinical applications. Current methods of generating transgene-free, genetically pristine iPS cells include transfection with synthetic mRNA, or self-replicating episomal vectors, or infection with non-integrating viruses such as Sendai (54-56). These approaches have proven efficacious in human cells, and work is ongoing to determine which of these will become the mainstay of clinical grade reprogramming techniques.
Once the murine iPS cells had been generated, one copy of the mutant hemoglobin genes was repaired by homologous recombination. This gene targeting technique is more robust in mouse cells than human cells, but many laboratories are working on improving its efficiency in human cells (see Sidebar). Currently, gene targeting in human cells is only effective if a sequence-specific nuclease can be designed to target the locus of interest (57). Before this technique can be moved into the clinic, the off-target effects of these nucleases must be measured and minimized.
The gene-corrected mouse iPS cells were then differentiated into hematopoietic progenitors. As discussed above, mouse pluripotent stem cells can be converted into engraftable hematopoietic progenitors by first allowing them to self-assemble into cystic ball-like structures that are composed of many different tissues, including hematopoietic cells, and then subsequently infecting the blood components with a virus encoding the HoxB4 gene, which is known to expand blood cells. The resulting hematopoietic progenitors were transplanted into an irradiated sickle cell mouse, and the recipient mice showed remarkable clinical recovery: a decrease in reticulocytosis and anisocytosis, fewer misshapen cells on blood smears, and improved kidney function (53). This improvement was observed at up to 12 weeks after transplantation.
Of course, it will not be possible to perform a similar trial in humans until researchers address the many challenges. For example, given their origins in pluripotent cells, the nature of all transplanted cells must be stringently assessed to ensure no undifferentiated cells remain which could form teratomas or other neoplasms. For treatment of non-malignant disorders such as sickle cell anemia, ideal transplantation preparative regimens must be determined that ensure engraftment while minimizing the dangerous side effects of bone marrow transplant. Finally, clinicians must monitor for any aberrant behavior of the transplanted cells, whether due to renegade pluripotent cells or enduring deleterious epigenetic memory. For this purpose, some researchers advocate inclusion of a “suicide gene” into transplanted cells, rendering them uniquely sensitive to a drug that could be applied in the case of a transplant gone awry. This and many other issues must be addressed before iPS cells will be used to treat hematological disease.
Learning about diseases using stem cell-based models: an overview
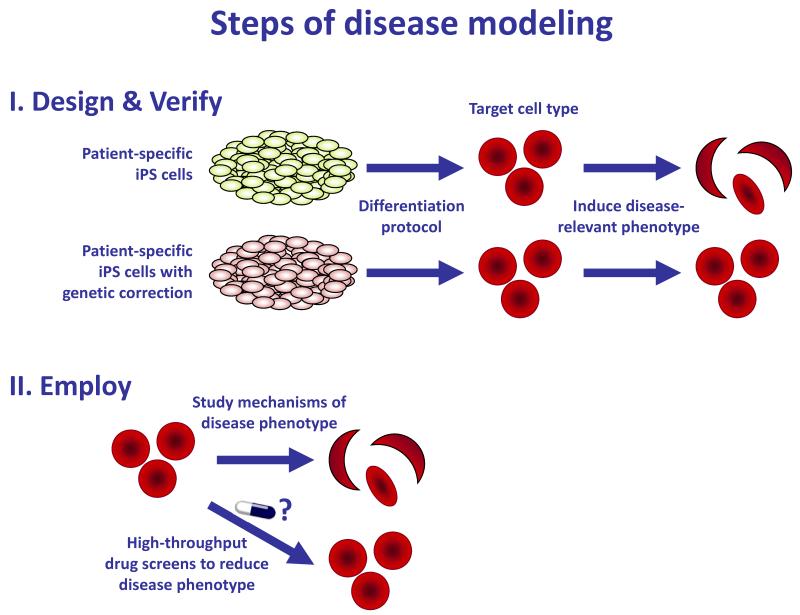
In addition to cellular therapeutics, researchers hope to create disease-specific iPS cells to enable research into disease mechanisms and screening for novel drugs, outlined in Figure 3. Very few papers to date have successfully drawn conclusions about diseases, largely because the process of establishing and verifying disease models is so demanding.
Figure 3. Steps of disease modeling.
I) Design and verify the disease model. Because of the large variability in behavior of pluripotent cell lines (23, 58, 59), disease models should be established using only genetically identical, disease-corrected controls. Robust protocols for differentiating iPS cells into the target tissues must be established, and a cellular phenotype must be identified that is both specific to the cells bearing the disease genotype and relevant to the disorder. II) Employ the disease model. Once a disease-specific cellular phenotype has been identified, the mechanism of the disease can be investigated. The model can also be assayed after experimental manipulations such as drug screens, in order to identify potential therapeutic agents.
Designing a disease model
A number of considerations go into design and testing of a disease model. During the process of reprogramming, a cell’s epigenome is reset and all environmental conditions are normalized, so that the only disease risk factors carried by the resulting iPS cells are those encoded in its genome (22). With this in mind, most researches have chosen to create their first disease models either from strictly genetic diseases (i.e. trisomy 21, Shwachman-Diamond syndrome, and dyskeratosis congenita) or from a genetic version of a more complex disease (i.e. the SOD1-mutant familial amyotrophic lateral sclerosis, the PINK1-mutant familial Parkinson’s disease, and the APP-mutant familial Alzheimer’s disease). By ensuring that the iPS cells carry the predisposition to disease, researchers are hoping they will be able to identify a disease-related phenotype when they differentiate the cells.
The goal of an iPS cell-based disease model is to identify a phenotype that distinguishes the disease-bearing cells from the control cells. However, it is often not clear what cells are the appropriate controls. Independent iPS or ES cell lines often show variation in their differentiation tendencies and abilities (58, 59). Some inconsistency may be due to underlying genetic variations between the patient and the control, but some is likely due to the particular nature of a given pluripotent cell line. Thus, the best control for any disease model is to genetically correct the disease genotype in the patient’s affected iPS line. Gene correction can be achieved by viral rescue, homologous recombination, or by genome editing (see Sidebar). The disease allele-carrying and the repaired lines are then isogenic, distinguishable only by the status of the disease gene. Any phenotypic discordance observed in cells differentiated from these lines can be immediately ascribed to the disease genotype.
In order to create an effective disease model, scientists must choose a cell type to investigate, develop or adopt a differentiation protocol to create that cell type, acquire a genetically-matched control, and identify a disease-related cellular phenotype.
Utilizing a disease model
iPS cell-based disease models are expected to contribute to disease research in two broad ways: as research tools for mechanistic studies and as the basis of drug screens. Scientists developing iPS cell-based models are just beginning to progress to the point of utilizing their models, and here we will describe an attempt in our lab to employ an iPS cell-based model of the genetic disorder Shwachman-Diamond syndrome (SDS).
Attributable to mutations in the SBDS gene, patients with SDS suffer from bone marrow failure and exocrine pancreas insufficiency. The mechanisms by which mutations in the SBDS gene lead to these phenotypes are unknown. To model the disease, we have followed the steps described above. We differentiated our iPS cells into both hematopoietic and pancreatic cell types, using protocols for each cell type previously published by other labs. Gene-correction was performed to obtain healthy, otherwise-identical iPS lines for controls, and remarkably, we found a disease-specific phenotype shared by both the hematopoietic and pancreatic cells bearing the disease mutation. Compared to the gene-corrected cells, cultures of cells containing the SBDS mutation were laden with granules—azurophilic granules for the myeloid lineage, and zymogen granules in the exocrine pancreatic cells. We hypothesized that because the granules in both myeloid and exocrine cells normally contain toxic proteases, perhaps the SBDS protein was required for successful granule maturation or expulsion. If this were the case, an SBDS mutation would be predicted to cause a buildup of granules and eventual poisoning of the affected cells by protease autodigestion, which could explain these organs’ failure in patients with SDS. Indeed, measurement of the protease content of SDS cell cultures revealed a marked excess in the culture media compared to gene-repaired controls. We then tested a number of protease inhibitor compounds, and observed that treatment restored the SBDS-mutant cellular phenotypes back to the levels seen in the gene-corrected cells. This suggests that the described disease model has identified a novel class of drugs to be tested for efficacy in SDS.
Thus, a validated disease model can be used to both learn about the biology underlying a pathologic condition and identify new approaches to drug therapy. Although the work discussed here only assayed a few compounds, researchers are starting to use iPS-derived cells as the basis for high-throughput screens. This work is anticipated to lead to novel drugs for myriad conditions.
Conclusions
Scientists are exploiting iPS cell technology to create models of dozens of genetic diseases. The insights that arise from these models will include theories of disease mechanisms and novel inspiration for therapeutic drugs. These advances will accrue for conditions that affect all organ systems, but our bias is to believe that the disease area likely to benefit soonest from cellular therapies is hematology. For hematopoietic cell therapy, the biggest challenge remains to convert human pluripotent stem cells into hematopoietic stem cells. Once this is accomplished, reprogramming-based cellular therapy will be a powerful strategy to attack any of dozens of genetic disorders of the blood that are currently treated inadequately by bone marrow transplantation. Thus, bold new approaches to regenerative medicine in hematology may, in time, be expanded to many other areas of medicine.
Sidebar: Genetic correction in patient cells
The goal of gene therapy is to remedy genetic deficiencies in patients by providing a healthy copy of the affected gene into a relevant cell or tissue. Three broad approaches are currently available to accomplish genetic correction: 1) virus-mediated gene transfer; 2) homologous recombination (HR)-based gene targeting; 3) nuclease-mediated genome editing.
Virus-mediated gene transfer takes advantage of a virus’s ability to permanently integrate a transgene into the host cell’s genome. But because viruses integrate at random locations, this method is prone to insertional mutagenesis and transgene misregulation (60). A cleaner approach would be to repair the mutation directly by converting the patient’s mutated DNA code into a healthy sequence.
HR-based gene targeting uses the cell’s DNA repair machinery to replace the cell’s mutant sequence with a wild-type sequence from a healthy template (61). Using traditional techniques, target cells successfully repaired by HR are so rare that this technique can only be applied to immortalized that can be expanded indefinitely in vitro. Unlike hematopoietic stem cells, which are not readily expanded in culture, iPS cells can be successfully altered using HR-based gene targeting (57). Recent papers have demonstrated successful gene targeting in iPS cells from patients with sickle cell anemia, progeria, and paroxysmal nocturnal hemoglobinuria (62-65).
Genome editing is an emerging technology that takes advantage of site-specific nucleases that introduce DNA strand breaks within the target gene of interest, thereby markedly enhancing the frequency of HR (57, 65-68). Genome editing promises to revolutionize the capacity for gene correction in vitro and possibly in vivo.
Acknowledgements
GQD is supported by grants from the NIH (UO1-HL100001, RC4DK090913, R24DK092760, and special funds from the ARRA stimulus package-RC2-HL102815), Alex’s Lemonade Stand, Ellison Medical Foundation, and the Doris Duke Medical Foundation. GQD is an affiliate member of the Broad Institute, and an investigator of the Howard Hughes Medical Institute and the Manton Center for Orphan Disease Research.
Acronyms and definitions list
- Pluripotent
Able to differentiate into cell types of all three germ layers, ectoderm (e.g. skin, nerves), endoderm (e.g. gut, liver), and mesoderm (e.g. blood, muscle, bone).
- Differentiate
Transition towards a more specialized cellular identity.
- Reprogram
Transition from a somatic cell into a pluripotent stem cell.
- Yamanaka factors
The four factors originally used by Shinya Yamanaka of Kyoto University in Japan to reprogram fibroblasts: Oct3/4, Sox2, Klf4, and c-Myc.
- Teratomas
Tumors that are comprised of cells from all three germ layers. They are formed by pluripotent stem cells.
- Bone marrow or hematopoietic stem cell transplant (BMT or HSCT)
Transplantation of hematopoietic stem cells from a donor into a recipient in order to reconstitute the blood system.
- Graft vs. host disease (GvHD)
After a BMT, the new immune system often recognizes the recipient’s tissues as “non-self” and attacks them, causing GvHD.
- Home
The process by which a stem cell migrates to and incorporates within the appropriate niche after transplant.
- Engraftment
When a transplanted stem cell makes a measurable contribution to organ function after transplant.
- Hematopoietic stem cell (HSC)
A single cell capable of reconstituting the entire blood system.
- Disease-related phenotype
A measurable cellular phenotype that is associated with the disease.
Works Cited
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–47. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Gottweis H, Prainsack B. Emotion in political discourse: contrasting approaches to stem cell governance in the USA, UK, Israel and Germany. Regenerative Medicine. 2006;1:823–29. doi: 10.2217/17460751.1.6.823. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K,, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Davis RL, Weintraub H, Lassar AB. EXPRESSION OF A SINGLE TRANSFECTED CDNA CONVERTS FIBROBLASTS TO MYOBLASTS. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 5.Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–U2. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 6.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–U1. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 7.Zhao XY, Li W, Lv Z, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–U88. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 8.Boland MJ, Hazen JL, Nazor KL, et al. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–U94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 10.Yu JY, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 11.Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–U1. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 12.Dimos JT, Rodolfa KT, Niakan KK, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–21. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 13.Park IH, Arora N, Huo H, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–86. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nature Reviews Cancer. 2011;11:268–77. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- 15.Zhao R, Daley GQ. From Fibroblasts to iPS Cells: Induced Pluripotency by Defined Factors. Journal of Cellular Biochemistry. 2008;105:949–55. doi: 10.1002/jcb.21871. [DOI] [PubMed] [Google Scholar]
- 16.Nishikawa S, Goldstein RA, Nierras CR. The promise of human induced pluripotent stem cells for research and therapy. Nature Reviews Molecular Cell Biology. 2008;9:725–29. doi: 10.1038/nrm2466. [DOI] [PubMed] [Google Scholar]
- 17.Lerou PH, Daley GQ. Therapeutic potential of embryonic stem cells. Blood Reviews. 2005;19:321–31. doi: 10.1016/j.blre.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Gore A, Li Z, Fung HL, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–U76. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussein SM, Batada NN, Vuoristo S, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–U67. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 20.Alikani M, Munne S. Nonviable human pre-implantation embryos as a source of stem cells for research and potential therapy. Stem Cell Reviews. 2005;1:337–43. doi: 10.1385/SCR:1:4:337. [DOI] [PubMed] [Google Scholar]
- 21.Tropel P, Tournois J, Come J, et al. High-efficiency derivation of human embryonic stem cell lines following pre-implantation genetic diagnosis. In Vitro Cellular & Developmental Biology-Animal. 2010;46:376–85. doi: 10.1007/s11626-010-9300-8. [DOI] [PubMed] [Google Scholar]
- 22.Cherry ABC, Daley GQ. Reprogramming Cellular Identity for Regenerative Medicine. Cell. 2012;148:1110–22. doi: 10.1016/j.cell.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–U60. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polo JM, Liu S, Figueroa ME, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nature Biotechnology. 2010;28:848–U130. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin MH, Mason MJ, Xie W, et al. Induced Pluripotent Stem Cells and Embryonic Stem Cells Are Distinguished by Gene Expression Signatures. Cell Stem Cell. 2009;5:111–23. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doi A, Park IH, Wen B, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nature Genetics. 2009;41:1350–U123. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guenther MG, Frampton GM, Soldner F, et al. Chromatin Structure and Gene Expression Programs of Human Embryonic and Induced Pluripotent Stem Cells. Cell Stem Cell. 2010;7:249–57. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas ED, Lochte HL, Cannon JH, et al. SUPRALETHAL WHOLE BODY IRRADIATION AND ISOLOGOUS MARROW TRANSPLANTATION IN MAN. Journal of Clinical Investigation. 1959;38:1709–16. doi: 10.1172/JCI103949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas ED, Storb R, Clift RA, et al. BONE-MARROW TRANSPLANTATION .1. New England Journal of Medicine. 1975;292:832–43. doi: 10.1056/NEJM197504172921605. [DOI] [PubMed] [Google Scholar]
- 30.Copelan EA. Medical progress: Hematopoietic stem-cell transplantation. New England Journal of Medicine. 2006;354:1813–26. doi: 10.1056/NEJMra052638. [DOI] [PubMed] [Google Scholar]
- 31.Martino R, Iacobelli S, Brand R, et al. Retrospective comparison of reduced-intensity conditioning and conventional high-dose conditioning for allogeneic hematopoietic stem cell transplantation using BLA-identical sibling donors in myelodysplastic syndromes. Blood. 2006;108:836–46. doi: 10.1182/blood-2005-11-4503. [DOI] [PubMed] [Google Scholar]
- 32.Goerner M, Gooley T, Flowers MED, et al. Morbidity and mortality of chronic GVHD after hematopoietic stem cell transplantation from HLA-identical siblings for patients with aplastic or refractory anemias. Biology of Blood and Marrow Transplantation. 2002;8:47–56. doi: 10.1053/bbmt.2002.v8.pm11858190. [DOI] [PubMed] [Google Scholar]
- 33.Stern M, Passweg JR, Locasciulli A, et al. Influence of donor/recipient sex matching on outcome of allogeneic hematopoietic stem cell transplantation for aplastic anemia. Transplantation. 2006;82:218, 26. doi: 10.1097/01.tp.0000226156.99206.d1. [DOI] [PubMed] [Google Scholar]
- 34.Bai LY, Chiou TJ, Liu JH, et al. Hematopoietic stem cell transplantation for severe aplastic anemia - experience of an institute in Taiwan. Annals of Hematology. 2004;83:38–43. doi: 10.1007/s00277-003-0781-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maury S, Bacigalupo A, Anderlini P, et al. Improved outcome of patients older than 30 years receiving HLA-identical sibling hematopoietic stem cell transplantation for severe acquired aplastic anemia using fludarabine-based conditioning: a comparison with conventional conditioning regimen. Haematologica-the Hematology Journal. 2009;94:1312–15. doi: 10.3324/haematol.2009.006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medvinsky A, Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- 37.Muller AM, Medvinsky A, Strouboulis J, et al. DEVELOPMENT OF HEMATOPOIETIC STEM-CELL ACTIVITY IN THE MOUSE EMBRYO. Immunity. 1994;1:291–301. doi: 10.1016/1074-7613(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 38.Chen MJ, Yokomizo T, Zeigler BM, et al. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–91. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertrand JY, Chi NC, Santoso B, et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–U20. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–U25. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- 41.Kumaravelu P, Hook L, Morrison AM, et al. Quantitative developmental anatomy of definitive haematopoietic stem cells long-term repopulating units (HSC/RUs): role of the aorta-gonad-mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Development. 2002;129:4891–99. doi: 10.1242/dev.129.21.4891. [DOI] [PubMed] [Google Scholar]
- 42.Gekas C, Dieterlen-Lievre F, Orkin SH, Mikkola HKA. The placenta is a niche for hematopoietic stem cells. Developmental Cell. 2005;8:365–75. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 43.Christensen JL, Wright DE, Wagers AJ, Weissman IL. Circulation and chemotaxis of fetal hematopoietic stem cells. Plos Biology. 2004;2:368–77. doi: 10.1371/journal.pbio.0020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ledran MH, Krassowska A, Armstrong L, et al. Efficient hematopoietic differentiation of human embryonic stem cells on stromal cells derived from hematopoietic niches. Cell Stem Cell. 2008;3:85–98. doi: 10.1016/j.stem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Ng ES, Davis RP, Azzola L, et al. Forced aggregation of defined numbers of human embryonic stem cells into embryoid bodies fosters robust, reproducible hematopoietic differentiation. Blood. 2005;106:1601–03. doi: 10.1182/blood-2005-03-0987. [DOI] [PubMed] [Google Scholar]
- 46.Kyba M, Perlingeiro RCR, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 47.Pilat S, Carotta S, Schiedlmeier B, et al. HOXB4 enforces equivalent fates of ES-cell-derived and adult hematopoietic cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12101–06. doi: 10.1073/pnas.0505624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Yates F, Naveiras O, et al. Embryonic stem cell-derived hematopoietic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:19081–86. doi: 10.1073/pnas.0506127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bowles KM, Vallier L, Smith JR, et al. HOXB4 overexpression promotes hematopoietic development by human embryonic stem cells. Stem Cells. 2006;24:1359–69. doi: 10.1634/stemcells.2005-0210. [DOI] [PubMed] [Google Scholar]
- 50.Wang LS, Menendez P, Shojaei F, et al. Generation of hematopoietic repopulating cells from human embryonic stem cells independent of ectopic HOXB4 expression. Journal of Experimental Medicine. 2005;201:1603–14. doi: 10.1084/jem.20041888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji JF, Vijayaragavan K, Bosse M, et al. OP9 Stroma Augments Survival of Hematopoietic Precursors and Progenitors During Hematopoietic Differentiation from Human Embryonic Stem Cells. Stem Cells. 2008;26:2485–95. doi: 10.1634/stemcells.2008-0642. [DOI] [PubMed] [Google Scholar]
- 52.Rideout WM, Hochedlinger K, Kyba M, et al. Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy. Cell. 2002;109:17–27. doi: 10.1016/s0092-8674(02)00681-5. [DOI] [PubMed] [Google Scholar]
- 53.Hanna J, Wernig M, Markoulaki S, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–23. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 53b.Campbell KHS, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 54.Warren L, Manos PD, Ahfeldt T, et al. Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified mRNA. Cell Stem Cell. 2010;7:618–30. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okita K, Matsumura Y, Sato Y, et al. A more efficient method to generate integration-free human iPS cells. Nature Methods. 2011;8:409–U52. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 56.Fusaki N, Ban H, Nishiyama A, et al. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proceedings of the Japan Academy Series B-Physical and Biological Sciences. 2009;85:348–62. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hockemeyer D, Soldner F, Beard C, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nature Biotechnology. 2009;27:851–U110. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osafune K, Caron L, Borowiak M, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nature Biotechnology. 2008;26:313–15. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 59.Choi KD, Yu J, Smuga-Otto K, et al. Hematopoietic and Endothelial Differentiation of Human Induced Pluripotent Stem Cells. Stem Cells. 2009;27:559–67. doi: 10.1634/stemcells.2008-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Howe SJ, Mansour MR, Schwarzwaelder K, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. Journal of Clinical Investigation. 2008;118:3143–50. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Capecchi MR. ALTERING THE GENOME BY HOMOLOGOUS RECOMBINATION. Science. 1989;244:1288–92. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 62.Sebastiano V, Maeder ML, Angstman JF, et al. In Situ Genetic Correction of the Sickle Cell Anemia Mutation in Human Induced Pluripotent Stem Cells Using Engineered Zinc Finger Nucleases. Stem Cells. 2011;29:1717–26. doi: 10.1002/stem.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zou J, Mali P, Huang X, et al. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118:4599–608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu GH, Suzuki K, Qu J, et al. Targeted Gene Correction of Laminopathy-Associated LMNA Mutations in Patient-Specific iPSCs. Cell Stem Cell. 2011;8:688–94. doi: 10.1016/j.stem.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zou JZ, Maeder ML, Mali P, et al. Gene Targeting of a Disease-Related Gene in Human Induced Pluripotent Stem and Embryonic Stem Cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun N, Liang J, Abil Z, Zhao HM. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Molecular Biosystems. 2012;8:1255–63. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- 67.Christian M, Cermak T, Doyle EL, et al. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics. 2010;186:757–U476. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cermak T, Doyle EL, Christian M, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research. 2011;39 doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]