Summary
To determine the role of telomere dysfunction and telomerase reactivation in generating pro-oncogenic genomic events and in carcinoma progression, an inducible telomerase reverse transcriptase (mTert) allele was crossed onto a prostate cancer-prone mouse model null for Pten and p53 tumor suppressors. Constitutive telomerase deficiency and associated telomere dysfunction constrained cancer progression. In contrast, telomerase reactivation in the setting of telomere dysfunction alleviated intratumoral DNA damage signaling and generated aggressive cancers with rearranged genomes and new tumor biological properties (bone metastases). Comparative oncogenomic analysis revealed numerous recurrent amplifications and deletions of relevance to human prostate cancer. Murine tumors show enrichment of the TGFβ/SMAD4 network and genetic validation studies confirmed the cooperative roles of Pten, p53 and Smad4 deficiencies in prostate cancer progression including skeletal metastases. Thus, telomerase reactivation in tumor cells experiencing telomere dysfunction enables full malignant progression and provides a mechanism for acquisition of cancer-relevant genomic events endowing new tumor biological capabilities.
Introduction
Many genome instability mechanisms can contribute to somatic events present in human cancer genomes, particularly epithelial cancers (DePinho, 2000; Storchova and Pellman, 2004). Genetic studies illuminated a key role for telomere dysfunction in driving cancer initiation and shaping cancer genomes (Artandi and DePinho, 2010). When combined with p53 mutation which deactivates p53-dependent DNA damage signaling (Chin et al., 1999), telomere dysfunction engenders DNA double-strand breaks which produce non-reciprocal translocations, amplifications and deletions which promotes epithelial carcinogenesis (Artandi et al., 2000; O'Hagan et al., 2002). Telomere dynamics contribute to human epithelial cancers as evidenced by coincidental telomere erosion, anaphase bridging, and chromosomal instability in early stages of carcinogenesis in the colon (Rudolph et al., 2001), prostate (Meeker et al., 2002), breast (Chin et al., 2004), and pancreas (Feldmann et al., 2007). Human carcinoma sequencing has provided additional evidence that a period of telomere dysfunction generates chromosomal rearrangements (Stratton et al., 2009). In human prostate cancer, cancer cells possess shorter telomeres (Sommerfeld et al., 1996) as a result of telomere erosion early in disease evolution (Meeker et al., 2002; Vukovic et al., 2003). Curiously, prostate cancers do not arise spontaneously in mice with telomere dysfunction and p53 deficiency (Artandi et al., 2000). While casting uncertainty as to relevance of telomeres in prostate cancer pathogenesis and in shaping its complex genome, it is also possible that mice may not possess key genetic or environmental factors required to harness telomere dysfunction as a mechanism to promote the neoplastic process in the prostate.
While telomere dysfunction serves to drive early stages of cancer development, subsequent telomerase activation and restoration of telomere function appears to be critical for full malignant progression. This hypothesis is supported by frequent activation of telomerase in diverse human cancers (Shay and Wright, 2006) and enablement of enforced TERT in oncogene-induced malignant transformation of human primary cells (Hahn et al., 1999). Accordingly, low telomerase activity in normal prostate tissues is markedly elevated in human prostate tumors (Kallakury et al., 1997; Lin et al., 1997; Sommerfeld et al., 1996; Koeneman et al., 1998; Zhang et al., 1998).
Genetic events associated with human prostate cancer, the most common cancer and second leading cause of cancer death in American men (Jemal et al., 2010), include ETS family member translocation (Tomlins et al., 2005; Rubin, 2008) and (epi)genetic alterations of PTEN, p27Kip1, NKX3.1, c-MYC, FGFRs, EZH2/MIR101, p53, SMAD4, among others (Li et al., 1997; Guo et al., 1997; Abate-Shen et al., 2008; Jenkins et al., 1997; Acevedo et al., 2009; Ding et al., 2011). Genomic analysis of human prostate cancers has revealed numerous recurrent amplifications and deletions (Taylor et al., 2010), pointing to existence of many new prostate cancer-relevant genes. Identification and validation of these genes in amplifications and deletions is challenged by involvement of a significant fraction of the genome, marked intratumoral heterogeneity, and paucity of human cell-based model systems. In this regard, comparison of human and mouse cancer genomes has proven highly effective in facilitating cancer gene discovery (Kim et al., 2006; Zender et al., 2006), particularly with use mouse models with telomere dysfunction which promotes regional amplifications and deletions of cancer-relevant loci (Maser et al., 2007; O'Hagan et al., 2002). Here, we sought to understand the role of telomere dysfunction and telomerase reactivation in prostate cancer progression and in generating genomic events that may promote new tumor biological properties of this common malignancy.
Results
Telomerase reactivation in genome-unstable mouse prostate cancer model drives metastatic progression
In this study, we employed two distinct telomerase reverse transcriptase (mTert) alleles to study how telomere dysfunction and subsequent telomerase reactivation influences the genomes and biology of prostate cancer. The first allele is a conventional mTert knockout resulting in constitutive telomerase deficiency and telomere dysfunction upon successive generational mTert−/− intercrosses. The second allele is a novel inducible mTert knock-in containing an intronic Lox-Stop-Lox cassette (LSL) (Fig. 1A) that, upon Cre-mediated excision of LSL, mTert is re-expressed under endogenous control mechanisms.
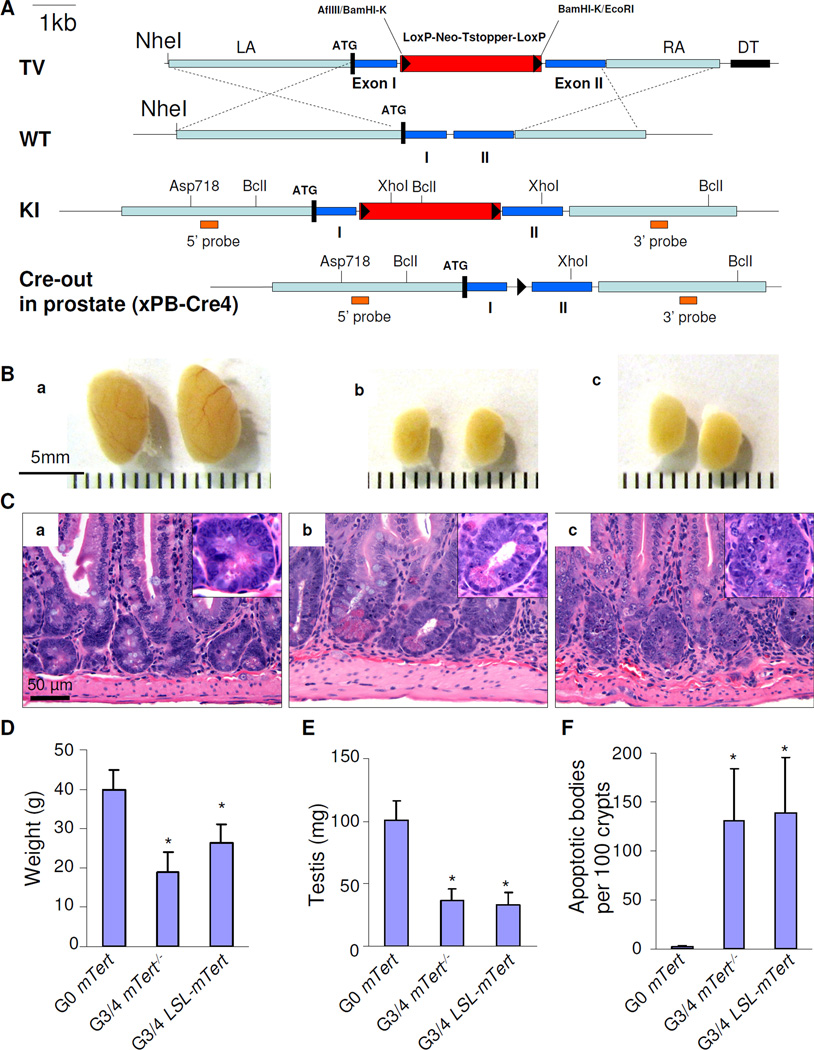
Figure 1. A novel inducible mTert knock-in containing a Lox-Stop-Lox (LSL) cassette in the first intron.
(A) LSL-mTert knock-in strategy. PB-Cre4–redirected Cre-mediated recombination removes the LSL cassette in prostate epithelium to restore endogenous mTert expression. (B,C) Later generations (G3, G4) of mTert−/− PB-Pten/p53 (panel b) or LSL-mTert PB-Pten/p53 alleles (panel c) displayed telomere dysfunction as demonstrated by decreased testis weight, compared to G0 mTert PB-Pten/p53 (panel a) (B), and increased apoptotic bodies in intestine (C). (D–F) Quantification of body weight (D), testis weight (E), and apoptotic bodies per 100 intestinal crypts of G0 PB-Pten/p53 (denoted as G0 mTert) (n=20), G3/4 mTert−/− PB-Pten/p53 denoted as G4 mTert−/− (n=31), and G3/4 LSL-mTert PB-Pten/p53 (denoted as G3/4 LSL-mTert) (n=20) mice (F). Error bars represent standard deviation (s.d.), * indicates p<0.05. See also Figure S1.
Mice possessing probasin (PB)-driven Cre transgene (Wu et al., 2001) and p53/pten conditional knockout alleles (hereafter PB-Pten/p53) characteristically develop locally invasive non-metastatic prostate adenocarcinoma with high penetrance and short latency (Chen et al., 2005). Alleles were backcrossed a minimum of 4 generations onto C57Bl/6. Upon successive generational intercrosses of LSL-mTert mice, late generations show classical constitutional signs of telomere dysfunction including reduced body weight (Fig. 1D), widespread organ atrophy (Fig. 1B, E), diminished proliferation and increased apoptosis in highly proliferative tissues (Fig. 1C, F), among other phenotypes as reported previously (Lee et al., 1998). Of note, PB-driven Cre expression is restricted to prostate epithelium and becomes active at sexual maturity. Thus, Cre-mediated deletion of LSL and mTert re-expression (telomerase reactivation) can occur in prostate epithelium experiencing telomere dysfunction.
PB-Pten/p53 alleles were carried through successive generational mating of LSL-mTert mice (Fig. S1), generating ‘telomere-intact’ controls (wildtype and LSL-mTert heterozygous mice, designated ‘G0 PB-Pten/p53’) and ‘telomere dysfunctional’ experimental mice (third and fourth generation LSL-mTert, designated G3/G4 LSL-mTert PB-Pten/p53). In parallel, we generated control and experimental cohorts of PB-Pten/p53 mice harboring the conventional mTert null allele (mTert−) (Farazi et al., 2006), producing analogous G3/4 mTert−/− PB-Pten/p53 to study the impact of telomere dysfunction only.
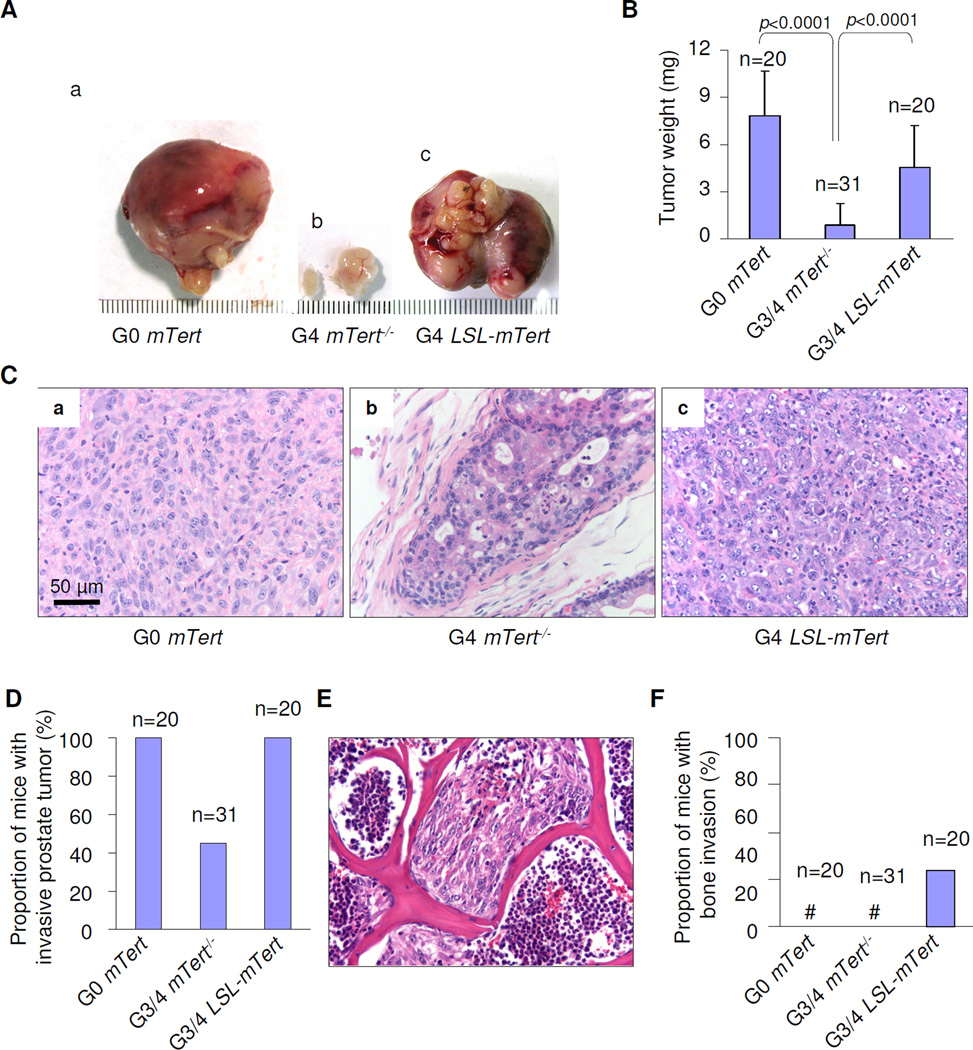
Consistent with previous report (Chen et al., 2005), G0 PB-Pten/p53 mice with mTert+/+ or mTert+/− allele (i.e. without telomere dysfunction) developed rapidly progressive locally invasive prostate adenocarcinomas with 100% penetrance, whereas G3/4 mTert−/− PB-Pten/p53 mice (i.e. experienced ongoing genome instability due to telomere dysfunction) had smaller poorly progressive tumors (Fig. 2A–D). Serial histological analyses revealed high-grade prostate intraepithelial neoplasia (HPIN) by age 9 weeks in both cohorts (Fig. S2A). However, most G3/4 mTert−/− PB-Pten/p53 tumors failed to progress beyond HPIN through age 24 weeks (Fig. 2C–D, Table S1), while G0 PB-Pten/p53 tumors evolved to invasive adenocarcinoma by age 24 weeks with 100% penetrance (Fig. 2C–D). In comparison, G3/4 LSL-mTert PB-Pten/p53 mice (i.e. experienced baseline telomere dysfunction prior to telomerase reactivation at sexual maturity) also initiated with high-grade prostate intraepithelial neoplasia (HPIN) by age 9 weeks (Fig. S2A), but unlike the G3/4 mTert−/− mice, they developed bulky lethal tumors by age 24 weeks (Fig. 2A–D). These observations reinforced the established role of telomere dysfunction in facilitating cancer initiation yet constraining full malignant progression (Rudolph et al., 2001; Gonzalez-Suarez et al., 2000; Chin et al., 1999), whereas telomerase reactivation enabled rapidly progressive disease. Importantly, a new phenotype emerged among some of the G3/4 LSL-mTert PB-Pten/p53 mice that is not observed in the genome-stable G0 PB-Pten/p53 mice, namely lumbar spine metastases (5/20, 25%) (Fig. 2E–F, Fig. S2B). The prostate cancer origin of these metastases was confirmed by genotype analysis of Cre-deleted Pten, p53, LSL sequences (Fig. S2C–D; not shown). This new phenotype suggests that its evolution requires not just stable genome but antecedent instability induced by telomere dysfunction.
Figure 2. Telomere dysfunction and telomerase impact on prostate cancer progression.
(A) Gross anatomy of representative prostates at 24 weeks of age. (B) Quantification of the prostate tumor of G0 mTert (n=20), G3/4 mTert−/− (n=31), and G3/4 LSL-mTert (n=20) mice. Error bars represent s.d. (C) Representative H&E sections of the prostate tumors from G0 mTert (panel a), G3/4 mTert−/− (panel b), and G3/4 LSL-mTert (panel c) at 24 weeks of age. (D) Quantification of locally invasive prostate tumors of G0 mTert (n=20), G3/4 mTert−/− (n=31), and G3/4 LSL-Tert (n=20) mice at 24 weeks of age. (E) H&E sections of the prostate tumors of G4 LSL-Tert in spinal bone at 24 weeks of age. (F) Quantification of mice with prostate tumors with spread lumbar spine of G0 mTert (n=20), G3/4 mTert−/− (n=31), and G4 LSL-Tert (n=20) mice. See also Figure S2 and Table S1.
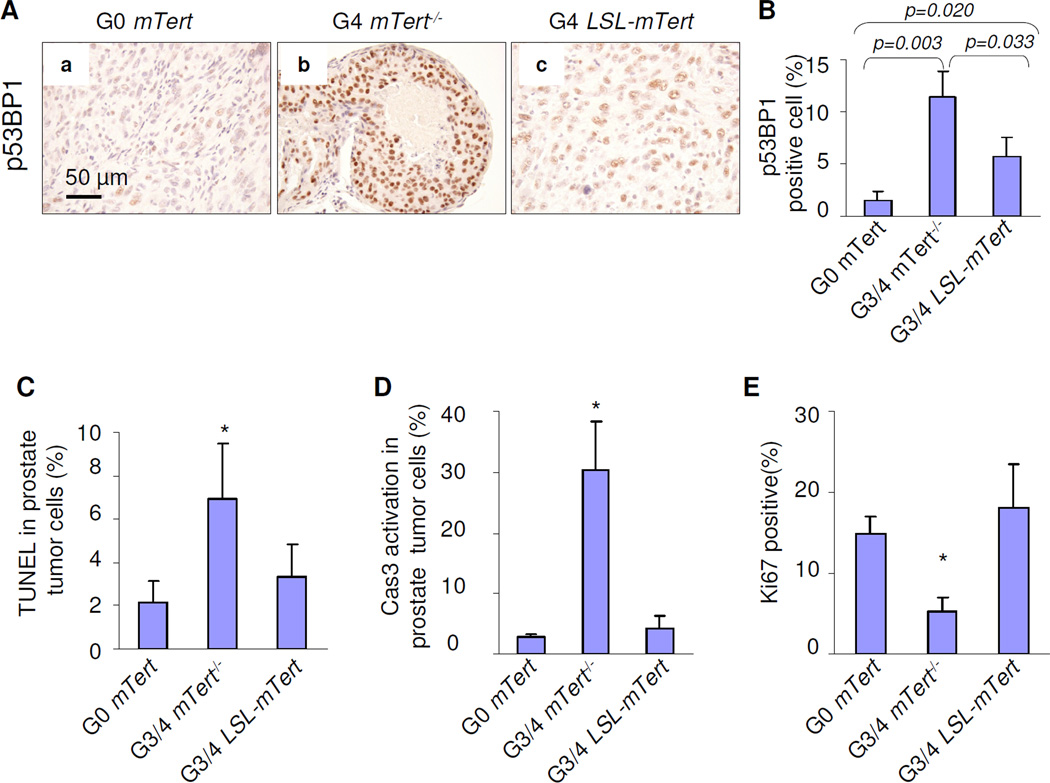
Next, the impact of telomerase reactivation was assessed in age-matched prostate tumors from each model. Quantitative Telomere-FISH revealed decreased telomere reserves in G3/4 mTert−/− PB-Pten/p53 samples relative to G0 PB-Pten/p53 samples, while G3/4 LSL-mTert PB-Pten/p53 samples showed longer telomere lengths relative to G3/4 mTert−/− PB-Pten/p53 (Fig. S3). As eroded dysfunctional telomeres generate a DNA damage response (Takai et al., 2003; d'Adda et al., 2003), functional restoration of telomeres was assessed by examination of DNA damage signaling (p53BP1 foci) in the various models. Strong anti-p53BP1 signal was detected in G3/4 mTert−/− PB-Pten/p53 tumor cells and was greatly reduced in G0 PB-Pten/p53 and G3/4 LSL-mTert PB-Pten/p53 tumor cells (Fig. 3A–B; n=3 each). Correspondingly, there was markedly increased apoptosis and decreased proliferation in G3/4 mTert−/− PB-Pten/p53 tumor cells compared with G0 PB-Pten/p53 and G3/4 LSL-mTert PB-Pten/p53 controls (Fig. 3C–E). Thus, telomerase activation alleviates telomere dysfunction checkpoints in G3/4 LSL-mTert PB-Pten/p53 cancers.
Figure 3. Telomerase reactivation maintains telomere length and allows tumor cells to proliferate.
(A) Strong telomere dysfunction-induced p53BP1 foci were presented in G3/4 mTert−/− cells (panel b), but not in G4 LSL-Tert cells (panel c). (B) Quantification of p53BP1 positive prostate tumor cells. Error bars represent s.d. for a representative experiment performed in triplicate. (C–E) Telomere dysfunction induced apoptosis and blockage of proliferation. Quantification of TUNEL positive (C), Caspase-3 activation positive (D), and Ki67 positive prostate tumor cells (E). Error bars represent s.d. for a representative experiment performed in triplicate. See also Figure S3.
Taken together, the molecular and phenotypic characterization of these three models demonstrated that telomerase reactivation not only enables the bypass of the progression block conferred by telomere dysfunction by quelling the DNA damage signals, but also engenders the acquisition of new tumor biological properties (bony tumor growth) not observed in tumors which did not experience a period of telomere dysfunction with subsequent telomerase reactivation in their evolution. This thus provides the first genetic proof in support of the thesis that telomerase reactivation and genome stabilization is necessary to drive full malignant progression in epithelial cancers.
Genome stabilization by telomerase reactivation selects for copy number aberrations of human relevance
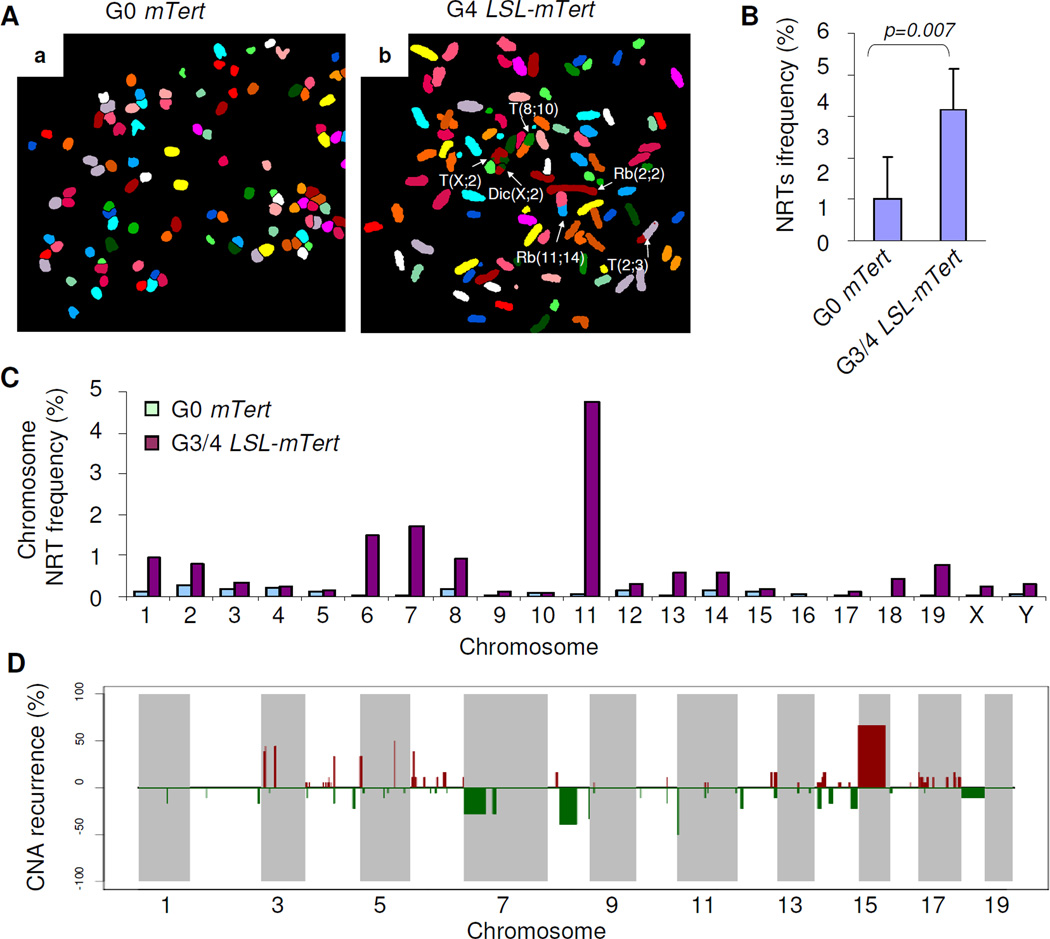
We hypothesized that baseline telomere dysfunction in G3/4 LSL-mTert PB-Pten/p53 cells followed by telomerase re-activation in the prostate upon sexual maturity at 5–7 weeks of age (Chen et al., 2005; Wu et al., 2001) would be permissive of the accumulation of genomic events in established tumors. Indeed, spectral karyotyping (SKY) analyses of G0 PB-Pten/p53 and G3/4 LSL-mTert PB-Pten/p53 prostate tumors revealed increased number of chromosomal structural aberrations in G3/4 mTert LSL-mTert PB-Pten/p53 tumor samples (n=5) relative to G0 PB-Pten/p53 controls (n=4) (Fig. 4A–C; 3.2 versus 1.0 per 100 chromosomes, respectively, P<0.05, t-test). These cytogenetic aberrations included multi-centric chromosomes, non-receiprocal translocations (NTRs), in addition to p-p, p-q and q-q chromosome arm fusions involving homologous and/or non-homologous chromosomes (Fig. 4B).
Figure 4. Oncogenomic alterations that occur in G3/4 LSL-mTert prostate tumors.
(A) Representative SKY images from metaphase spreads from G0 (panel a) and G3/G4 (panel b) prostate tumors. (B) Quantification of cytogenetic aberrations (recurrences) detected by SKY in G0 mTert and G3/G4 LSL-mTert prostate tumors. (C) Quantification of cytogenetic aberrations (recurrences) detected by SKY in G0 mTert (green) and G3/G4 LSL-mTert (purple) prostate tumors. (D) Recurrence plot of CNAs defined by aCGH for 18 mouse prostate tumors. The x axis shows physical location of each chromosome. The percentage of prostate tumors harboring gains (bright red, log2>0.6, losses (green, log2<−0.3), and deletions (dark green, log2<−0.6) for each locus is depicted. See also Figure S4 and Table S2–3.
Telomere dysfunction and associated bridge-fusion-breakage process are known to create DNA double-strand breaks, leading to regional amplifications and deletions at the sites of breakage (O’Hagan et al., 2002). Under biological selection, this process can result in copy number aberrations (CNAs) at cancer-relevant loci (Maser et al., 2007; O'Hagan et al., 2002). Prompted by this, we performed array-based CGH and transcriptional profile analyses of 18 G3/G4 LSL-mTert PB-Pten/p53 tumors. Compared to matched germline DNA in each case, 94 recurrent somatic copy number alterations (sCNAs) were defined by MCR analysis, encompassing 2183 amplified and 3531 deleted genes (Fig. S4, Table S2).
To assess human relevance of these murine sCNAs, we analyzed genome-wide copy number profiles of 194 human prostate tumors (Taylor et al., 2010) by GISTIC2 (Beroukhim et al., 2007) which defined 55 recurrent focal sCNAs and 10 recurrent chromosomal arm-level gains or losses (Fig. S4). Cross-species comparisons by orthologs revealed that twenty-two of the 94 murine sCNAs harbored synteny to either focal or broad sCNAs in humans. Consistent with previous studies, resident genes (300 amplified genes and 441 deleted genes) in these syntenic sCNAs were indeed significantly enriched for cancer-relevant genes (Fig. S4, Table S2). In particular, one of the cross-species conserved CNAs involving mouse chromosome 15 and human chromosome 8 was notable for high recurrence in both species (mouse: 12/18, 67% and human: 43/194, 22%) (Table S2, Fig. 4D). This syntenic region contains the prostate cancer-relevant MYC oncogene as well as other known cancer genes not previously implicated in prostate cancer such as WNT pathway regulator FDZ6 (Table S2).
Recognizing that not all genes resident in sCNAs are drivers, we next implemented a series of integrative analyses (as outlined in Fig. S4) designed to cull passengers. Briefly, for genes resident in regions of syntenic amplifications or gains, we prioritized those with copy number-correlated expression in mouse and human samples; for genes in regions of loss, we prioritized those with documented non-synonymous mutations in COSMIC (Forbes et al., 2011) or in NCBI PubMed, as well as evidence of promoter DNA hypermethylation in the published literature (Ongenaert et al., 2008). Furthermore, we posit that drivers targeted by sCNAs with functional consequences are more likely to be selected for during progression. Thus, we utilized the expression profile data in metastatic versus primary tumors from 6 prostate cancer cohorts on Oncomine (Lapointe et al., 2004; LaTulippe et al., 2002; Vanaja et al., 2003; Varambally et al., 2005; Yu et al., 2004; Holzbeierlein et al., 2004) to define the subset of genes exhibiting a progression-correlated expression pattern in human; namely genes resident in regions of loss in the mouse tumors are downregulated in human metastasis compared to primary, or conversely genes in regions of gains are upregulated in human metastases. This multi-dimensional integrative analysis narrowed our cross-species conserved gene list down to 228 genes (77 amplified and 151 deleted) (Fig. S4, Table S3).
TGFβ/Smad4 pathway in prostate tumors with bone metastasis
As a first step to identify molecular events capable of driving metastasis to the bone, we asked whether a subset of the above 228 candidate genes are subjected to consistent amplification/deletion in the 14 bone metastasis in the cohort reported by Taylor et al (Taylor et al., 2010). Specifically, we interrogated each of the 77 amplified or 151 deleted candidates for evidence that it is more likely to be amplified or deleted in bone metastasis, respectively. The resultant 113 gene list (comprising of 37 amplified and 76 deleted genes associated with bone-metastasis) was then enlisted into knowledge-based pathway analysis. Interestingly, TGFβ signaling genes represented the most significantly enriched network among the 9 significant pathways with FDR < 0.1 (Table S4; Fig. S5A). Corroborating with this pathway analysis result is the observation that Smad4 is encompassed by genomic loss in 2 of the 18 (11%) G3/G4 LSL-mTert PB-Pten/p53 tumor genomes, suggesting that TGFβ signaling and SMAD4 specifically may be targeted during prostate cancer skeletal metastasis. This is consistent with recent reports on the pathogenetic and prognostic roles of SMAD4 in human prostate cancer (Ding et al., 2011) and its frequent epigenetic silencing in advanced disease (Aitchison et al., 2008).
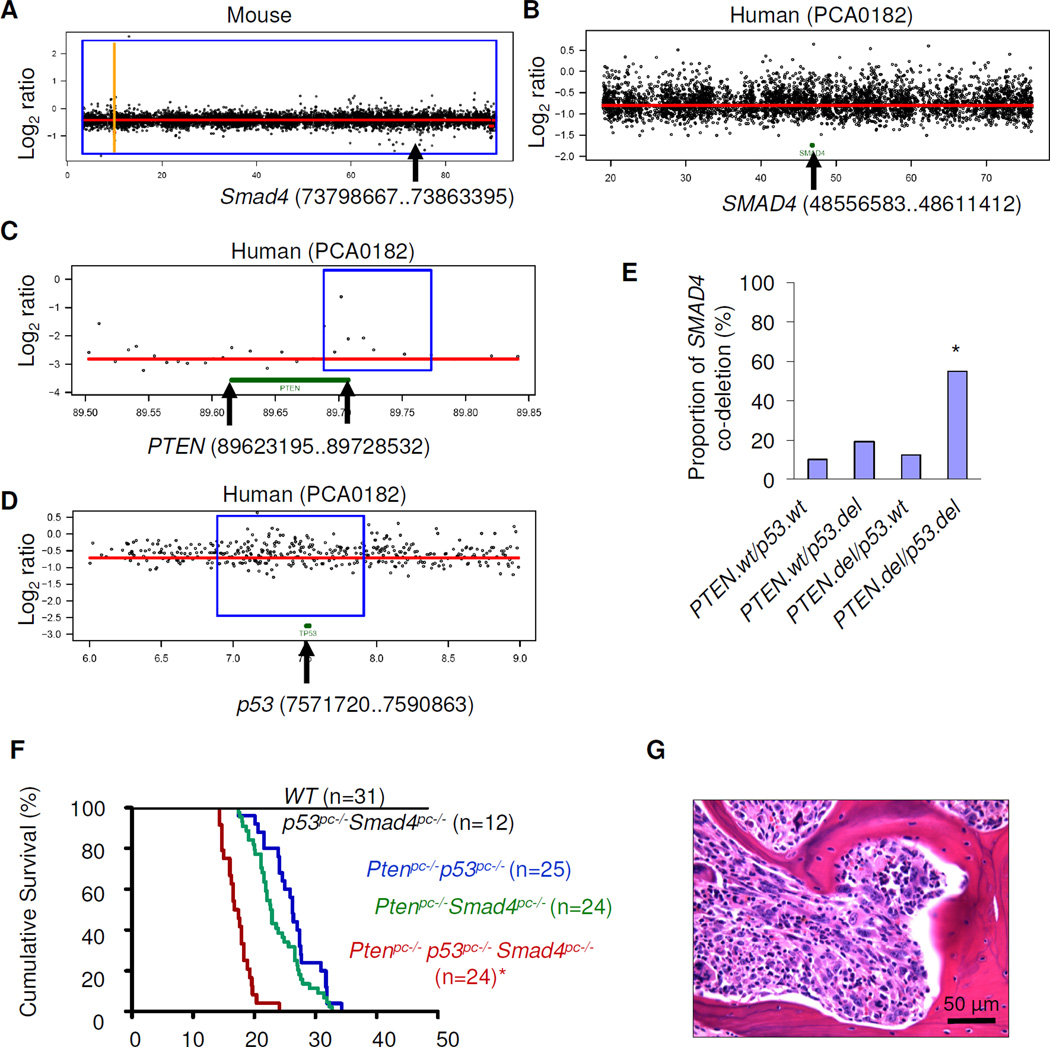
To genetically validate the above hypothesis, we crossed a prostate-specific probasin-driven Smad4 conditional knockout allele onto the prostate-specific p53/Pten double null model (PB-p53/Pten). The rationale for using the prostate-specific p53/pten knockout model is based on the fact that this mouse model does not develop bone metastasis, and, in human prostate cancers (Taylor et al., 2010), loss of SMAD4 as part of a large regional CNA is frequent (18% = 35/194) and often co-occurred with p53 and PTEN loss (Fig. 5B–D) (Fig. 5E, p=2.9e-6 by Fisher’s exact test). Consistent with the genomic data, prostate specific deletion of all 3 tumor suppressors led to more aggressive tumor phenotype and shorter overall survival. The median survival time of 17.05 weeks in Pten/p53/Smad4 was significantly shorter than Pten/p53 (26.3 weeks) or Pten/Smad4 (22.8 weeks) models (Fig. 5F; P<0.0001, logrank test). Most importantly, 3/24 (12.5%) of the triple knockout mice developed spontaneous bone metastasis (Fig. 5G).
Figure 5. Co-deletion of SMAD4 together with PTEN and p53 lead to aggressive prostate cancer progression.
(A–B) Log2 ratio of array-CGH plots showing conserved deletion of SMAD4 in both mouse G3/G4 LSL-mTert tumor (A) and human prostate tumor samples (B). The y axis shows log2 of copy number ratio (normal, log2 = 0); amplifications are above and deletions are below this axis; x axis is chromosome position, in Mbp. (C–D) Log2 ratio of array-CGH plots showing co-deletion of PTEN (C) and p53 (D) in that same human prostate sample with the SMAD4 deletion. (E) Co-deletion analysis of PTEN, p53 and SMAD4 in human prostate cancer samples (n=194). The P-value (Fisher’s exact test) = 2.9e-6 (asterisk). (F) Survival curves showing significant decrease in lifespan in PB-Pten/p53/Smad4 (n=24) (asterisk) compared with PB-Pten/p53 (n=25) or PB-Pten/Smad4 (n=44) cohorts by Kaplan-Meier overall cumulative survival analysis (P<0.0001). (G) H&E sections of prostate tumors of PB-Pten/p53/Smad4 in spinal bone at age19 weeks. See also Figure S5 and Table S4.
In summary, pathway analysis of the cross-species conserved gene list triangulated with the biological phenotype in human prostate cancers led to the hypothesis that TGFβ/SMAD4 signaling is an important driver of bone metastasis in the context of Pten and p53 deficiencies. Utilizing the combined Pten/p53/Smad4 GEM model, we demonstrate the new tumor biological properties (skeletal metastases) of this GEM model is not present in Pten/p53 or Pten/Smad4 telomere-intact GEM models. This study establishes, in a genetic manner, that telomerase reactivation in tumor cells experiencing telomere dysfunction provides a mechanism for selection of cooperative events required to progress fully and manifest the tumor biological properties governed by such genomic events.
Evolutionarily conserved genomically altered genes correlating with bone metastasis are prognostic in human
The in vivo genetic experiment above proving a driver role for Smad4 in bone metastasis suggests that additional genes on our bone metastasis-associated gene list may have functional importance as well. Since SMAD4 has also been shown to carry prognostic significance (Ding et al., 2011), we reasoned that prognostic relevance may serve as a surrogate for biological importance. As a proof of concept, we focused on the 14 genes that are represented in the 9 pathways found to be significantly enriched in the bone-metastasis associated gene list (Table S4). Specifically, we assessed how robustly these 14 genes can stratify risk for biochemical recurrence (BCR >0.2 ng/ml) among the 140 patients with outcome annotation (Taylor et al., 2010). Gratifyingly, the overall risk score based on the 14-gene signature was significantly prognostic of BCR with hazard ratio of 13 (P-value <10−14, overall C-index = 0.93, see Fig. S5B) by multivariate Cox regression analysis. Further support for these 14 genes as likely drivers of bone metastasis phenotype derived from the observation that they provided independent prognostic value to the previously reported 4-gene signature (comprising of PTEN/SMAD4/CCND1/SPP1) derived from the Pten/Smad4 model (Ding et al., 2011), consistent with the fact that bone metastasis was not observed in the Pten/Smad4 GEM model (hazard ratio = 8.7, P=2.16×10−13, and overall C-index = 0.93, see Fig. S5C). In particular, combination of 14-gene with the 4-gene signature increases the predictive power of either gene set alone (hazard ratio = 20, P<10−14, and overall C-index = 0.96, see Fig. S5D).
Taken together, the prognostic correlation of these 14 genes represented in the 9 functional pathways enriched in the bone-metastasis associated gene set provides the correlative evidence for biological relevance of these genes to human prostate cancers, although their individual contribution and mechanism of actions will require further exploration. Additionally, these results serve as validation of the integrative approaches adopted by this study which leverages the clear genotype-phenotype correlation in model systems with the power of genomic and bioinformatic analyses to elucidate molecular mechanisms driving bone metastasis in human prostate cancers.
Discussion
We explored the role of telomere dysfunction and telomerase reactivation in shaping the genomes and impacting on the biology of prostate cancer. These genetic studies in vivo, together with human and mouse prostate cancer genomic data, provide evidence that telomere dysfunction plays a critical role in prostate cancer initiation and progression, permitting acquisition of and selection for cancer-relevant genomic events upon telomerase reactivation. In addition, our studies establish first formal proof that telomere dysfunction and subsequent telomerase activation enables evolving cancers to progress fully and acquire new tumor biological properties including cardinal features of advanced human prostate cancer. Finally, comparative oncogenomic analysis of gene copy number and expression profiles with genotype-phenotype correlation resulted in identification of genes associated with progression to bone metastasis, highlighting the potential utility of this integrative approach for cancer gene discovery in prostate cancer.
Previous telomere and cytogenetic studies have documented chromosomal instability and telomere loss in early stage human prostate cancers (Sommerfeld et al., 1996; Meeker et al., 2002; Vukovic et al., 2003), implicating this mechanism in driving chromosomal instability and intratumoral heterogeneity in these emerging malignancies. These findings, along with work in the telomerase knockout mouse model (Artandi and DePinho, 2010), support a model where telomere dysfunction provides a mechanism fueling the early acquisition of somatic genomic events in prostate cancer as well as other epithelial cancers. Consistent with previous work in other epithelial cancers, we observed that constitutive telomere dysfunction in the G3/4 mTert−/− PB-Pten/p53 model enabled cancer initiation; however, these malignancies exhibited a constrained progression phenotype which was associated with activation of DNA damage signaling and increased apoptosis and decreased proliferation. Together, these data suggest that, while telomere dysfunction may enable cancer initiation, ongoing telomere dysfunction and cellular checkpoints impedes full malignant progression of HPIN to invasive and metastatic disease and that activation of telomere maintenance mechanisms may be needed to quell both rampant chromosomal instability and residual cellular checkpoints. In this regard, it is notable that there was no discernable inhibition in the progression from HPIN to invasive disease in G2 mTert−/− PB-Pten/p53 mice which do not express telomerase yet maintain adequate telomere reserves (data not shown). These data establish that telomerase activity per se is not essential for prostate cancer progression, providing tumor cells possess functional telomeres. Correspondingly, current cell-based evidence in human and mouse systems, together with the consistent expression of telomerase in human prostate cancers (Kallakury et al., 1997; Lin et al., 1997; Sommerfeld et al., 1996; Koeneman et al., 1998; Zhang et al., 1998) supports the view that telomerase reactivation plays a critical role in overcoming apoptotic and proliferative blocks needed for full malignant progression (Chang et al., 2003; Hahn et al., 1999; Chin et al., 1999).
Here, our inducible telomerase model system enabled genetic analysis of the impact of physiological endogenous telomerase reactivation in a naturally arising solid tumor with short dysfunctional telomeres. These studies established that telomerase reactivation enabled rapidly progressive disease in all cases. At the same time, we established that antecedent telomere dysfunction enabled the acquisition of genomic events including those capable of endowing tumors with new biological properties such as bone metastases, a phenotype not observed in G0 PB-Pten/p53 tumors (telomere intact). Thus, we conclude that a period of telomere dysfunction is a mechanism for the development of chromosomal aberrations targeting genes involved in prostate cancer development including bone metastasis.
The recurrent nature of amplifications and deletions of human prostate cancer raised the possibility that, along with a handful of known genetic lesions, there remain many uncharacterized genes governing genesis and progression of this cancer (Taylor et al., 2010). Beyond restoring genome stability and eliminating DNA damage signals, we and others have shown that genomic alterations acquired in genome-unstable mouse tumor genomes are not random as they show statistical significant overlap between mouse (which is telomerase-deficient/unstable) and human (which is telomerase-reactivated) (Maser et al., 2007). This observation leads us to hypothesize that reactivation of telomerase in setting of pre-existing genome instability can be a genomic mechanism for selection of cooperative events required for ultimate progression – in other words, it is not merely a permissive step by removing DNA damage, but telomerase reactivation is instead an active driver of progression. This study provides formal genetic proof for this thesis. By triangulating the list of genes resident in syntenic sCNAs in mouse and human prostate cancers with biological phenotype in human (e.g. documented bone metastasis), we have defined a prioritized list of bone-metastasis associated genes. Pathway analysis with this list revealed dominance of TGFβ/SMAD4 network, coupled with the observation of spontaneously acquired Smad4 genomic loss in two of the mouse tumors, led to the hypothesis that TGFβ signaling and SMAD4 inactivation is a driver for bone metastasis in prostate cancers. Again, leveraging the power of genetic engineering in the mouse, we went on to perform the definitive genetic validation experiment proving the cooperativity of p53/Pten/Smad4 co-deletion in driving prostate tumorigenesis and progression to bone metastasis in vivo.
It is however important to note that the penetrance of bone metastasis in mice with triple inactivation of p53/Pten/Smad4 was far from 100%, suggesting that additional events beyond Smad4 in the tumor and/or stromal cells can drive or are required for bone metastasis. Indeed, many other candidates with likely relevance have also been identified through our cross-species comparative oncogenomic analysis. For example, among the amplified genes, several have been implicated in cancer progression. Metadherin (MTDH), a gene with dual activity to promote metastasis and chemoresistance (Hu et al., 2009), is overexpressed in human prostate cancer and known to act as an activator of AKT and a suppressor of FOXO3a (Kikuno et al., 2007). Protein tyrosine kinase 2 (PTK2, a.k.a. FAK), a gene located in the frequently amplified 8q24 region, is well known for its role in cell motility and proliferation (Chang et al., 2007) and in promoting human prostate cancer cell invasiveness (Johnson et al., 2008) Notable deleted genes include APC (20% in human prostate cancer, 11% in mouse prostate cancer) and Smad2/Smad4 (20% in human prostate cancer, 11% in mouse prostate cancer), highlighting the importance of activation of Wnt signaling pathway and deactivation of TGFβ pathway in prostate cancer progression (Ding et al., 2011).
Lastly, since not all of the evolutionarily conserved candidates identified in this study likely represent true drivers of prostate cancer progression in human, definitive demonstration of biological activity and elucidation of mechanisms of action for each will require significant downstream activities by many. On the other hand, the prognostic significance of the 14 pathway-representative genes is a strong correlative support for functional veracity of the candidate list, even though we recognize that BCR is only a surrogate for aggressiveness of human prostate cancers. Further prognostic studies for lethality will be necessarily to validate the true utility of the 14-gene signature in human.
In summary, this study provides in vivo genetic evidence confirming the long-held hypothesis that telomerase reactivation quells DNA damage signaling and stabilizes the genome of an initiated cancer to permit cancer progression. This study also provides the first genetic proof in naturally occurring and initiated cancer in vivo that telomere dysfunction followed by telomerase re-activation serves as a mechanism for the generation of and selection for cancer-relevant genomic alterations to drive progression and new tumor biological hallmarks such as metastasis to bone. Thus, telomerase serves as an active driver of cancer progression in the setting of telomere-based crisis. Furthermore, the validation of telomere dysfunction as a relevant genome instability mechanism in prostate cancer, the generation of highly rearranged genomes with syntenic events, and the in silico documentation that altered genes are enriched for cancer-relevance collectively provide a system to enhance the mining of complex human prostate cancer genomes to identify genetic events governing prostate cancer progression.
METHODS
mTert knockout allele, Pten and Trp53 (p53) conditional alleles
mTert knockout allele and the PtenloxP conditional knockout alleles have been described elsewhere (Zheng et al., 2008; Farazi et al., 2006). p53loxP strain was generously provided by A. Berns (Marino et al., 2000). Prostate epithelium-specific deletion was effected by the PB-Cre4 (Wu et al., 2001) and was obtained from MMHCC (http://mouse.ncifcrf.gov/search_results.asp).
Generation of the LSL-mTERTloxP allele
We knocked in a LSL cassette into the first intron of the mouse Tert gene (Fig. 1A). The presence of the LSL cassette produces a null mTert allele and its removal by PB-Cre expression restores activity under the control of the native mTert promoter. This mouse model scheme allows for excellent specific control of telomerase reconstitution in prostate epithelia cells. Following introduction the knockin construct into ES cells, screening ES cells, confirming germline transmission, and NeoR cassette deletion via EIIa-Cre, the LSL-mTert allele has been backcrossed 4 generations onto the C57Bl/6 background.
Mating scheme
As depicted in Fig. S1, the LSL-mTertL/+ p53L/LPtenL/L mice were crossed with G0 mTert+/−p53L/LPtenL/LPB-Cre4 mice to generate G0 mTert+/− p53L/LPtenL/LPB-Cre4, G0 LSL-mTertL/+p53L/LPtenL/LPB-Cre4, and G1 mTert −LSL-mTertLp53L/LPtenL/LPB-Cre4 and G1 mTert −LSL-mTertLp53L/LPtenL/L mice. These mice were then intercrossed to generate G2 mTert−−/−p53L/LPtenL/LPB-Cre4, G2 mTert −LSL-mTertLp53L/LPtenL/LPB-Cre4, G2 LSL-mTertL/Lp53L/LPtenL/LPB-Cre4, G2 mTert−−/− p53L/LPtenL/L, G2 mTert −LSL-mTertLp53L/LPtenL/L, G2 LSL-mTertL/Lp53L/LPtenL/L. G2 mice were then intercrossed to generate G3 and G4 mice
Tissue analysis
Normal and tumor tissues were fixed in 10% neutral-buffered formalin overnight then processed, paraffin-embedded, sectioned and stained with hematoxylin and eosin according to standard protocol. Immunohistochemistry analysis was done as previously described (Ding et al., 2011) Antibodies used for IHC include anti-Ki67 (Dako), anti-53BP1 (Bethyl Labs), and anti-Cleaved Caspase 3 (Cell Signaling). Cells with the 53BP1 nuclear punctuate foci was considered as 53BP1-positive, whereas the absence of 53BP1 nuclear foci signal was considered 53BP1-negative. Western blot analysis was done as previously described (Ding et al., 2011).
Laser Capture Microdissection and DNA Extraction
Laser capture microdissection was done as previously described (Emmert-Buck et al., 1996). Genomic DNA of microdissected prostate tumor cells was extracted with phenol-chloroform prior to PCR analysis.
TUNEL assay
To determine apoptosis in prostate tumor cells, TUNEL staining was performed was done as previously described (Ding et al., 2011).
Cytogenetics, quantitative telomere FISH and spectral karyotyping analysis
We prepared metaphase chromosomes from early passage prostate tumor cells was done as previously described (Maser et al., 2007).
Establishment of mouse prostate tumor cell lines
Tumors were dissected from prostates of G0 PtenL/L Trp53L/LPB-Cre4+, G3 and G4 mice. Cell lines were established as described previously (Ding et al., 2011).
RNA isolation and real-time PCR
Total RNA was extracted and real-time PCR were done as previously described (Ding et al., 2011).
Array-CGH profiling and analysis of mouse prostate tumors
Murine G3/G4 LSL-mTert PB-Pten/p53 tumors were profiled against matched normal tail DNA. Labeled DNAs were hybridized onto Agilent mouse 244K CGH arrays and scanning was performed as per manufacturer's protocol. Data were processed using Agilent software. The array-CGH data of 18 G3/G4 LSL-mTert PB-Pten/p53 tumors were analyzed with the MCR algorithm (Maser et al., 2007) to detect focal genomic regions with copy number alteration (CNA) events in at least two tumors. Mouse genome data build mm9 was used in the analysis. A total of 2183 genes from 57 amplified regions (Supplemental Table 2) and 3531 genes from 38 deletion regions were detected by the MCR algorithm.
Array-CGH analysis for recurrent focal and arm-level chromosomal copy number alterations in human prostate tumors with the GISTIC2 algorithm
The array-CGH data of 194 human prostate tumors (Taylor et al., 2010) were analyzed with the GISTIC2 algorithm (Beroukhim et al., 2007) to detect focal genomic regions with copy number alteration (CNA) events. Focal regions with q-values smaller than 0.25 are considered significant, which resulted in 16 amplified and 39 deleted regions. Arm-level changes with q-values smaller than 0.005 are considered significant, which suggested chromosome 7p, 7q, and 8q amplification and 6q, 8p, 12p, 13q, 16q, 17p, and 18q deletion.
Homolog mapping for CNA synteny regions cross human and mouse tumors
We used NCBI homologene database (version 39.2) to map human and mouse homolog genes and detect synteny CNA regions. The homologene analysis characterized 300 amplified genes and 441 deleted genes that commonly recurred in human and mouse prostate tumors.
Co-deletion analysis in human clinical samples
Based on the results of GISTIC2 analysis, 194 human prostate tumors (Taylor et al., 2010) were classified into 4 groups according to PTEN and p53 focal deletion status (Fig. 5E). The numbers of SMAD4 deletion events in each group were used to estimate the significance P value of co-deletion enrichment by Fisher’s exact test in R environment.
Correlation analysis between gene copy numbers and gene expression changes
The Spearman correlation coefficients between individual gene copy numbers and expression levels in matching samples were calculated in R environment. P values less than 0.05 were considered significant.
Oncomine Consensus Analysis
Six prostate cancer cohorts (Holzbeierlein et al., 2004; Lapointe et al., 2004; LaTulippe et al., 2002; Vanaja et al., 2003; Varambally et al., 2005; Yu et al., 2004) in the Oncomine database (www.oncomine.com) were used to filter our candidate marker gene lists. We tested the following hypotheses: if genes in the amplified regions are related to metastatic phenotypes in any of the 6 cohorts, or if genes in the deleted regions are related to indolent phenotypes in any of the 6 cohorts.
Bone metastasis related copy number changes
We tested if genes recurrently amplified or deleted in the whole prostate cancer cohort of Taylor et al. showed consistent copy number alteration patterns in tumors with documented bone metastasis (Taylor et al., 2010). For each candidate gene, we counted the number of gene gain (copy number > 0.3 in log-2 scale) and loss (copy number < −0.3 in log-2 scale) in 14 bone metastasis tumors. Consistent changes are defined if an amplified CNA gene is more likely to have gain than loss or a deleted CNA gene more likely to be lost than gained in bone metastatic tumors.
Survival Analysis
We applied Cox proportional hazard regression on biomarkers of interest to get a multivariate linear regression model that best predict the biochemical recurrence of prostate cancer. Tumors were subsequently divided into high-risk and low-risk groups according to the scores. Kaplan-Meier curves were plotted by R software, and the statistical significance was estimated by log-rank test.
Supplementary Material
Acknowledgements
We thank S. Zhou and J. P. Morin for excellent mouse husbandry and care, J. Paik, M. Kim, and H. Ying for helpful discussion. We thank W. Hahn for shRNA constructs. Z.D was supported by Damon Runyon Cancer Research Foundation. D.H was supported by a graduate fellowship from the National Science Foundation. YAW was supported by Multiple Myeloma Research Foundation. This work is supported by NCI R01CA084628 (R.A.D) U01CA141508 (L.C. and R.A.D.), Prostate Cancer Foundation (R.A.D and Z.D), DOD W81XWH-07-PCRP-IDA (R.A.D and Z.D). R.A.D. was an American Cancer Society Research Professor and supported by the Robert A. and Renee E. Belfer Foundation. The gene signature and technology developed in this paper have been licensed by Metamark GENETICS (http://www.metamarkgenetics.com/). L.C. and R.A.D. are the founders of Metamark GENETICS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abate-Shen C, Shen MM, Gelmann E. Integrating differentiation and cancer: The Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Differentiation. 2008 doi: 10.1111/j.1432-0436.2008.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo VD, Ittmann M, Spencer DM. Paths of FGFR-driven tumorigenesis. Cell Cycle. 2009;8:580–588. doi: 10.4161/cc.8.4.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison AA, Veerakumarasivam A, Vias M, Kumar R, Hamdy FC, Neal DE, Mills IG. Promoter methylation correlates with reduced Smad4 expression in advanced prostate cancer. Prostate. 2008;68:661–674. doi: 10.1002/pros.20730. [DOI] [PubMed] [Google Scholar]
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Khoo CM, Naylor ML, Maser RS, DePinho RA. Telomere-based crisis: functional differences between telomerase activation and ALT in tumor progression. Genes Dev. 2003;17:88–100. doi: 10.1101/gad.1029903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YM, Kung HJ, Evans CP. Nonreceptor tyrosine kinases in prostate cancer. Neoplasia. 2007;9:90–100. doi: 10.1593/neo.06694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, Kuo WL, Ljung BM, Chew K, Myambo K, Miranda M, Krig S, Garbe J, Stampfer M, Yaswen P, Gray JW, Lockett SJ. In situ analyses of genome instability in breast cancer. Nat. Genet. 2004;36:984–988. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- d'Adda dF, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- DePinho RA. The age of cancer. Nature. 2000;408:248–254. doi: 10.1038/35041694. [DOI] [PubMed] [Google Scholar]
- Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, Hoshida Y, Hiller D, Hu B, Jiang S, Zheng H, Stegh AH, Scott KL, Signoretti S, Bardeesy N, Wang YA, Hill DE, Golub TR, Stampfer MJ, Wong WH, Loda M, Mucci L, Chin L, DePinho RA. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Farazi PA, Glickman J, Horner J, DePinho RA. Cooperative interactions of p53 mutation, telomere dysfunction, and chronic liver damage in hepatocellular carcinoma progression. Cancer Res. 2006;66:4766–4773. doi: 10.1158/0008-5472.CAN-05-4608. [DOI] [PubMed] [Google Scholar]
- Feldmann G, Beaty R, Hruban RH, Maitra A. Molecular genetics of pancreatic intraepithelial neoplasia. J Hepatobiliary. Pancreat. Surg. 2007;14:224–232. doi: 10.1007/s00534-006-1166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Samper E, Flores JM, Blasco MA. Telomerase-deficient mice with short telomeres are resistant to skin tumorigenesis. Nat. Genet. 2000;26:114–117. doi: 10.1038/79089. [DOI] [PubMed] [Google Scholar]
- Guo Y, Sklar GN, Borkowski A, Kyprianou N. Loss of the cyclin-dependent kinase inhibitor p27(Kip1) protein in human prostate cancer correlates with tumor grade. Clin. Cancer Res. 1997;3:2269–2274. [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Holzbeierlein J, Lal P, LaTulippe E, Smith A, Satagopan J, Zhang L, Ryan C, Smith S, Scher H, Scardino P, Reuter V, Gerald WL. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am. J. Pathol. 2004;164:217–227. doi: 10.1016/S0002-9440(10)63112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, Reiss M, Au JL, Haffty BG, Kang Y. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010 doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Jenkins RB, Qian J, Lieber MM, Bostwick DG. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res. 1997;57:524–531. [PubMed] [Google Scholar]
- Johnson TR, Khandrika L, Kumar B, Venezia S, Koul S, Chandhoke R, Maroni P, Donohue R, Meacham RB, Koul HK. Focal adhesion kinase controls aggressive phenotype of androgen-independent prostate cancer. Mol. Cancer Res. 2008;6:1639–1648. doi: 10.1158/1541-7786.MCR-08-0052. [DOI] [PubMed] [Google Scholar]
- Kallakury BV, Brien TP, Lowry CV, Muraca PJ, Fisher HA, Kaufman RP, Jr, Ross JS. Telomerase activity in human benign prostate tissue and prostatic adenocarcinomas. Diagn. Mol. Pathol. 1997;6:192–198. doi: 10.1097/00019606-199708000-00003. [DOI] [PubMed] [Google Scholar]
- Kikuno N, Shiina H, Urakami S, Kawamoto K, Hirata H, Tanaka Y, Place RF, Pookot D, Majid S, Igawa M, Dahiya R. Knockdown of astrocyte-elevated gene-1 inhibits prostate cancer progression through upregulation of FOXO3a activity. Oncogene. 2007;26:7647–7655. doi: 10.1038/sj.onc.1210572. [DOI] [PubMed] [Google Scholar]
- Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, Flotte TJ, Duncan LM, Granter SR, Chin L. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Koeneman KS, Pan CX, Jin JK, Pyle JM, III, Flanigan RC, Shankey TV, Diaz MO. Telomerase activity, telomere length, and DNA ploidy in prostatic intraepithelial neoplasia (PIN) J. Urol. 1998;160:1533–1539. [PubMed] [Google Scholar]
- Lapointe J, Li C, Higgins JP, van de RM, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, Ekman P, DeMarzo AM, Tibshirani R, Botstein D, Brown PO, Brooks JD, Pollack JR. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- Lee HW, Blasco MA, Gottlieb GJ, Horner JW, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Lin Y, Uemura H, Fujinami K, Hosaka M, Harada M, Kubota Y. Telomerase activity in primary prostate cancer. J. Urol. 1997;157:1161–1165. [PubMed] [Google Scholar]
- Marino S, Vooijs M, van der GH, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, O'Neil J, Gutierrez A, Ivanova E, Perna I, Lin E, Mani V, Jiang S, McNamara K, Zaghlul S, Edkins S, Stevens C, Brennan C, Martin ES, Wiedemeyer R, Kabbarah O, Nogueira C, Histen G, Aster J, Mansour M, Duke V, Foroni L, Fielding AK, Goldstone AH, Rowe JM, Wang YA, Look AT, Stratton MR, Chin L, Futreal PA, DePinho RA. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker AK, Hicks JL, Platz EA, March GE, Bennett CJ, Delannoy MJ, De Marzo AM. Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis. Cancer Res. 2002;62:6405–6409. [PubMed] [Google Scholar]
- O'Hagan RC, Chang S, Maser RS, Mohan R, Artandi SE, Chin L, DePinho RA. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2002;2:149–155. doi: 10.1016/s1535-6108(02)00094-6. [DOI] [PubMed] [Google Scholar]
- Ongenaert M, Van Neste L, De Meyer T, Menschaert G, Bekaert S, Van Criekinge W. PubMeth: a cancer methylation database combining text-mining and expert annotation. Nucleic Acids Res. 2008;36:D842–D846. doi: 10.1093/nar/gkm788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin MA. Targeted therapy of cancer: new roles for pathologists--prostate cancer. Mod. Pathol. 2008;21(Suppl 2):S44–S55. doi: 10.1038/modpathol.2008.11. [DOI] [PubMed] [Google Scholar]
- Rudolph KL, Millard M, Bosenberg MW, DePinho RA. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat. Genet. 2001;28:155–159. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nat. Rev. Drug Discov. 2006;5:577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- Sommerfeld HJ, Meeker AK, Piatyszek MA, Bova GS, Shay JW, Coffey DS. Telomerase activity: a prevalent marker of malignant human prostate tissue. Cancer Res. 1996;56:218–222. [PubMed] [Google Scholar]
- Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63:3877–3882. [PubMed] [Google Scholar]
- Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Vukovic B, Park PC, Al Maghrabi J, Beheshti B, Sweet J, Evans A, Trachtenberg J, Squire J. Evidence of multifocality of telomere erosion in high-grade prostatic intraepithelial neoplasia (HPIN) and concurrent carcinoma. Oncogene. 2003;22:1978–1987. doi: 10.1038/sj.onc.1206227. [DOI] [PubMed] [Google Scholar]
- Wu X, Wu J, Huang J, Powell WC, Zhang J, Matusik RJ, Sangiorgi FO, Maxson RE, Sucov HM, Roy-Burman P. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech. Dev. 2001;101:61–69. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, Michalopoulos G, Becich M, Luo JH. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J. Clin. Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, Mu D, Lucito R, Powers S, Lowe SW. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Kapusta LR, Slingerland JM, Klotz LH. Telomerase activity in prostate cancer, prostatic intraepithelial neoplasia, and benign prostatic epithelium. Cancer Res. 1998;58:619–621. [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, DePinho RA. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.