Abstract
OBJECTIVE
To determine the utility of an antibiogram in predicting the susceptibility of Pseudomonas aeruginosa isolates to targeted antimicrobial agents based on the day of hospitalization the specimen was collected.
DESIGN
Single-center retrospective cohort study.
SETTING
A 750-bed tertiary care medical center.
PATIENTS AND METHODS
Isolates from consecutive patients with at least 1 clinical culture positive for P. aeruginosa from January 1, 2000, to June 30, 2007, were included. A study antibiogram was created by determining the overall percentages of P. aeruginosa isolates susceptible to amikacin, ceftazidime, ciprofloxacin, gentamicin, imipenem-cilastin, piperacillin-tazobactam, and tobramycin during the study period. Individual logistic regression models were created to determine the day of infection after which the study antibiogram no longer predicted susceptibility to each antibiotic.
RESULTS
A total of 3,393 isolates were included. The antibiogram became unreliable as a predictor of susceptibility to ceftazidime, imipenem-cilastin, piperacillin-tazobactam, and tobramycin after day 10 and ciprofloxacin after day 15 but longer for gentamicin (day 21) and amikacin (day 28). Time to unreliability of the antibiogram varied for antibiotics based on location of isolation. For example, the time to unreliability of the antibiogram for ceftazidime was 5 days (95% confidence interval [CI], <1–8) in the intensive care unit (ICU) and 12 days (95% CI, 7–21) in non-ICU hospital wards (P=.003).
CONCLUSIONS
The ability of the antibiogram to predict susceptibility of P. aeruginosa decreases as duration of hospitalization increases.
Pseudomonas aeruginosa is a common and potentially lethal etiology of gram-negative infections.1,2 In fact, P. aeruginosa has become the most common etiology of gram-negative bloodstream infections (BSI) among hospitalized patients and the third most common etiology of BSI in hospitalized and community-dwelling patients.3,4 Infections due to P. aeruginosa are associated with a high rate of crude mortality, ranging from 28% to 48% for non–intensive care unit (ICU) and ICU patients, respectively.5 Unfortunately, the increasing prevalence of multidrug-resistant P. aeruginosa complicates treatment decisions and leads to potential delays in appropriate empiric antimicrobial therapy.6 Importantly, patients who receive inappropriate empiric antimicrobial therapy for gram-negative sepsis have mortality rates of 14%–38%.5,7,8
Clinicians often make empiric treatment decisions regarding initial antimicrobial therapy based on institution-specific antibiograms. These antibiograms frequently provide a summary of in vitro activity of antimicrobials at a specific institution. Most antibiograms are collated and reported annually in order to detect changes and trends in antibiotic resistance in a specific location (eg, a hospital or unit).
Infectious Disease Society of America guidelines regarding antibiotic stewardship recommend using institutional anti-biograms in the development of empiric antibiotic therapy guidelines.9 However, the ability of an antibiogram to predict antimicrobial susceptibility in individual patients (and therefore guide empiric therapy) can be limited by several factors, including sampling bias, inclusion of multiple samples of the same isolate, inclusion of surveillance (ie, nonclinical) isolates, and differences in resistance patterns based on the patient population, infection site, and healthcare location.10,11
Antibiograms also do not take into account the timing of the onset of infection. Infections occurring later in the hospital course are more likely to be caused by resistant pathogens than infections diagnosed early in the course of hospitalization,12–14 but it is not clear how this trend affects the value of the antibiogram as a tool to guide empiric antibiotic choice. Thus, our primary objective was to determine the utility of an antibiogram in predicting the susceptibility of P. aeruginosa isolates to anti-pseudomonal antimicrobial agents based on the day of hospitalization the specimen was collected. Our secondary objective was to describe the impact of the location of isolation on the predictive capability of the study antibiogram.
METHODS
This single-center retrospective cohort study was reviewed and approved by the Duke University Hospital (DUH) Institutional Review Board. Potential subjects were identified by querying the DUH Microbiology Laboratory and Duke Health Technology Solutions administrative databases. First, we reviewed all positive clinical cultures for P. aeruginosa from January 1, 2000, to June 30, 2007. Cultures obtained in either the outpatient setting or the inpatient setting were included. Second, only the first isolate from each admission or encounter was included to minimize the potential influence of duplicate isolates (independent of number of cultures or source). Patients could be included multiple times in our sample if they had more than 1 independent admission and/or encounter with a pseudomonal infection. Patients with cystic fibrosis were excluded. Data (including age, admission date, culture date, presence/absence of cystic fibrosis, and susceptibility results) were extracted from electronic medical records.
In vitro susceptibility testing was performed on all isolates and interpreted by the Duke University Clinical Microbiology Laboratory according to criteria published by the Clinical Laboratory Standards Institute15 for the following antibacterials: amikacin, ceftazidime, ciprofloxacin, gentamicin, imipenem-cilastin, meropenem, piperacillin-tazobactam, and tobramycin. Intermediately susceptible and resistant strains were classified as nonsusceptible. A study antibiogram was then produced by determining the overall percentages of P. aeruginosa isolates susceptible to each antimicrobial during the entire study period. Due to the high (>99.8%) similarity to imipenem-cilastin data, susceptibility data for meropenem were ultimately not included in our analyses.
For the primary analysis, we identified the day of hospitalization after which the study antibiogram no longer reliably predicted susceptibility to the targeted antibiotics. First, the day of infection was calculated for all isolates based on the day of hospitalization. Isolates obtained on the first day of admission and outpatient isolates were assigned a day of infection of 1. Logistic regression models were then created for each antibiotic, comparing percent susceptible (dependent variable) to day of infection (independent variable). The percent susceptible value for each antibiotic from the study antibiogram was then compared with results from the logistic regression models in order to identify the day of infection after which the study antibiogram no longer reliably predicted susceptibility to the antibiotic (ie, the average percent susceptible to that antibiotic was lower than the value calculated for the study antibiogram). This value is hereafter labeled as “time to unreliability” of the antibiogram. Importantly, this descriptive term is not intended to imply statistical reliability.
Simple logistic regression models were created as reference models for each antibiotic, with day of infection as the independent variable and susceptibility of each isolate as the dependent variable. Quadratic (day of infection2) and cubic (day of infection3) variables were created and added to each model in stepwise fashion and included if significant. Ultimately, simple logistic models were created for ceftazidime, imipenem-cilastin, tobramycin, and piperacillin-tazobactam. Logistic models with quadratic terms were created for ciprofloxacin, gentamicin, and amikacin. No models included a cubic term. Susceptibility data from isolates obtained more than 30 days after admission were not included in the models due to sporadic and decreasing numbers of isolates.
In order to determine the impact of the location of isolation on the predictive capability of the study antibiogram, we repeated the process described above after first stratifying isolates into outpatient and inpatient locations. For inpatient isolates, we further stratified into ICU and non-ICU ward locations. The χ2 test was used to compare susceptibilities by location. Differences in time to unreliability of the antibio-gram based on location were determined using logistic regression by including a binary variable for location (ICU vs non-ICU) in each of the models created above. Interaction terms (eg, ICU × day) were also evaluated for inclusion. Outpatient specimens were excluded from these models.
Data were maintained in a Microsoft Access database. All statistical analyses were performed using SAS v9.2.
RESULTS
We identified 8,078 P. aeruginosa isolates during the study period. After application of our inclusion/exclusion criteria, data from 3,393 isolates were included (Figure 1). The majority of clinical isolates were from respiratory, blood, or urine samples (Table 1). The median patient age was 57 years (range, 0–104).
FIGURE 1.
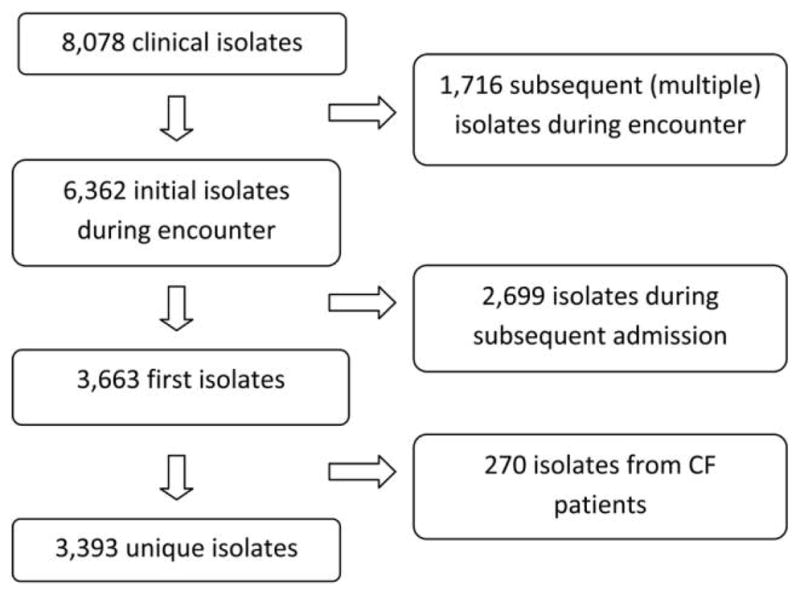
Selection of Pseudomonas aeruginosa isolates for inclusion in analyses.
TABLE 1.
Sources of 3,393 Pseudomonas aeruginosa Isolates Obtained at Duke University Hospital from January 1, 2000, to June 30, 2007
| Characteristics | n, % (N = 3,393) |
|---|---|
| Source of culture | |
| Urine | 1,161 (34) |
| Respiratory | 747 (22) |
| Blood | 706 (21) |
| Other | 376 (11) |
| ENT | 293 (9) |
| Eye | 60 (2) |
| Abdominal | 50 (2) |
NOTE. ENT, ear, nose, and throat.
The study antibiogram is presented in Table 2. Among the targeted antibiotics, amikacin exhibited the highest percent susceptibility (95%), while ciprofloxacin yielded the lowest (73%). In total, 2,302 (68%) isolates were obtained during hospitalization, while 1,091 (32%) were obtained in outpatient settings. Among the 2,302 inpatient isolates, 644 (28%) were from ICUs. Percent susceptibility to antibiotics changed based on location at the time of isolation (Table 2). In general, susceptibilities were lower among inpatient isolates than among outpatient isolates. For example, 1,034 (95%) out-patient isolates were susceptible to ceftazidime, while only 1,994 (87%) inpatient isolates were susceptible to ceftazidime (P< .0001). However, most antibiotic susceptibilities decreased by only 1%–6%. More notable decreases were observed when comparing isolates obtained in ICUs with isolates obtained in non-ICU hospital wards, though the decreases varied by antibiotic. For example, susceptibility to ceftazidime decreased by 12% and imipenem-cilastin decreased by 10%, while susceptibility to amikacin actually increased by 1%.
TABLE 2.
Susceptibility of Pseudomonas aeruginosa Isolates Susceptible to Targeted Antibiotics Based on Site of Care (Inpatient vs Outpatient and Inpatient Intensive Care Units [ICUs] vs Non-ICU)
| Antibiotic | Study antibiogram, % (n = 3,393)a | Inpatient isolates (n = 2,302)a | Outpatient isolates (n = 1,091)a | P | Isolates from ICUs (n = 644)b | Isolates from non-ICU inpatients (n = 1,658)b | P |
|---|---|---|---|---|---|---|---|
| Amikacin | 95.0 | 2,192 (95) | 1,023 (94) | .09 | 617 (96) | 1,575 (95) | .55 |
| Ceftazidime | 89.3 | 1,994 (87) | 1,034 (95) | <.0001 | 504 (78) | 1,490 (90) | <.0001 |
| Ciprofloxacin | 72.8 | 1,439 (71) | 734 (77) | .0004 | 404 (65) | 1,035 (73) | <.0001 |
| Gentamicin | 80.4 | 1,831 (80) | 892 (82) | .10 | 497 (77) | 1,334 (81) | .07 |
| Imipenem-cilastin | 90.8 | 2,043 (89) | 1,024 (94) | <.0001 | 525 (82) | 1,518 (92) | <.0001 |
| Piperacillin-tazobactam | 93.3 | 2,107 (92) | 1,059 (97) | <.0001 | 561 (87) | 1,546 (93) | <.0001 |
| Tobramycin | 92.6 | 2,103 (92) | 1,035 (95) | .0002 | 565 (88) | 1,538 (93) | <.0001 |
NOTE. All data are no. (%) unless otherwise indicated.
Susceptibility data were missing for the following antibiotics: amikacin (n =7), ceftazidime (n =1), ciprofloxacin (n =407), gentamicin (n =5), imipenem-cilastin (n =14), and tobramycin (n =5).
Susceptibility data were missing for the following antibiotics: amikacin (n =4), ceftazidime (n =1), ciprofloxacin (n =269), gentamicin (n =2), imipenem-cilastin (n =8), and tobramycin (n =3).
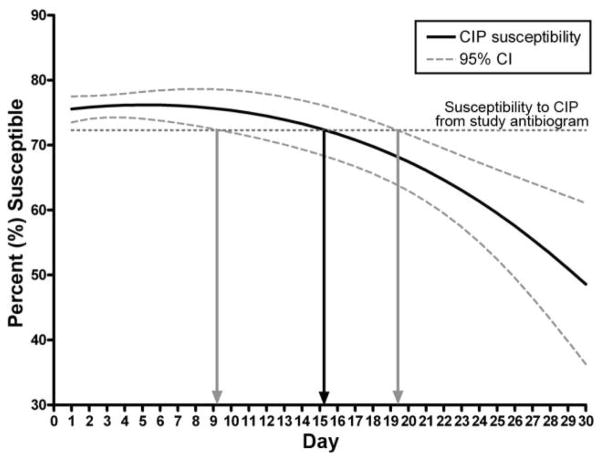
Time to unreliability of the antibiogram also varied for each antibiotic (Table 3). For example, the antibiogram became unreliable as a predictor of tobramycin on day 9; cef-tazidime, imipenem-cilastin, and piperacillin-tazobactam on day 10; and ciprofloxacin on day 15 but remained reliable for gentamicin (day 21) and amikacin (day 28) for longer. Figure 2 demonstrates the output from the quadratic logistic regression model created for ciprofloxacin (time to unreliability, 15 days; 95% confidence interval [CI], 9–19).
TABLE 3.
Time to Unreliability of the Antibiogram for Pseudomonas aeruginosa Isolates Based on Location of Isolation
| Antibiotic | Overall day (95% CI) | Non-ICU hospital ward day (95% CI) | ICU day (95% CI) | P a |
|---|---|---|---|---|
| Amikacin | 28 (21–>30) | 24 (<1–>30) | >30 (<1–>30) | .69 |
| Ceftazidime | 10 (8–11) | 12 (7–21) | 5 (<1–8) | .003 |
| Ciprofloxacin | 15 (9–19) | 19 (13–22) | 10 (<1–>8) | .66 |
| Gentamicin | 21 (12–27) | 21 (12–29) | 22 (<1–130) | .26 |
| Imipenem-cilastin | 10 (8–14) | >30 (13–>30) | 2 (<1–8) | <.0001 |
| Piperacillin-tazobactam | 10 (7–13) | 11 (2–>30) | 5 (<1–10) | .07 |
| Tobramycin | 9 (5–14) | 13 (1–>30) | 1 (<1–8) | .03 |
NOTE. CI, confidence interval; ICU, intensive care unit.
Comparison of time to unreliability of the antibiogram for isolates obtained in the ICU vs non-ICU hospital wards.
FIGURE 2.
Time to unreliability of the antibiogram as a predictor for Pseudomonas aeruginosa susceptibility to ciprofloxacin (CIP).
As before, time to unreliability of the antibiogram varied for several antibiotics based on location of isolation (Table 3). For example, the time to unreliability of the antibiogram for ceftazidime was 5 days (95% CI, <1–8) in the ICU and 12 days (95% CI, 7–21) in non-ICU hospital wards (P= .003).
DISCUSSION
Our study is the first to demonstrate that the reliability of data presented in the antibiogram decreases as length of hospitalization increases. In general, our study antibiogram became unreliable as a predictor for P. aeruginosa susceptibility to ceftazidime, imipenem-cilastin, piperacillin-tazobactam, and tobramycin after approximately 1.5 weeks of hospitalization, to ciprofloxacin after approximately 2 weeks, and to gentamicin and amikacin after 3 or more weeks. The reliability was even shorter for P. aeruginosa isolates obtained in ICUs. In contrast, the antibiogram was completely reliable for predicting susceptibility of P. aeruginosa isolates obtained in outpatient settings.
Antibiograms are often used by clinicians as an aid in selecting initial empiric antibiotic therapy and for monitoring changes in local antimicrobial-resistant patterns over time.16–19 The utility of an institution’s antibiogram to predict antimicrobial susceptibility in individual patients (and therefore guide empiric antimicrobial therapy), however, can be limited by several factors. Sampling bias may result when clinicians submit samples for patients with more severe infections or longer hospital stays or, conversely, from predominantly outpatient settings.10 In addition, duplicate isolates may be included if provisions are not in place to identify multiple samples obtained from the same patient. Similarly, provisions must also be in place to avoid reporting of susceptibility testing from isolates obtained as part of infection control surveillance rather than from clinical specimens. The origin of the pathogen (ie, community associated vs health-care associated), patient age group, prior antimicrobial exposure, infection site, or patient location at the time of isolation (ICU vs intermediate care) are usually not considered.11
Based on our findings, it is evident that the utility of the antibiogram decreases as the length of hospital stay increases. Thus, clinicians must be aware of this limitation and seek additional guidance when choosing empiric antimicrobial therapy for a patient with a prolonged hospitalization. There are numerous explanations for why this observation may occur. Of likely primary significance is the interaction between known trends: (1) organisms isolated from patients later in the hospitalization are more likely to represent infections acquired during the hospitalization20 and (2) infections occurring later in the hospital course are more likely to be caused by resistant pathogens than infections diagnosed early in the course of hospitalization.12–14
There are limitations to our retrospective observational study. First, we did not include data on potential patient-specific confounders (such as prior antibiotics, severity of illness, and comorbidities). Thus, we were unable to measure the potential impact of healthcare exposure (eg, nursing home or hemodialysis) in this analysis. While we excluded cystic fibrosis patients in an attempt to minimize the impact of multidrug-resistant P. aeruginosa infections, such patients would normally have been included in the antibiogram data. As such, our results are not generalizable to this specific population. Next, we assumed that patients with positive clinical cultures represented infection. Data from clinical specimens, regardless of whether the culture represents infection or colonization, however, are typically included in standard anti-biograms. Finally, our results likely require further validation, as we were unable to test our models in an independent sample of patients.
Our results must be interpreted in the context of local epidemiology. While we suspect that the same trends we described are present in other hospitals, our models are specific to our setting, location, and patient population. As such, we encourage other institutions to perform similar analyses to determine time to unreliability of the antibiogram in light of local epidemiology. These data could potentially be used to modify institution-specific guidelines for the empiric treatment of hospital-acquired infections where Pseudomonas spp. are likely pathogens.
Clinicians should be aware of methodologies considered in the formulation of the institution’s antibiogram. The antibiogram is an important tool to help guide clinicians in choosing appropriate empiric antimicrobial agents for suspected infection. Based on our findings, we believe clinicians should be cautious when using antibiogram data to predict the likelihood of susceptibility of P. aeruginosa isolates in patients with prolonged hospitalization.
Acknowledgments
Financial support. D.J.A. received grant support from the Robert Wood Johnson Foundation Physician Faculty Scholars Program and the National Institutes of Health (NIAID 1K23AI095357-01).
Footnotes
Presented in part: 49th Annual Interscience Conference on Antimicrobial Agents and Chemotherapeutics; San Francisco, California; September 12, 200977.
Potential conflicts of interest. D.J.A. has received research funding from Merck and Pfizer, has participated on the speaker’s bureau for Merck, and has received publication royalties from UpToDate Online. R.D. has received research funding from Merck/Schering-Plough, has participated on the speaker’s bureaus for Cubist and Merck/Schering-Plough, and has received publication royalties from UpToDate Online. All other authors report no conflicts of interest relevant to this article. All authors submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest, and the conflicts that the editors consider relevant to this article are disclosed here.
References
- 1.Wisplinghoff H, Bischoff T, Tallent S, Seifert H, Wenzel R, Ed-mond M. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 2.Pittet D, Li N, Woolson RF, Wenzel RP. Microbiological factors influencing the outcome of nosocomial bloodstream infections: a 6-year validated, population-based model. Clin Infect Dis. 1997;24:1068–1078. doi: 10.1086/513640. [DOI] [PubMed] [Google Scholar]
- 3.Neuhauser MM, Weinstein RA, Rydman R, Danziger LH, Karam G, Quinn JP. Antibiotic resistance among gram-negative bacilli in US intensive care units: implications for fluoroquinolone use. JAMA. 2003;289:885–888. doi: 10.1001/jama.289.7.885. [DOI] [PubMed] [Google Scholar]
- 4.Richards MJ, Edwards JR, Culver DH, Gaynes RP. Nosocomial infections in medical intensive care units in the United States. National Nosocomial Infections Surveillance System. Crit Care Med. 1999;27:887–892. doi: 10.1097/00003246-199905000-00020. [DOI] [PubMed] [Google Scholar]
- 5.Diekema DJ, Pfaller MA, Jones RN, et al. Survey of bloodstream infections due to gram-negative bacilli: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, and Latin America for the SENTRY Antimicrobial Surveillance Program, 1997. Clin Infect Dis. 1999;29:595–607. doi: 10.1086/598640. [DOI] [PubMed] [Google Scholar]
- 6.Obritsch MD, Fish DN, MacLaren R, Jung R. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob Agents Chemother. 2004;48:4606–4610. doi: 10.1128/AAC.48.12.4606-4610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang C, Kim S, Park W. Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother. 2005;49:760–766. doi: 10.1128/AAC.49.2.760-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marschall J, Agniel D, Fraser V, Doherty J, Warren D. Gram-negative bacteraemia in non-ICU patients: factors associated with inadequate antibiotic therapy and impact on outcomes. J Antimicrob Chemother. 2008;61:1376–1383. doi: 10.1093/jac/dkn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dellit T, Owens R, McGowan J., Jr Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clin Infect Dis. 2007;44:159–177. doi: 10.1086/510393. [DOI] [PubMed] [Google Scholar]
- 10.Pakyz A. The utility of hospital antibiograms as tools for guiding empiric therapy and tracking resistance insights from the society of infectious diseases pharmacists. Pharmacotherapy. 2007;27:1306–1312. doi: 10.1592/phco.27.9.1306. [DOI] [PubMed] [Google Scholar]
- 11.Banter C, Alcazar G, Franco D. Are laboratory-based antibio-grams reliable to guide the selection of empirical antimicrobial treatment in patients with hospital-acquired infections? J An-timicrob Chemother. 2007;59:140–143. doi: 10.1093/jac/dkl434. [DOI] [PubMed] [Google Scholar]
- 12.Carmeli Y, Troillet N, Eliopoulos GM, Samore MH. Emergence of antibiotic-resistant Pseudomonas aeruginosa: comparison of risks associated with different antipseudomonal agents. Anti-microb Agents Chemother. 1999;43(6):1379–1382. doi: 10.1128/aac.43.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodise TP, Miller CD, Graves J, et al. Clinical prediction tool to identify patients with Pseudomonas aeruginosa respiratory tract infections at greatest risk for multidrug resistance. Anti-microb Agents Chemother. 2007;51(2):417–422. doi: 10.1128/AAC.00851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasink LB, Fishman NO, Weiner MG, Nachamkin I, Bilker WB, Lautenbach E. Fluoroquinolone-resistant Pseudomonas aerugi-nosa: assessment of risk factors and clinical impact. Am J Med. 2006;119(6):e519–e525. doi: 10.1016/j.amjmed.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute (CLSI) Development of In Vitro Susceptibility Testing Criteria and Quality Control Parameters: Approved Guideline. 3. Wayne, PA: CLSI; 2008. CLSI document M23-A3. [Google Scholar]
- 16.Mizuta M, Linkin DR, Nachamkin I, et al. Identification of optimal combinations for empirical dual antimicrobial therapy of Pseudomonas aeruginosa infection: potential role of a combination antibiogram. Infect Control Hosp Epidemiol. 2006;27:413–415. doi: 10.1086/503175. [DOI] [PubMed] [Google Scholar]
- 17.Bryce EA, Smith JA. Focused microbiological surveillance and gram-negative beta-lactamase-mediated resistance in an intensive care unit. Infect Control Hosp Epidemiol. 1995;16:331–334. doi: 10.1086/647120. [DOI] [PubMed] [Google Scholar]
- 18.Stratton CW, Ratner H, Johnston PE, Schaffner W. Focused microbiologic surveillance by specific hospital unit as a sensitive means of defining antimicrobial resistance problems. Diagn Microbiol Infect Dis. 1992;15:11S–18S. [PubMed] [Google Scholar]
- 19.Binkley S, Fishman NO, LaRosa LA, et al. Comparison of unit-specific and hospital-wide antibiograms: potential implications for selection of empirical antimicrobial therapy. Infect Control Hosp Epidemiol. 2006;27:682–687. doi: 10.1086/505921. [DOI] [PubMed] [Google Scholar]
- 20.El Amari EB, Chamot E, Auckenthaler R, Pechere JC, Van DC. Influence of previous exposure to antibiotic therapy on the susceptibility pattern of Pseudomonas aeruginosa bacteremic isolates. Clin Infect Dis. 2001;33:1859–1864. doi: 10.1086/324346. [DOI] [PubMed] [Google Scholar]