Abstract
Bin3 is a conserved RNA methyltransferase found in eukaryotes ranging from fission yeast to humans. It was originally discovered as a Bicoid-interacting protein in Drosophila, where it is required for anterior-posterior and dorso-ventral axis determination in the early embryo. The mammalian ortholog of Bin3 (BCDIN3), also known as methyl phosphate capping enzyme (MePCE), plays a key role in repressing transcription. In transcription, MePCE binds the non-coding 7SK RNA, which forms a scaffold for an RNA-protein complex that inhibits P-TEFb, an RNA polymerase II elongation factor. MePCE uses S-adenosyl methionine to transfer a methyl group onto the γ-phosphate of the 5′ guanosine of 7SK RNA generating an unusual cap structure that protects 7SK RNA from degradation. Bin3/MePCE also has a role in translation regulation. Initial studies in Drosophila indicate that Bin3 targets 7SK RNA and stabilizes a distinct RNA-protein complex that assembles on the 3′ UTR of caudal mRNAs to prevent translation initiation. Much remains to be learned about Bin3/MeCPE function, including how it recognizes 7SK RNA, what other RNA substrates it might target, and how widespread a role it plays in gene regulation and embryonic development.
INTRODUCTION
Bin3 was first identified in the 1990’s by a graduate student as part of her thesis project aimed at understanding how Bicoid directs anterior development in the fruit fly.1 At the time, it was a novel protein, of large size (1368 aa) whose function was completely unknown. Bin3 is highly conserved and turns out to be very important; it is required for axis formation during Drosophila embryonic development2 and plays critical roles in transcription and translation regulation. What helped set the field in motion was the discovery in 2007 of the human homolog (BCDIN3/MePCE; 689 aa), and the demonstration that it uses RNA rather than a protein as a substrate for methylation.3 Its main target, the ubiquitous non-coding 7SK RNA, was already known to be a critical component of a regulatory complex that inhibits transcription elongation by P-TEFb. In this review, we describe the important features of the Bin3/MePCE protein from Drosophila and humans, its major substrate (7SK RNA), functional partners (HEXIMs, LARPs, P-TEFb, Bicoid), the structure of its catalytic domain, and the role of Bin3/MePCE in gene regulation and embryonic development. Finally, we speculate on additional roles of Bin3/MePCE and mention some major challenges that lie ahead.
BIN3 IS A CONSERVED RNA METHYLTRANSFERASE
Discovery of Bin3
Bin3 was discovered in a custom yeast two-hybrid screen using Drosophila melanogaster Bicoid as a bait protein.4 Bin3 also interacts directly with Bicoid in vitro in a GST pull-down assay.4 The bin3 gene is expressed during Drosophila oogenesis and in early embryos from maternal and zygotic promoters, respectively, and bin3 mRNA is distributed uniformly in embryos with a peak of expression at about 2 hours of development, coincident with Bicoid.2, 4 bin3 is also expressed during larval and pupal development, and in adults.5 Both transcripts encode the same 1368 residue protein. The protein is highly-conserved, and contains an S-adenosyl-L-methionine (SAM or AdoMet) binding domain, characteristic of DNA, RNA and protein (Arg/Lys) methyltransferases.6 Attempts to demonstrate protein methyltransferase activity were negative using histones, cytochromes and Bicoid as substrates.1
To understand Bin3, it is useful to introduce Bicoid. Bicoid is sequence-specific DNA binding (homeodomain) protein that is present in early Drosophila embryos in a steep anterior-to-posterior (A-P) concentration gradient.7 Bicoid determines cell fate by stimulating the transcription of target genes at discrete positions along the A-P axis.8, 9 Remarkably, Bicoid also binds a specific RNA sequence, the Bicoid Response Element (BRE), in the 3′ untranslated region (UTR) of caudal mRNA to repress its translation.10, 11 Bin3 is required for Bicoid to prevent Caudal protein from being expressed in the anterior of the embryo, a prerequisite for proper head development.12, 13 We will return to Bin3’s function in Drosophila development later in the review.
Conservation of Bin3 in eukaryotes
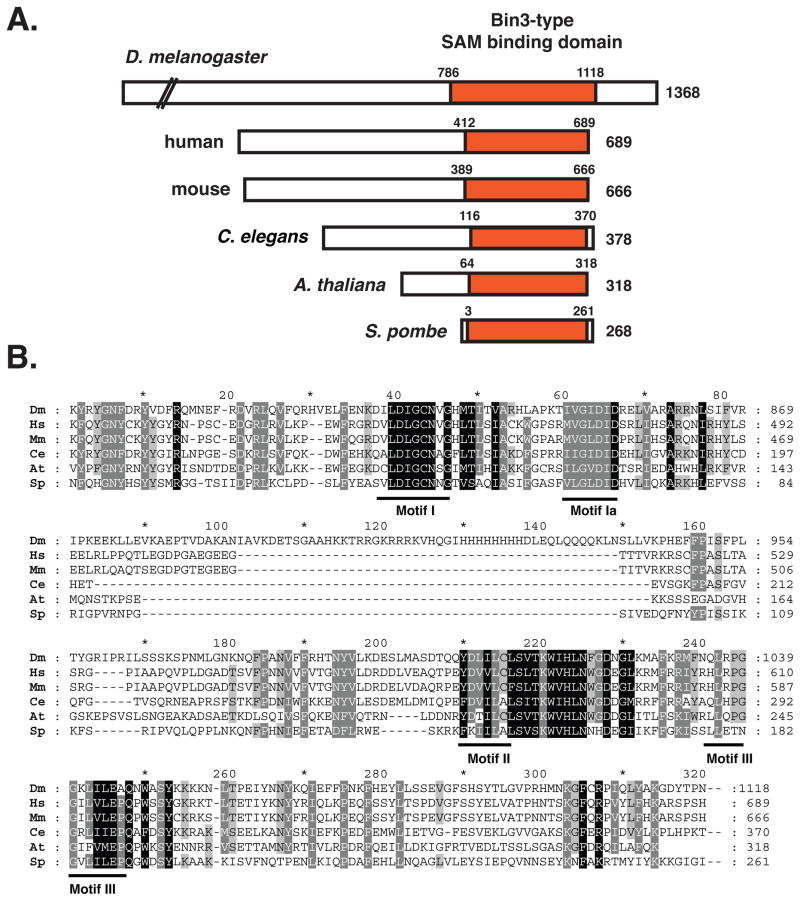
Putative Bin3 orthologs can be found in a wide range of eukaryotic organisms from Schizosaccharomyces pombe to humans.3, 4, 14 Phylogenetic analysis suggests an early evolutionary origin, with strong constraints on divergence limited to the AdoMet-binding domain (data not shown). A schematic of selected Bin3 orthologs and a sequence alignment of the conserved AdoMet binding domain, with signature motifs indicated are shown in Figure 1A, 1B. No additional conserved domains have been found. The previously noted “Bin3 domain”3, 15 is actually the second part of the bipartite AdoMet-binding (SAM) domain characteristic of these enzymes and slightly more conserved in Bin3-family members. The Drosophila Bin3 protein is much longer than Bin3 in other organisms; the function of the additional sequences is not known. No ortholog of Bin3/MePCE appears to be present in Saccharomyces cerevisiae. It should be noted that there is another Bin3 protein in the literature, Bridging-Integrator-3 (also known as Tcp2), originally identified in S. cerevisiae16, but this is an unrelated protein. Orthologs of the Drosophila Bin3 protein (MePCE) are defined by the presence of the AdoMet-binding domain and are all putative methyltransferases.
Figure 1.
(A) Alignment of Bin3-related proteins from selected species. The conserved catalytic domain, AdoMet (SAM)-binding domain is indicated. We extended the region based on sequence similarities in these Bin3/MePCE orthologs to the indicated residues. (B) Sequence alignment of conserved AdoMet (SAM)-binding domain. Note the extended region present in the Drosophila protein that is absent in the other orthologs. Motif I, II, and III are generic motifs characteristic of all SAM-dependent methyltransferases including RNA, DNA and protein methyltransferases.6 Sequences are from D. melanogaster (NP_ 724468.1; aa786–1118), H. sapiens (NP_062552.2; aa412–689), M. musculus (NP_659162.3; aa389–666), C. elegans (NP_496573.1; aa116–370), A. thaliana (NP_568752; aa64–318) and S. pombe (NP_596220.1; aa3–261). The alignment was performed by using ClustalW (http://www.ch.embnet.org/software/ClustalW.html) with default parameters for all settings and formatted using GeneDoc (http://www.psc.edu/biomed/genedoc).
Bin3 is the 5′-γ-phosphate capping enzyme
Although Bin3 was first reported in 20004, it was not recognized as an RNA methyltransferase. In a tour-de-force proteomic study of transcription factor networks, Jeronimo et al.3 identified the human ortholog of Bin3 (BCDIN3) and showed it had RNA methyltransferase activity. In their study, BCDIN3 co-purified with components of a regulatory complex that inhibits P-TEFb, an RNAPII elongation factor. P-TEFb consists of a cyclin-dependent kinase, CDK9 and CyclinT1 or CyclinT2, and is down regulated by sequestration into an RNA-protein complex (snRNP) containing 7SK RNA, HEXIM1/2, and LARP7 (Table 1). Jeronimo et al. found BCDIN3 in a protein network that included all of these proteins, and made the critical connection that BCDIN3 was probably the long sought-after 7SK RNA 5′ methyltransferase17, 18 (Box 1). The sequence of human BCDIN3 contained the AdoMet-binding domain originally found in Drosophila Bin3, and indeed they showed that BCDIN3 directly and specifically mono-methylates 7SK RNA on its 5′ γ-phosphate (Figure 2). Moreover, depletion of BCDIN3 from cultured human embryonic kidney cells (HEK293) using shRNAs significantly lowered 7SK RNA levels, consistent with the idea that uncapped 7SK RNA, like uncapped U6 RNA is sensitive to exonucleolytic degradation.19
TABLE 1.
| Name | Notable motifs/homology | Function | Interacts w/Bin3/MePCE1 |
|---|---|---|---|
| Bin3/BCDIN3 (MePCE) | SAM (Ado-Met) binding domain | RNA methyltransferase, targets 7SK RNA | - |
| 7SK RNA | (RNAPIII transcript; 330–440 nt in length) | Non-coding scaffold RNA; forms snRNP that inhibits P-TEFb | yes.3 |
| Transcription Regulation | |||
| CDK9 (human) | kinase (yeast Bur1) | PTEFb kinase; stimulates transcription elongation | no |
| CyclinT 1/2 (human) | cyclin box (yeast Bur2) | P-TEFb regulatory subunit | no |
| HEXIM 1/2 (human) | Arg-rich motif (ARM) | binds 7SK RNA; inhibits P-TEFb | ? |
| LARP7 (human) | La-related protein 7; RNA recognition motif (RRM) | binds 7SK RNA; inhibits P-TEFb | yes55 |
| Translation Regulation | |||
| Bicoid (Drosophila) | homeodomain | anterior morphogen directs head development, activates transcription & represses translation | yes4 |
| dLarp1 (Drosophila) | La-related protein 1; RNA recognition motif (RRM) | RNA binding protein, translation repression | ? |
| AGO2 | Argonaut, PAZ domain, | RISC complex, RNA silencing; translation repression | ? |
| PABP | RNA recognition motif (RRM) | poly(A)-binding protein; stimulates (or inhibits) translation initiation | ? |
| eIF4E | translation initiation factor | ? |
direct interaction documented
BOX 1. The unusual methylated 5′-γ-phosphate cap structure.
There are three types of cap structures that characterize eukaryotic RNAs, each of which requires the activity of a distinct AdoMet-dependent methyltransferase.88 The 5′ cap of most RNA polymerase II (RNAPII) products such as mRNAs and many viral RNAs consists of a 7-methylguanosine (m7G) attached to the 5′ nucleoside via a “reverse” (5′-5′) triphosphate linkage (m7GpppN). The methyl group is added to the N7 of guanine by a guanylytransferase. The capping reactions occur co-transcriptionally, and the final structure is permissive for translation by the ribosome. For non-coding RNAPII products including most small nuclear and nucleolar RNAs (e.g. U1, U2, U4, U5 snRNAs), this m7G cap is further methylated by the enzyme Tgs1 to form a trimethylguanosine (TMG) cap.89 Finally, some RNA polymerase III (RNAPIII) products including U6 snRNA, 7SK RNA, a mouse transposon-associated RNA called B2, and plant U3 snRNA, carry an unusual cap structure that does not utilize a 5′-5′-linked guanosine.90 Instead, the cap simply consists of a methyl group added to the γ-phosphate of the terminally-encoded guanosine residue (see Figure 2).17, 40 This structure is not permissive for translation, and only occurs on small non-coding RNAs. Purification of the enzyme activity responsible for γ-methylation of U6 snRNA has been reported.91, but the enzyme that methylates 7SK RNA had been more elusive until the discovery of the human Bin3 ortholog, BCDIN3, which has since been renamed MePCE for methyl phosphate capping enzyme.3
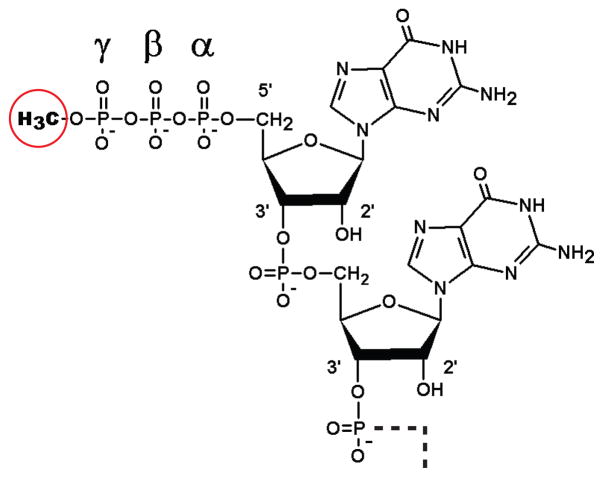
Figure 2.
Structure of the mono 5′ γ-monomethyl guanosine triphosphate cap of 7SK RNA. The terminal and penultimate residues (GG) of 7SK RNA are shown with the methyl group added to the 5′ γ-phosphate (circled). This is added by Bin3/MePCE from using S-adenosyl-L-methionine as a donor. This cap structure is different than the canonical m7G cap of most eukaryotic mRNAs (see Box 1).
As a result of this work, BCDIN3 was renamed as MePCE (Methyl Phosphate Capping Enzyme). Although we will refer to the original fly protein as Bin3, the human and other orthologs are most often referred to as MePCE. Interestingly, both 7SK and U6 RNAs associate with BCDIN3/MePCE in vivo, but the level of U6 snRNA was not reduced in BCDIN3/MePCE knockdown cells.3 Thus, it is not certain whether BCDIN3/MePCE targets only 7SK RNA or whether it carries out other 5′ γ-methylations in the cell.
Structure of the human Bin3/MePCE methyltransferase domain
The three-dimensional X-ray structure of the human MEPCE methyltransferase domain has been determined (PDB ID# 3G07) by the Structural Genomics Consortium in Toronto at 2.65 Å resolution.20 The model contains amino acids residues 431–487, corresponding to conserved motifs I and Ia; and residues 539–685, containing conserved motifs II and III. 52 of the less conserved residues between motifs I and II were disordered and not modeled. In addition, amino acids 665–675 were not modeled.
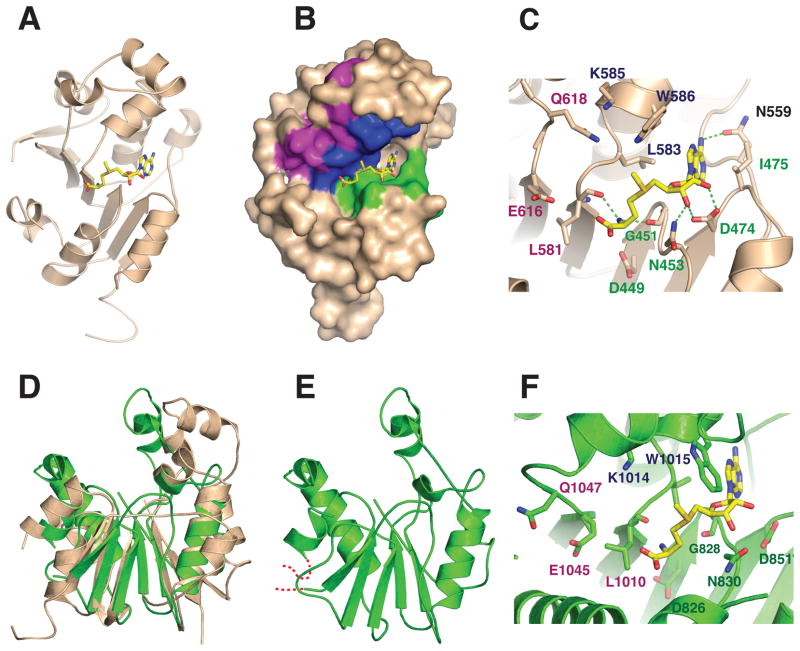
The structure contains a classical α/β methyltransferase fold, or “Rossman” fold, with a central parallel β-sheet containing a topological switch point in the center (Figure 3A). Like other nucleotide binding domains, S-adenosyl-L-methionine is bound in the cleft formed at the topological switch point and is lined with conserved residues from motifs I and II (Figure 3B). Binding is stabilized by numerous side chain and main chain hydrogen bonds (Figure 3C). Motif I interactions include a bidentate interaction between the carboxylate of D474 and the 2′- and 3′ hydroxyl atoms from the ribose moiety of AdoMet. Coenzyme binding is further stabilized by hydrogen bonds from the side chains of N453 and N559, and from the main chain carbonyls of G451 and L581 (Figure 3C). Residues I475, L583, and W586 form a hydrophobic pocket that surrounds the adenine moiety of S-adenosyl-L-methionine and the activated methyl group is oriented towards residues in Motifs II and III. We speculate that residues Q618 and E616 from Motif III and K585 from Motif II might bind 7SK RNA, positioning the γ-phosphate moiety of the 5′ guanosine of 7SK for nucleophilic attack. The side chains of these residues are within 5–7 Å of the activated methyl group of AdoMet and could hydrogen bond or form charge-charge interactions with the oxygen of the γ-phosphate. However, it is also possible that the active site is rearranged upon RNA binding and other residues may perform this function.
Figure 3.
(A) Cartoon diagram of the X-ray crystal structure of the catalytic domain of human Bin3/MePCE (residues 431–685) bound to S-adenosyl-L-methionine (AdoMet) (Yellow). The structure was drawn with PDB coordinates 3G07 using PyMol. (B) Surface representation of human MePCE showing the locations of the consensus AdoMet-binding Motifs I (green), II (blue) and III (purple).6 (C) Active site of human MePCE highlighting hydrogen bond contacts (green dashed lines) between conserved MePCE residues and AdoMet. The position of the donor methyl group of AdoMet suggests that residues from Motifs II and III may be involved in orienting the 7SK RNA substrate for methylation. (D) Superposition of human MePCE and Drosophila Bin3. (E) The Drosophila structure (green) is a homology model based on based on the human structure and refined with the Swiss Model server.21 Residues modeled include aa806–864 and 981–1092. The modeled residues are 47% identical between human and Drosophila Bin-3. The RMS deviation for Cα atom positions is 3.5 Å. (F) Putative active site of Drosophila Bin3 based on homology modeling. Shown are residues that could potentially interact with AdoMet and RNA as in (C).
Based on the structure of human Bin3/MePCE (Figure 3D) and the conservation of amino acids between human and fly orthologs, we created a homology model of D. melanogaster Bin3 using the Swiss Model server.21 (Figure 3E). The model is missing amino acid residues 864–967 (D. melanogaster numbering) which corresponds to the Drosophila-specific insert (Figure 1B, and red dashes in Figure 3E), the function of which is unknown. Superposition of human Bin3/MePCE with the Drosophila homology model of Bin3 (Figure 3D), indicates the overall fold is similar. Most of the conserved residues in the putative active site map to similar positions in the Drosophila homology model, suggesting that they will have similar functions. For example, substitutions in Drosophila residues L1010, D826, G828, N830 and D851 (corresponding to human MePCE residues L581, D449, G451, N453, and D474, respectively) should compromise AdoMet binding, while subsitutions in residues Q1047, E1045, K1014 (corresponding to residues Q618, E616 and K585 in humans) might affect RNA binding. The in vivo importance of these residues could be tested using transgenic Drosophila.
7SK RNA IS THE MAJOR TARGET OF BIN3/MEPCE
Basics of 7SK RNA
7SK is a small non-coding RNA discovered in human cells22 and transcribed by RNA polymerase III.23 7SK gene transcription is directed from an upstream promoter element, which at the time, was unusual for an RNAPIII product.24 In humans, 7SK is a 331 nucleotide GC-rich RNA. that is very abundant in cells (50–100,00 copies per cell), and localizes primarily to the nucleus, mostly within nuclear speckles, which contain mRNA processing factors.25,26 In addition to its 5′ γ-methylation, 7SK RNA is adenylated at its 3′-end, as are other RNAPIII products such as ribosomal 5S and U6 RNA.27, 28 In 7SK, a single adenosine is added 3′ to the terminal UUUU sequence.28 7SK RNA is highly conserved 29, and although initially thought to exist primarily in vertebrates, detailed sequence analyses revealed 7SK genes to be widespread in metazoans, including D. melanogaster and C. elegans.14, 30,31 Interestingly, organisms with 7SK RNA have almost always co-evolved with recognizable HEXIM, LARP7 and Bin3/MePCE orthologs.14 Exceptions include S. pombe, which has Bin3/MePCE but no recognizable 7SK RNA or HEXIMs, and nematodes, which have 7SK RNA, HEXIM and Bin3/MePCE but no LARP7.14 Several excellent reviews about 7SK RNA have been published.32–35
Structure of 7SK RNA
Two major themes have emerged from computational and structural studies of 7SK RNA. First, 7SK RNA is highly structured and the motifs formed as a result of folding serve as binding sites for HEXIMs, LARP7 and other proteins in the P-TEFb inhibitory complex.14, 25, 36, 37 Second, the structure is dynamic, and as part of P-TEFb regulation, structural changes occur in 7SK RNA that accompany protein exchange.38, 39 Most predicted structures display 4 major stem-loop regions, with alternate substructures forming as a result of protein interactions (see Peterlin et al. al for review).34 With respect to binding and 5′ γ-methylation by Bin3/MePCE, it is not known what sequences within 7SK RNA are required, although most models have the 5′ and 3′ regions of the RNA in close proximity. For U6 snRNA, the terminal guanine and a short stem-loop followed by the sequence AUAUAC was necessary and sufficient for γ-methylation by HeLa cell extracts40, although it is not known whether Bin3/MePCE was the enzyme responsible.
Drosophila 7SK RNA
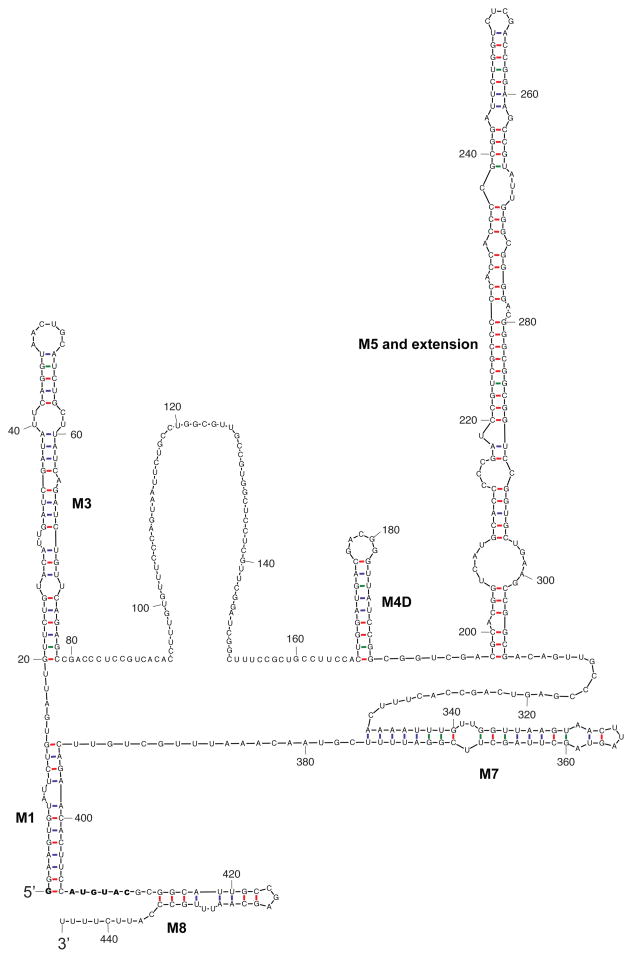
The Drosophila 7SK RNA transcript (444 nt) is significantly longer than its mammalian counterpart (330 nt)30, 41, and although the function of these extra nucleotides is not known, they were predicted to form an expanded loop between stems M4 and M5 that occur in the second major hairpin of mammalian 7SK RNA.14 To better understand Drosophila 7SK RNA, we utilized a computational analysis program known as Sfold42 to predict its secondary structure (Figure 4). Using Sfold and alignment information and the consensus structure model from Marz et al.14, we annotated the stems in our predicted structure (Figure 4). We found that M6 is absent as reported by Martz et al.14 In addition, none of M2a, M2b or M2c are present, and M4 is missing due to complete single-strandedness of the nucleotide block from U120 to C157, which was aligned to a human 7SK RNA sequence block containing the 5′ end nucleotides in M4. A downstream neighboring nucleotide block (C168 to G190) forms a new stem upstream of M5 with an extension. We named this new stem M4D (D for Drosophila). The stem-loops implicated in protein binding for human 7SK RNA, M1, M3 and M8 are retained in the Drosophila 7SK RNA, which in addition to M4D, also has a much larger M5 (Figure 4).
Figure 4.
Secondary structure prediction of D. melanogaster 7SK RNA. Methods: Sfold software is based on structure ensemble sampling, structure clustering and centroid representation of clusters.92, 93 It has been observed that the centroid structure of one of the clusters can often make an accurate structure prediction.93 However, the identity of the best performing centroid is unknown without additional information. For human 7SK RNA, the centroid structure for one of four structural clusters closely matched the proposed consensus structure.14 This indicated that, by using information from the consensus structure, we could identify the best performing centroid for structure prediction of 7SK RNA. For Drosophila 7SK RNA, we found that the centroid of one of two clusters closely resembles the consensus structure. This centroid is an informed predictor of the secondary structure for Drosophila 7SK RNA (Figure 4). The structure diagram was produced by the Sir_Graph program of the UNAFold package.94 Red = G-C; blue = A-U, green = G-U base pairing
The 5′G that is methylated by Bin3/MePCE is indicated in bold as is a putative recognition motif, AUGUAC (Figure 4), based on the AUAUAC sequence utilized in U6 RNA.40 Remarkably, this AUGUAC motif is located in the exact position relative to the 5′ G that is required for methylation in U6.40 Curiously, this sequence does not seem to be well-conserved in human 7SK RNA (AAAUGA). Alternatively, the 3′ stem loop whose presence (but not sequence) near the 5′G is conserved between Drosophila and humans could be important for recognition by Bin3/MePCE. In the long single-stranded “bowling pin” region, about two thirds of the 89 nucleotides are Drosophila-specific, suggesting that this region may contain regulatory sites for Drosophila-specific RNA binding proteins.
BIN3/MEPCE IN TRANSCRIPTION REGULATION
Regulation of P-TEFb activity by the Bin3/7SK snRNP
Positive-acting transcription elongation factor b, P-TEFb, is thought to be important for transcription of most if not all RNAPII-dependent genes (reviewed in refs.32–34). P-TEFb works by overcoming promoter-proximal pausing of RNAPII, which may be a common regulatory step of transcription in metazoans.43–45 The CDK9 catalytic subunit of P-TEFb phosphorylates Ser2 within the heptapeptide repeat (YSPTSPS)n of the carboxy-terminal domain of the large subunit of RNAPII, generating an elongation-competent form of the polymerase. P-TEFb also phosphorylates a negative elongation factor, NELF, leading to its release, and DSIF (DRB-sensitive inducing factor; aka Spt4/5), leading to its conversion into an elongating form. The action of P-TEFb is thus a key step in the conversion of initiated but stalled RNAPII into a form capable transcribing genes.
Where does Bin3/MePCE fit in? 7SK RNA, the target of Bin3/MePCE, was discovered to inhibit the activity of P-TEFb.46, 47 7SK RNA serves as a scaffold for a snRNP, later shown to contain two RNA-binding proteins, HEXIM (hexamethylene bisacetamide-inducible) proteins 1 and 248–50, LARP7 (La-related protein 7)28, 37, 51, as well as Bin3/MePCE.3 Bin3/MePCE and LARP7 are stably bound to 7SK RNA28, 37, while a HEXIM dimer binds reversibly and is required for inhibition of P-TEFb via interaction with the CyclinT subunits.49, 52 When HEXIM dissociates from 7SK, hnRNPs (hnRNPA1/2, hnRNPQ, hnRNPR) take its place.53, 54 Regulation of P-TEFb therefore occurs by a mechanism of sequestration and release from the 7SK snRNP (Figure 5). Knockdown of 7SK RNA was reported to disrupt the organization of the nuclear speckle, and cause upregulation of reporter genes, most likely because P-TEFb is released from the inhibitory 7SK snRNP.26
Figure 5.
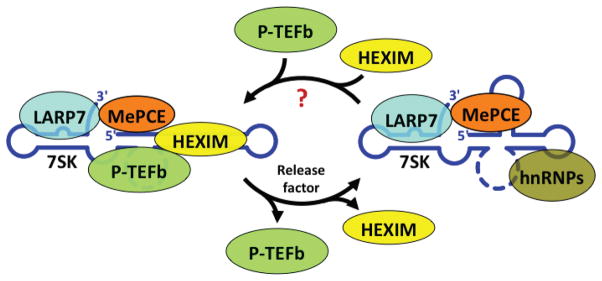
Dynamic association of P-TEFb with the inhibitory 7SK snRNP. Figure summarizes work from a number of laboratories (reviewed in refs.32–34). The P-TEFb transcription elongation factor is composed of CDK9 and CyclinT1 or T2. 7SK RNA forms a scaffold for an snRNP containing 7SK, LARP7, MePCE and hnRNPs. When P-TEFb, along with a HEXIM1/2 dimer is incorporated into the 7SK snRNP, it displaces hnRNPs and inactivates P-TEFb. P-TEFb inactivation involves interaction between HEXIMs and the cyclinT subunit of P-TEFb. 7SK RNA folding is different in the two complexes, as it is known to undergo structural rearrangements. LARP7 and HEXIMs are shown in the approximate positions they bind to 7SK RNA based on chemical protection experiments. See text for details. Figure is from Peterlin et al. (2011).34
Bin3/MePCE plays at least two critical roles. First, it covalently modifies 7SK RNA via 5′ methylation, which is likely to protect it from degradation (and from translation). Second, Bin3/MePCE interacts directly with LARP7, and together these proteins bind cooperatively to 7SK RNA, further stabilizing the complex.55 Interactions were also detected between Bin3/MePCE and both CDK9 and HEXIM1, but it is not known if these were direct or indirect.3 In the absence of Bin3/MePCE, 7SK RNA levels are drastically reduced.2, 3 Without a functional 7SK snRNP, P-TEFb activity is unleashed and gene expression goes unregulated, which can lead to cancer and other disease states.28, 56–58.
Several important cellular and viral signaling pathways converge on P-TEFb, resulting in phosphorylation/dephosphorylation of CDK9, CyclinT, HEXIMs and Brd4, a protein that recruits P-TEFb to genes, thus controlling the activity of this critical regulator.59–61 P-TEFb is a direct target of HIV Tat protein, which recruits P-TEFb to the TAR RNA element allowing expression of viral genes.62, 63 Regulation of P-TEFb activity is a very active area of research and has been reviewed elsewhere.32, 64 The human MePCE sequence contains 13 potential MAPK/CDK kinase sites (S/T-P).65 It seems plausible that Bin3/MeCPE is a target for modification and/or regulation that would impact P-TEFb. For example, stimulation of ERK (extracellular-signal-regulated kinase; a MAPK) via T-cell receptor-mediated activation disrupts the 7SK snRNP complex, freeing P-TEFb to enter the nucleus and promote HIV provirus elongation (independent of Tat protein)60. It is possible that ERK targets Bin3/MePCE for phosphoryation, as well as other components of the inhibitory snRNP to cause release and activation of P-TEFb.
Conservation of the P-TEFb system in Drosophila
While P-TEFb was originally discovered in Drosophila cell extracts66, it was unclear whether 7SK snRNP control of P-TEF was operational in this organism. In fact, only recently was it even possible to identify a 7SK gene in Drosophila. This was done by a novel strategy that took into account the 7SK gene’s upstream RNAPIII promoter.30 Singh et al.2 used qRT-PCR to show that 7SK RNA is present in ovaries and embryos and is destabilized in bin3 mutants, consistent with this transcript being a target for 5′ γ-methylation as in human cells.3 They also found that a 7SK insertion allele was embryonic lethal (Singh and Hanes, unpublished). Nguyen et al. then showed that all the key players in the 7SK RNA/P-TEFb regulatory network are present in Drosophila cells. In addition to identifying dHEXIM and dLARP7 orthologs, they showed these proteins bind to 7SK RNA, and are present in DRB- and flavopiridol-sensitive complexes along with CyclinT, as expected if they regulate P-TEFb by a conserved mechanism. These findings open the door to study of 7SK snRNP regulatory functions in a developmental context. Indeed, a series of UAS-GAL4 RNAi knockdown lines were used to show that loss of dHEXIM caused pronounced mutant phenotypes in the tissues in which it was reduced.41 Further work using insertion and excision alleles of 7SK snRNP components will be useful for studying maternal and early embryonic functions.
Additional roles for Bin3/MePCE in transcription and RNA processing
RNA splicing occurs co-transcriptionally, and RNAPII elongation rates can effect splicing efficiency and even splice-site choice.67–70 Not surprisingly, defects in splicing are observed when regulation of P-TEFb by the 7SK snRNP is disrupted.71 For example, RNAi depletion of Bin3/MePCE or LARP7 in HeLa cells and zebrafish embryos reduced 7SK RNA levels and altered the splicing patterns on selected mRNAs.72 Interestingly, depletion of Bin3/MePCE in zebrafish lead to developmental defects in the anterior (brain) regions. No splicing defects were observed, however, in early Drosophila embryos2, perhaps because the mRNAs were maternally-derived and elongation might not be subject to P-TEFb regulation during oogenesis.
Another role for 7SK RNA has been proposed.73 This was based on the observation that 7SK RNA knockdown affected the expression of a larger set of genes than P-TEFb was known to regulate. Eilebrecht et al.73 used a structural loop (L2) of 7SK RNA that is not involved in binding HEXIMs, LARP7 or MePCE, and biochemically identified HMG (high mobility group) protein A1 as a direct binder of 7SK RNA. HMGA1 normally binds DNA and functions as a chromatin architectural protein affecting the expression of a large number of genes.74 They found that 7SK RNA competes for DNA binding with HMGA1 and that overexpression of 7SK RNA (L2) altered the expression of HMGA1-dependent genes, separate from P-TEFb effects.75 For common targets it was proposed that HMGA1 recruits P-TEFb to genes via their interactions with the 7SK snRNP.75 Specific roles for Bin3/MePCE in this function of the 7SK snRNP have not been described.
BIN3/MEPCE IN TRANSLATION REGULATION
Bin3 is critical for embryonic axis formation
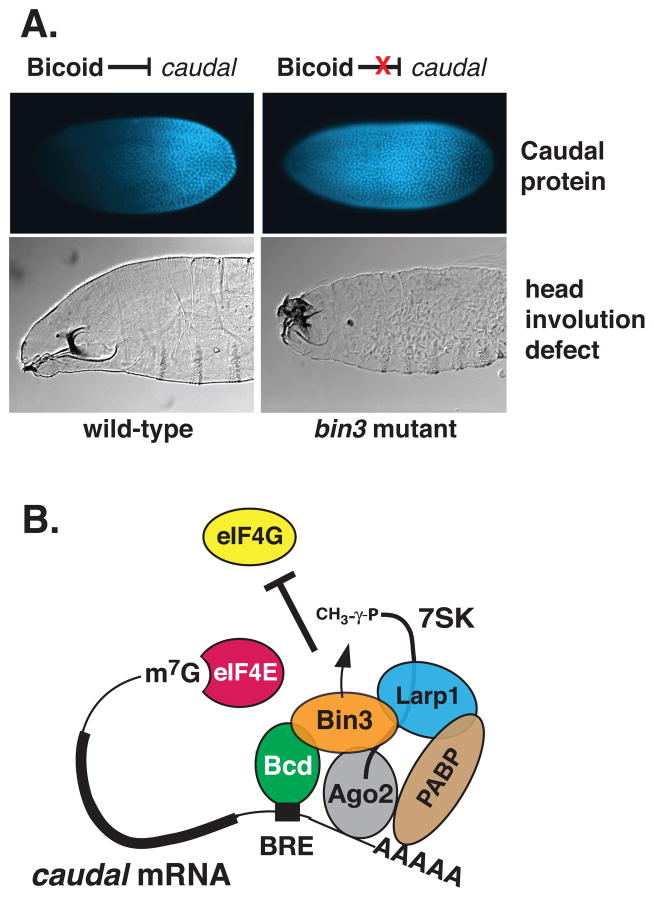
To study Bin3/MePCE’s role in Drosophila development, Singh et al.2 used site-specific recombination to generate precise excision alleles in the bin3 gene that removed most exons including the entire AdoMet (SAM)-binding domain. The surprise result was that although Bin3 was identified as a Bicoid-interacting protein, bin3 null mutants showed no effect on Bicoid-dependent gene transcription. None of the Bicoid target genes examined had detectable changes in abundance or spatial/temporal expression patterns. Instead, embryos from bin3 null mothers failed to repress caudal mRNA translation in the anterior region of the embryo, leading to severe head involution defects and lethality (Figure 6A). 2 Notably, 7SK RNA levels were reduced dramatically in bin3 mutants, 140-fold in ovaries and 50-fold embryos, respectively.2 As in MePCE knockdowns in human cells, there was no effect on U6 snRNA levels in bin3 mutants (Singh and Hanes, unpublished). Perhaps a different enzyme methylates U6, or uncapped U6 is not subject to degradation. There are two predicted Bin3-like proteins in Drosophila that might have capping activity (CG11342, CG1239).
Figure 6.
(A) Bin3/MePCE is required for translation repression in early development. Blastoderm-staged embryos (0–2 hr) are stained for Caudal protein (upper panels). Embryos and larvae are oriented with the anterior-left, dorsal-up. In wild-type embryos, Bicoid represses translation of caudal mRNA preventing accumulation of Caudal protein in the anterior (left panel). In bin3 loss-of-function mutant embryos, Bicoid is unable to repress caudal translation and Caudal protein accumulates throughout the embryo (right panel), resulting in failure to undergo proper head involution (lower, right panel), as visualized in first instar larvae. Data are from Singh et al. (2011).2 (B) Model for Bin3-Bicoid repression of caudal mRNA translation. Bin3 stabilizes Bicoid binding to the BRE in the caudal 3′ UTR. Bin3 does so by methylating and remaining bound to 7SK RNA which serve as scaffold for binding of other proteins, including the La-related protein, Larp1 and Ago2 and PABP which contribute to negative regulation of initiation. Although Bin3 is drawn as methylating the 5′ end of 7SK RNA as part of this repression complex, this modification may instead take place early, e.g. during transcription of 7SK RNA synthesis.55 See text for details. Model based on Singh et al. (2011).2
A model for how Bin3 augments Bicoid’s function in translation repression is shown in Figure 6B. In this model, Bin3 directly binds to Bicoid (Bcd), which targets the 3′ UTR of caudal mRNA at the BRE. Bin3 methylates 7SK RNA and remains bound to it, protecting it from degradation. 7SK RNA functions as a scaffold to stabilize the formation of a repressive snRNP complex that contains poly(A) binding protein (PABP), La-related protein 1 (Larp1) and Argonaut 2 (Ago2), and inhibits initiation by preventing binding of required factors to eIF4E. Each of these proteins has been shown to play roles in translation repression in Drosophila.76–80 The model is supported by double mutant analyses as well as RNA co-immunoprecipitation data.2 These studies were the first indication that a distinct 7SK snRNP, which includes Bin3/MePCE plays a role outside of transcription.
A general role for Bin3/MePCE in translation regulation?
It is unlikely that the role of Bin3/MePCE in early embryos is strictly dedicated to Bicoid function in A-P axis formation. In fact, oocytes from bin3 mutant mothers also show dorso-ventral (D-V) defects. Loss of bin3 results in dorsalized oocytes, whereas overexpression of bin3 leads to ventralized oocytes.2 These defects are likely due to faulty repression of gurken mRNA translation, which also requires a repression complex to assemble on its 3′ UTR.81, 82 It is possible that Bin3/7SK RNA forms a scaffold that stabilizes this complex. Thus, we suspect that in early Drosophila embryos, before cellularization occurs, Bin3/MePCE might play a global role in translation regulation. This is the time of development, prior to the onset of zygotic transcription, that is directed by stores of maternal RNAs and proteins that have been deposited in the oocyte. During this time, transport, localization, and translation of individual mRNAs is carefully regulated to allow spatial and temporal control of protein production within the embryo.82, 83 By contrast, in mammalian embryos, where the influence of maternal gene products is far less important, Bin3/MePCE may have little or no role(s) in translation regulation.
Additional roles of Bin3/MePCE during development
Analysis of bin3 mutants, and HEXIM and LARP7 knockdown flies clearly indicates additional roles for Bin3/MePCE and the 7SK snRNP in late embryonic, larval and pupal development.2, 41 During these stages, transcription regulation is even more critical and the 7SK snRNP probably plays a major role in P-TEFb regulation of developmental genes such as those in the HOX cluster.84 Bin3/MePCE has even been implicated in adult function in sleep regulation.85 The recent identification of the genes encoding components of the 7SK complex in Drosophila provides an excellent opportunity for future studies.
CONCLUSION
Much attention has focused on non-coding RNAs discovere d in genome-wide transcriptome analysis, but the biological roles of these RNAs remain poorly understood.86, 87 The 7SK RNA provides a clear example of how a longer non-coding RNA (i.e. not a microRNA) can play critical roles in gene regulation. Bin3, or MePCE as it will probably become known, plays a key role in the metabolism and activity of 7SK RNA in both transcription and translation regulation. Thus, Bin3/MePCE is of central importance for understanding regulatory mechanisms at the heart of embryonic developmental and cellular regulation. Many questions remain, however.
It is currently not understood how Bin3/MePCE can play dual roles in transcription and translation, and how 7SK RNA might serve as a scaffold for distinct complexes. While there is no direct biochemical evidence to indicate which proteins are present in the putative 7SK translation regulatory complex other than Bicoid and Bin3 (e.g. are HEXIMS, LARP7 present?), it is clear that Bicoid, PABP and Ago2 and Larp1 are not present in the P-TEFb transcription regulatory snRNP. Therefore, how would distinct 7SK RNP complexes form? Perhaps transcription- and translation-specific RNPs assemble at distinct times in development (i.e. are temporally regulated), or their formation is driven by nuclear vs. cytoplasmic localization of the respective protein components. For Bin3/MePCE, it is possible that post-translational modifications could direct distinct forms of the enzyme to the nucleus and cytoplasm. It will also be interesting to know if the different complexes contain alternatively-folded forms of 7SK RNA. Additional biochemical and cell biological studies are required to address these important issues.
No mechanistic studies have been done on Bin3/MePCE. The sequence-specificity of Bin3/MePCE and the mechanism of binding to RNA have not been defined, nor has the catalytic mechanism been studied for this class of RNA methyltransferase. The spectrum of substrates is not known. It has not even been rigorously determined whether Bin3/MePCE methylates U6, plant U3 or other RNAPIII products that contain a 5′ γ-methyl-phosphate cap structure. Only about half of Bin3/MePCE was reported to be associated with P-TEFb28, 37, suggesting alternative activities.
From an evolutionary standpoint it is intriguing that Bin3/MePCE seems to be present in some organisms that lack 7SK RNA, S. pombe, for example, suggesting it may have other cellular targets.14 Assuming the targets are RNAs, might they also serve a scaffold-type function to assemble additional regulatory snRNPs? For Drosophila Bin3, it is also not clear why this protein is so much larger (1368 aa) than its human MePCE counterpart (689aa), nor what the extra 100 or so nucleotides in Drosophila 7SK RNA are doing. No information is available on the function or relevance of Bin3-related proteins encoded in many genomes. Finally, given the link between P-TEFb and an number of human diseases, including cancer and HIV infection, its seems reasonable to think that targeting of Bin3/MePCE might have future therapeutic applications. An ample research frontier exists for study of this interesting enzyme.
Acknowledgments
The authors apologize for any citations that were overlooked. We thank David Price (U. Iowa) and the Toronto Structural Genomics Consortium for sharing unpublished data, and David Atencio, Cassandra Barnes and Navjot Singh, for help with the figures and/or comments on the manuscript. We are grateful for research support from the March of Dimes (1-FY07-527) to SDH, and the National Institutes of Health to SDH (R01-GM55108), MC (R01-CA140522), and YD (R01-GM099811), and from the National Science Foundation to YD (DBI-0650991).
Footnotes
The authors declare no Conflict of Interest.
References
- 1.Zhu W. PhD Thesis. Department of Biomedical Sciences, School of Public Health, State University of New York; 2000. Isolation and Characterization of Bicoid-interacting Proteins: Bin1 a Homolog of Human SAP18 and Bin3, a putative Protein Methyltransferase. [Google Scholar]
- 2.Singh N, Morlock H, Hanes SD. The Bin3 RNA methyltransferase is required for repression of caudal translation in the Drosophila embryo. Dev Biol. 2011;352:104–115. doi: 10.1016/j.ydbio.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, Chabot B, Poirier GG, Hughes TR, Blanchette M, Price DH, Coulombe B. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu W, Hanes SD. Identification of Drosophila Bicoid-interacting proteins using a custom two-hybrid selection. Gene. 2000;245:329–339. doi: 10.1016/s0378-1119(00)00048-2. [DOI] [PubMed] [Google Scholar]
- 5.http://flybase.org/
- 6.Kagan RM, Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch Biochem Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- 7.Driever W, Nusslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- 8.Driever W, Nusslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- 9.Driever W, Nusslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- 10.Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 11.Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring WJ, Jackle H. RNA binding and translational suppression by bicoid. Nature. 1996;379:746–749. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 12.Mlodzik M, Gehring WJ. Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell. 1987;48:465–478. doi: 10.1016/0092-8674(87)90197-8. [DOI] [PubMed] [Google Scholar]
- 13.Mlodzik M, Gibson G, Gehring WJ. Effects of ectopic expression of caudal during Drosophila development. Development. 1990;109:271–277. doi: 10.1242/dev.109.2.271. [DOI] [PubMed] [Google Scholar]
- 14.Marz M, Donath A, Verstraete N, Nguyen VT, Stadler PF, Bensaude O. Evolution of 7SK RNA and its protein partners in metazoa. Mol Biol Evol. 2009;26:2821–2830. doi: 10.1093/molbev/msp198. [DOI] [PubMed] [Google Scholar]
- 15.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Sullivan DS, Huffaker TC. Two yeast genes with similarity to TCP-1 are required for microtubule and actin function in vivo. Proc Natl Acad Sci USA. 1994;91:9111–9115. doi: 10.1073/pnas.91.19.9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shumyatsky GP, Tillib SV, Kramerov DA. B2 RNA and 7SK RNA, RNA polymerase III transcripts, have a cap-like structure at their 5′ end. Nucleic Acids Res. 1990;18:6347–6351. doi: 10.1093/nar/18.21.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta S, Busch RK, Singh R, Reddy R. Characterization of U6 small nuclear RNA cap-specific antibodies. Identification of gamma-monomethyl-GTP cap structure in 7SK and several other human small RNAs. J Biol Chem. 1990;265:19137–19142. [PubMed] [Google Scholar]
- 19.Hamm J, Darzynkiewicz E, Tahara SM, Mattaj IW. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990;62:569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- 20.http://www.sgc.utoronto.ca/pmwiki/pmwiki.php?n=Crystallography.HomePage (PDB ID 3G07)
- 21.http://swissmodel.expasy.org/
- 22.Zieve G, Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976;8:19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]
- 23.Zieve G, Benecke BJ, Penman S. Synthesis of two classes of small RNA species in vivo and in vitro. Biochemistry. 1977;16:4520–4525. doi: 10.1021/bi00639a029. [DOI] [PubMed] [Google Scholar]
- 24.Murphy S, Tripodi M, Melli M. A sequence upstream from the coding region is required for the transcription of the 7SK RNA genes. Nucleic Acids Res. 1986;14:9243–9260. doi: 10.1093/nar/14.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy R, Henning D, Subrahmanyam CS, Busch H. Primary and secondary structure of 7-3 (K) RNA of Novikoff hepatoma. J Biol Chem. 1984;259:12265–12270. [PubMed] [Google Scholar]
- 26.Prasanth KV, Camiolo M, Chan G, Tripathi V, Denis L, Nakamura T, Hubner MR, Spector DL. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol Biol Cell. 2010;21:4184–4196. doi: 10.1091/mbc.E10-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Sinha K, Perumal K, Gu J, Reddy R. Accurate 3′ end processing and adenylation of human signal recognition particle RNA and alu RNA in vitro. J Biol Chem. 1998;273:35023–35031. doi: 10.1074/jbc.273.52.35023. [DOI] [PubMed] [Google Scholar]
- 28.He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell. 2008;29:588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ullu E, Esposito V, Melli M. Evolutionary conservation of the human 7 S RNA sequences. J Mol Biol. 1982;161:195–201. doi: 10.1016/0022-2836(82)90286-8. [DOI] [PubMed] [Google Scholar]
- 30.Gruber AR, Kilgus C, Mosig A, Hofacker IL, Hennig W, Stadler PF. Arthropod 7SK RNA. Mol Biol Evol. 2008;25:1923–1930. doi: 10.1093/molbev/msn140. [DOI] [PubMed] [Google Scholar]
- 31.Copeland CS, Marz M, Rose D, Hertel J, Brindley PJ, Santana CB, Kehr S, Attolini CS, Stadler PF. Homology-based annotation of non-coding RNAs in the genomes of Schistosoma mansoni and Schistosoma japonicum. BMC Genomics. 2009;10:464. doi: 10.1186/1471-2164-10-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNABiology. 2009;6:122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- 33.Kohoutek J. P-TEFb- the final frontier. Cell Division. 2009;4:19. doi: 10.1186/1747-1028-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. WIRES RNA. 2012;3:92–103. doi: 10.1002/wrna.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blencowe BJ. Transcription: surprising role for an elusive small nuclear RNA. Curr Biol. 2002;12:R147–149. doi: 10.1016/s0960-9822(02)00711-x. [DOI] [PubMed] [Google Scholar]
- 36.Wassarman DA, Steitz JA. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol Cell Biol. 1991;11:3432–3445. doi: 10.1128/mcb.11.7.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, Coulombe B, Price DH. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krueger BJ, Varzavand K, Cooper JJ, Price DH. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PloS One. 2010;5:e12335. doi: 10.1371/journal.pone.0012335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lebars I, Martinez-Zapien D, Durand A, Coutant J, Kieffer B, Dock-Bregeon AC. HEXIM1 targets a repeated GAUC motif in the riboregulator of transcription 7SK and promotes base pair rearrangements. Nucleic Acids Res. 2010;38:7749–7763. doi: 10.1093/nar/gkq660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh R, Gupta S, Reddy R. Capping of mammalian U6 small nuclear RNA in vitro is directed by a conserved stem-loop and AUAUAC sequence: conversion of a noncapped RNA into a capped RNA. Mol Cell Biol. 1990;10:939–946. doi: 10.1128/mcb.10.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen D, Krueger BJ, Sedore SC, Brogie JE, Rogers JT, Rajendra TK, Saunders A, Matera AG, Lis JT, Uguen P, Price DH. The Drosophila 7SK snRNP and the essential role of dHEXIM in development. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding Y, Chan CY, Lawrence CE. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 48.Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, Bensaude O. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol Cell Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 50.Ouchida R, Kusuhara M, Shimizu N, Hisada T, Makino Y, Morimoto C, Handa H, Ohsuzu F, Tanaka H. Suppression of NF-kappaB-dependent gene expression by a hexamethylene bisacetamide-inducible protein HEXIM1 in human vascular smooth muscle cells. Genes to Cells. 2003;8:95–107. doi: 10.1046/j.1365-2443.2003.00618.x. [DOI] [PubMed] [Google Scholar]
- 51.Markert A, Grimm M, Martinez J, Wiesner J, Meyerhans A, Meyuhas O, Sickmann A, Fischer U. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Reports. 2008;9:569–575. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, Bensaude O. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Herreweghe E, Egloff S, Goiffon I, Jady BE, Froment C, Monsarrat B, Kiss T. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26:3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol Cell Biol. 2007;27:6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue Y, Yang Z, Chen R, Zhou Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2010;38:360–369. doi: 10.1093/nar/gkp977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conaway JW, Conaway RC. Transcription elongation and human disease. Ann Rev Biochem. 1999;68:301–319. doi: 10.1146/annurev.biochem.68.1.301. [DOI] [PubMed] [Google Scholar]
- 57.Romano G, Giordano A. Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle. 2008;7:3664–3668. doi: 10.4161/cc.7.23.7122. [DOI] [PubMed] [Google Scholar]
- 58.Espinoza-Derout J, Wagner M, Salciccioli L, Lazar JM, Bhaduri S, Mascareno E, Chaqour B, Siddiqui MA. Positive transcription elongation factor b activity in compensatory myocardial hypertrophy is regulated by cardiac lineage protein-1. Circulation Res. 2009;104:1347–1354. doi: 10.1161/CIRCRESAHA.108.191726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ai N, Hu X, Ding F, Yu B, Wang H, Lu X, Zhang K, Li Y, Han A, Lin W, Liu R, Chen R. Signal-induced Brd4 release from chromatin is essential for its role transition from chromatin targeting to transcriptional regulation. Nucleic Acids Res. 2011;39:9592–9604. doi: 10.1093/nar/gkr698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim YK, Mbonye U, Hokello J, Karn J. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J Mol Biol. 011, 410:896–916. doi: 10.1016/j.jmb.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Contreras X, Barboric M, Lenasi T, Peterlin BM. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathogens. 2007;3:1459–1469. doi: 10.1371/journal.ppat.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cho S, Schroeder S, Ott M. CYCLINg through transcription: posttranslational modifications of P-TEFb regulate transcription elongation. Cell Cycle. 2010;9:1697–1705. doi: 10.4161/cc.9.9.11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang HD, Lee TY, Tzeng SW, Horng JT. KinasePhos: a web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res. 2005;33:W226–229. doi: 10.1093/nar/gki471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 67.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 68.Bird G, Zorio DA, Bentley DL. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol Cell Biol. 2004;24:8963–8969. doi: 10.1128/MCB.24.20.8963-8969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kornblihtt AR. Coupling transcription and alternative splicing. Adv Expt Med and Biol. 2007;623:175–189. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 70.Dutertre M, Sanchez G, De Cian MC, Barbier J, Dardenne E, Gratadou L, Dujardin G, Le Jossic-Corcos C, Corcos L, Auboeuf D. Cotranscriptional exon skipping in the genotoxic stress response. Nat Struct Mol Biol. 2010;17:1358–1366. doi: 10.1038/nsmb.1912. [DOI] [PubMed] [Google Scholar]
- 71.Lenasi T, Barboric M. P-TEFb stimulates transcription elongation and pre-mRNA splicing through multilateral mechanisms. RNA Biology. 2010;7:145–150. doi: 10.4161/rna.7.2.11057. [DOI] [PubMed] [Google Scholar]
- 72.Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci USA. 2009;106:7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eilebrecht S, Brysbaert G, Wegert T, Urlaub H, Benecke BJ, Benecke A. 7SK small nuclear RNA directly affects HMGA1 function in transcription regulation. Nucleic Acids Res. 2011;39:2057–2072. doi: 10.1093/nar/gkq1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions (Review) Intern J Oncology. 2008;32:289–305. [PubMed] [Google Scholar]
- 75.Eilebrecht S, Becavin C, Leger H, Benecke BJ, Benecke A. HMGA1-dependent and independent 7SK RNA gene regulatory activity. RNA Biology. 2011;8:143–157. doi: 10.4161/rna.8.1.14261. [DOI] [PubMed] [Google Scholar]
- 76.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burrows C, Latip NA, Lam SJ, Carpenter L, Sawicka K, Tzolovsky G, Gabra H, Bushell M, Glover DM, Willis AE, Blagden SP. The RNA binding protein Larp1 regulates cell division, apoptosis and cell migration. Nucleic Acids Res. doi: 10.1093/nar/gkq294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 79.Duncan KE, Strein C, Hentze MW. The SXL-UNR corepressor complex uses a PABP-mediated mechanism to inhibit ribosome recruitment to msl-2 mRNA. Mol Cell. 2009;36:571–582. doi: 10.1016/j.molcel.2009.09.042. [DOI] [PubMed] [Google Scholar]
- 80.Kawahara H, Imai T, Imataka H, Tsujimoto M, Matsumoto K, Okano H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J Cell Biol. 2008;181:639–653. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clouse KN, Ferguson SB, Schupbach T. Squid, Cup, and PABP55B function together to regulate gurken translation in Drosophila. Dev Biol. 2008;313:713–724. doi: 10.1016/j.ydbio.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kugler JM, Lasko P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly. 2009;3:15–28. doi: 10.4161/fly.3.1.7751. [DOI] [PubMed] [Google Scholar]
- 83.Lasko P. Posttranscriptional regulation in Drosophila oocytes and early embryos. Wiley interdisciplinary reviews. RNA. 2011;2:408–416. doi: 10.1002/wrna.70. [DOI] [PubMed] [Google Scholar]
- 84.Chopra VS, Hong JW, Levine M. Regulation of Hox gene activity by transcriptional elongation in Drosophila. Current biology: CB. 2009;19:688–693. doi: 10.1016/j.cub.2009.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Harbison ST, Carbone MA, Ayroles JF, Stone EA, Lyman RF, Mackay TF. Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep. Nat Genet. 2009;41:371–375. doi: 10.1038/ng.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wery M, Kwapisz M, Morillon A. Noncoding RNAs in gene regulation. WIRES Systems Biology and Medicine. 2011;3:728–738. doi: 10.1002/wsbm.148. [DOI] [PubMed] [Google Scholar]
- 87.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shuman S. Transcriptional networking cap-tures the 7SK RNA 5′-gamma-methyltransferase. Mol Cell. 2007;27:517–519. doi: 10.1016/j.molcel.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 89.Mattaj IW. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- 90.Epstein P, Reddy R, Henning D, Busch H. The nucleotide sequence of nuclear U6 (4. 7 S) RNA. J Biol Chem. 1980;255:8901–8906. [PubMed] [Google Scholar]
- 91.Shimba S, Reddy R. Purification of human U6 small nuclear RNA capping enzyme. Evidence for a common capping enzyme for gamma-monomethyl-capped small RNAs. J Biol Chem. 1994;269:12419–12423. [PubMed] [Google Scholar]
- 92.Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]