Abstract
Aspergillus fumigatus has been shown to form biofilms that are associated with adaptive antifungal resistance mechanisms. These include multidrug efflux pumps, heat shock proteins, and extracellular matrix (ECM). ECM is a key structural and protective component of microbial biofilms and in bacteria has been shown to contain extracellular DNA (eDNA). We therefore hypothesized that A. fumigatus biofilms also possess eDNA as part of the ECM, conferring a functional role. Fluorescence microscopy and quantitative PCR analyses demonstrated the presence of eDNA, which was released phase dependently (8 < 12 < 24 < 48 h). Random amplification of polymorphic DNA (RAPD) PCR showed that eDNA was identical to genomic DNA. Biofilm architectural integrity was destabilized by DNase treatment. Biochemical and transcriptional analyses showed that chitinase activity and mRNA levels of chitinase, a marker of autolysis, were significantly upregulated as the biofilm matured and that inhibition of chitinases affected biofilm growth and stability, indicating mechanistically that autolysis was possibly involved. Finally, using checkerboard assays, it was shown that combinational treatment of biofilms with DNase plus amphotericin B and caspofungin significantly improved antifungal susceptibility. Collectively, these data show that eDNA is an important structural component of A. fumigatus ECM that is released through autolysis, which is important for protection from environmental stresses, including antifungal therapy.
INTRODUCTION
The opportunistic mold Aspergillus fumigatus is a saprophytic filamentous fungus associated with life-threatening pulmonary infections in individuals with an immunocompromised status or those who are genetically predisposed. There is growing clinical and experimental evidence that these infections are characterized by multicellular biofilm structures (1, 2). For example, aspergilli have been reported to cause serious biomaterial-related biofilm infections of catheters, joint replacements, cardiac pacemakers, heart valves, and breast augmentation implants (3–6). Moreover, aspergillary bronchitis is characterized by bronchial casts containing mucus and mycelia (7), and bronchopulmonary lavage (BAL) fluid of some patients with aspergillosis reveals the presence of numerous hyphae in the form of a complex multicellular mycetoma structure (8). Collectively, these reports confirm that A. fumigatus does maintain the capacity to form biofilms, which are characterized by aggregates of multicellular hyphae enclosed within extracellular matrix (ECM) and which are clinically significant because they are resistant to host defenses and, particularly, antifungal drugs (2).
ECM is a fundamental characteristic of biofilms, providing protection from hostile factors, including antimicrobial agents (9, 10). In bacterial biofilms, the typical ECM consists of exopolysaccharides, proteins, surfactants, lipids, water, and nucleic acids. In A. fumigatus biofilms, the hydrophobic ECM is composed of galactomannan, galactosaminogalactan, alpha-1,3 glucans, monosaccharides, polyols, melanin, and proteins (11, 12). Whereas in vivo studies suggested distinct chemical alterations, with galactosaminogalactan and galactomannan identified as the major ECM polysaccharides (12), independent investigations showed that biofilm cells attaching to epithelial cells had increased levels of ECM, coincidental with a decreased sensitivity to antifungal drugs (13). While the precise role of the ECM is not known, it is hypothesized that it plays a significant role in antifungal resistance by adsorbing the molecule and preventing its diffusion (11). This is supported by data emerging from the Candida albicans biofilm field, where it was demonstrated that ECM expression (specifically beta-glucans) sequesters antifungal agents and reduces susceptibility (14). Previous work from our group demonstrated phase-dependent antifungal activity against A. fumigatus biofilms and an association of efflux pumps (15, 16). This work suggested that early resistance phenotypes were coincident with increased efflux pump activity. This correlation was not evident in mature biofilms, so we hypothesize that ECM may play a greater role in the later phases of biofilm growth.
Recent studies have shown that extracellular DNA (eDNA) is another important component of biofilm ECM (9). eDNA is a key component of both fungal and bacterial biofilms (17, 18) and is proposed to improve overall structural integrity. In Pseudomonas aeruginosa, for example, it was shown to contribute <1 to 2% of the ECM composition. Moreover, bacterial biofilm studies have suggested that eDNA has a multifactorial purpose, namely, as a nutrient source (19), facilitator of genetic information exchange (20), contributor to biofilm stability and dispersal (18, 21–23), and antimicrobial resistance factor (24, 25). Mechanistically, cell death and lysis (26), quorum sensing (27), and excretion from outer membrane vesicles (28) have been proposed to be involved in its release from bacteria (21, 28).
Despite the growing body of knowledge regarding eDNA in bacterial biofilms, there is scant knowledge concerning its existence and role in fungal biofilms. Recent studies have shown that eDNA is also an important component of Candida biofilms, including those of C. albicans, Candida tropicalis, and Candida parapsilosis (17, 29, 30). In C. albicans, eDNA has been shown to contribute to maintenance and stability of mature biofilms, but not to their establishment (17, 29), and as a regulator of biofilm cell antifungal resistance (31). These data suggest the potential therapeutic use of agents that affect the ECM. Remarkably, little is known about the presence and contribution of eDNA in A. fumigatus biofilms. The purpose of this study was to confirm the presence of eDNA in A. fumigatus biofilm ECM and to investigate its biological role. Here we show for the first time that A. fumigatus releases eDNA through an autolytic mechanism which contributes to biofilm stability and resistance to antifungal challenge.
MATERIALS AND METHODS
Strains and conidial preparation.
A. fumigatus Af293 clinical isolates (YHCF1 to YHCF5) were obtained from the Royal Hospital for Sick Children (Yorkhill Division), Glasgow, United Kingdom. An A. fumigatus transformant expressing green fluorescent protein (Af-GFP) was used during microscopy as described previously (32). Isolates were stored on Sabouraud dextrose slopes (Oxoid, Basingstoke, United Kingdom) at 4°C. All A. fumigatus strains were grown on Sabouraud dextrose agar at 37°C for 72 h. Conidia were then harvested by flooding the surfaces of plates with phosphate-buffered saline (PBS) (Oxoid, Cambridge, United Kingdom) containing 0.025% (vol/vol) Tween 80 while gently rocking them. Conidia were then counted on a Neubauer hemocytometer and adjusted to a standardized suspension of 5 × 105 conidia ml−1 in RPMI AutoMod medium (Sigma, United Kingdom) (33). All procedures were carried out in a laminar-flow cabinet (Hera Safe laminar-flow cabinet, model K515; Kendro).
Biofilm visualization.
Standardized conidia of Af293 or Af-GFP were inoculated in RPMI medium onto Thermanox coverslips (13 mm) within a 24-well tissue culture plate and then incubated for 24 h at 37°C. These were gently rinsed with PBS and stained, according to the manufacturers' instructions, with 5 μM calcofluor white (Af293 only) (Invitrogen), which binds chitin and beta-glucans of fungal cell walls, and 20 μM propidium iodide (PI) (Sigma), which stains the DNA present within a biofilm. Biofilm growth and accumulation of eDNA were visualized under a fluorescence microscope (Motic BA400 Colorview system) or a confocal laser scanning microscope (CLSM) (Leica SP5) at excitation and emission wavelengths, respectively, of 350 and 400 nm for calcofluor white, 540 and 525 nm for propidium iodide, and 480 and 490 nm for GFP. One representative from each group was digitally photographed. Quantification of the z stacks was performed using Volocity software (PerkinElmer). For scanning electron microscopy (SEM), representative biofilms grown and treated on Thermanox coverslips were processed as previously described (34). Briefly, the acrylic resin specimens were washed in PBS and fixed in 2% paraformaldehyde, 2% glutaraldehyde, and 0.15% [wt/vol] alcian blue in 0.15 M sodium cacodylate (pH 7.4). The biofilms were sputter coated with gold and viewed under a JEOL JSM-6400 scanning electron microscope.
DNA extraction.
ECM was extracted as previously described (35), with slight modifications. Briefly, standardized A. fumigatus conidia were inoculated into a 75-cm2 tissue culture flask (Nunc, Rochester, NY) containing RPMI medium, yeast extract-peptone-dextrose (YPD), and yeast nitrogen base (YNB) (Sigma, United Kingdom) at 37°C for 8, 12, 24, 48, and 72 h on a rocking platform. After incubation, the biofilm was removed from the flasks by use of sterile cell scrapers and then washed with PBS. The disaggregated biofilm was then treated with 0.2 M EDTA to extract the ECM. The samples were then centrifuged at 10,000 × g, and the EDTA supernatant was recovered and filtered using a 0.45-μm syringe filter (Millipore). The eDNA was then extracted from supernatants by using a MasterPure yeast DNA purification kit (Epicentre Biotechnologies, Madison, WI) following the manufacturer's instructions. Genomic DNAs from Af293 and C. albicans 3153A were extracted using a MasterPure yeast DNA purification kit (Epicentre Biotechnologies) according to the manufacturer's instructions.
Quantification of ECM-associated eDNA.
Different phases of biofilm (8, 12, and 24 h), with or without azetazolamide (128 mg/liter; Sigma), a chitinase inhibitor (36), were formed at 37°C either in microtiter plates or in tissue culture flasks as described above. The quantity of eDNA within the ECM was measured using a microplate fluorescence assay (MFA) with a DNA-binding dye (SYBR green I) as previously described (37). Briefly, SYBR green I (Invitrogen, Paisley, United Kingdom) was added to eDNA extract in a black-well microtiter plate (Costar, Corning, NY) at a ratio of 1:1. Binding of this dye produces fluorescence in direct proportion with the DNA concentration. The levels of eDNA were quantified using a fluorescence plate reader (Fluostar Optima; BMG Labtech, Aylesbury, United Kingdom) at excitation and emission wavelengths of 485 and 518 nm, respectively. The concentration of eDNA in the sample was quantified by using a DNA standard curve as previously described elsewhere (37).
RAPD PCR.
The PCR amplification of seven different genes was performed using Reddymix PCR master mix (Thermo Scientific). Primers used for random amplified polymorphic DNA (RAPD) PCR amplification of ANXC4, BGT1, CAT1, LIP, MAT1-2, SODB, and ZRF2 and the cycling conditions were previously described elsewhere (38). PCR was performed in a MyCycler PCR machine (Bio-Rad, Hertfordshire, United Kingdom). The amplified samples were then separated using horizontal gel electrophoresis, and the gel was observed under UV light excitation (Bio-Rad Gel Doc 2000; Bio-Rad Life Sciences, Hemel Hempstead, United Kingdom).
Evaluating biofilm formation and stability.
Biofilms were formed in 96-well microtiter plates as previously described (33). DNase I from bovine pancreas (Sigma), hereafter referred to as DNase for simplicity, was prepared in 0.15 M NaCl supplemented with 5 mM MgCl2 (31). To evaluate the effect of DNase treatment on preformed biofilms, Af293 biofilms were grown for 24 h and then treated with DNase (0.25, 1, and 4 mg/ml) at 37°C overnight, with an untreated buffered control included on each plate for comparison. For further controls, we investigated heat-inactivated DNase (0.25 mg/liter), RNase (0.25 mg/liter), and proteinase K (0.25 mg/liter) (Sigma, United Kingdom) under the same conditions. To assess the effect of DNase on biofilm formation, a standardized preparation of 5 × 105 Af293 conidia was incubated with DNase (0.25, 1, and 4 mg/ml) at 37°C overnight. Additionally, the effect of azetazolamide (128 mg/liter) on biofilm formation was assessed. Finally, to assess the effect of exogenous DNA on biofilm formation, we incubated standardized conidia with salmon sperm DNA (Sigma, United Kingdom) and C. albicans and A. fumigatus genomic DNAs (4, 8, and 20 mg/liter) at 37°C overnight. DNA was extracted as described above. After each treatment, the biofilms were washed in PBS and their biomass quantified using a crystal violet technique previously described by our group (33).
Quantifying chitinase activity.
The chitinase activity of A. fumigatus at different phases of biofilm formation was assessed using a fluorometric chitinase assay kit (Sigma, United Kingdom) per the manufacturer's instructions. In brief, supernatants were collected at different time points, and an appropriate volume of each sample was incubated with a substrate working solution (4-methylumbelliferyl N-acetyl-β-d-glucosaminide) at 37°C for 30 min. Fluorescence was then quantified at excitation and emission wavelengths of 360 nm and 450 nm, respectively. Appropriate positive and negative controls included in the kit were added to each plate. Finally, chitinase activity was calculated and expressed in U/ml. One unit of chitinase activity was defined as the activity resulting in the release of 1 μmol of 4-methylumbelliferone from the substrate per min at pH 5.0 and 37°C. We also used the chitinase inhibitor azetazolamide to act as a control for inhibition of activity in this assay.
Quantitative gene expression.
A. fumigatus Af293 and YHCF1 to YHCF4 biofilms were prepared in triplicate on tissue culture-treated flasks as described above. RNA was extracted by mechanical disruption in TRIzol (Invitrogen) and purified using an RNeasy MinElute cleanup kit (Qiagen, Crawley, United Kingdom) per the manufacturer's instructions. RNA was quantified and its quality determined using a NanoDrop spectrophotometer (ND-1000; Thermo Scientific, Loughborough, United Kingdom). At first, cDNA was synthesized with a high-capacity RNA-to-cDNA master mix (Applied Biosystems), using a MyCycler PCR machine (Bio-Rad, Hertfordshire, United Kingdom), and was stored at −20°C for expression analysis. The expression of a panel of class III chitinase genes (Chit1-5) was then assessed using quantitative reverse transcription-PCR (RT-PCR) with SYBR greenER (Invitrogen) according to the manufacturer's instructions. Primer sequences for these genes and PCR cycling conditions are described elsewhere (39). The individual gene expression levels were then calculated using the 2−ΔΔCT method for different phases and normalized to the TEF1 housekeeping gene.
Biofilm susceptibility testing.
DNase was used in combination with different classes of antifungals to determine whether antifungal efficacy could be enhanced. Amphotericin B and caspofungin were prepared according to Clinical and Laboratory Standards Institute recommendations and used throughout these experiments (40). For testing of antifungal susceptibility in combination with DNase, a checkerboard assay was performed with 24-h preformed biofilms as described previously (41). The checkerboard assay format involves two compounds serially double diluted perpendicularly to one another on a 96-well plate, which produces a combined concentration gradient of both compounds. An assay was prepared with DNase (0 to 256 mg/ml) and either amphotericin B (0 to 16 mg/liter) or caspofungin (0 to 512 mg/liter). The cell sessile MIC (sMIC50) was determined as the concentration giving a 50% reduction in metabolism compared to untreated controls in the 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)2H-tetrazolium-5-carboxanilide (XTT)-based reduction assay, as previously described (33). Following these initial experiments, a defined concentration of DNase was selected (128 mg/liter) for all subsequent microdilution testing on different phases of biofilms for the panel of strains (n = 6). To test the effect of chitinase inhibition on antifungal susceptibility, the biofilms (Af293 and 5 clinical isolates) formed in the presence of azetazolamide (256 mg/liter) were treated with amphotericin B (0 to 32 mg/liter) and assessed as described above.
Statistics.
Analysis of variance (ANOVA) and t tests were used to investigate independent sample data. Bonferroni's correction for multiple comparisons was applied to the data where appropriate. SPSS (version 11; SPSS, Chicago, IL) was used for these analysis, and GraphPad Prism (version 4; GraphPad, La Jolla, CA) was used for production of the figures. Spearman's rho correlation coefficient (R) was determined to investigate the relationships between the parameters measured. P values of <0.05 were considered significant.
RESULTS
Aspergillus fumigatus releases extracellular DNA in a phase-dependent manner.
The presence of eDNA in bacterial biofilms is known to be associated with biofilm formation, stability, and dispersal (18, 22, 23). Due to the key structural and functional roles of eDNA within bacterial biofilm ECM (21), and more recently in C. albicans biofilms (17), we hypothesized that eDNA might play similar roles in A. fumigatus biofilms. We first undertook a visual inspection, using CLSM, of GFP-labeled A. fumigatus (green fluorescence) stained with propidium iodide (red fluorescence) to detect the possible presence of eDNA. At low magnification, no localized areas of diffuse red fluorescence were observed for the 8- and 12-h biofilms (data not shown), but at 24 h (Fig. 1a) and 48 h (Fig. 1b), diffuse areas of red/yellow fluorescence were observed commensurate with increased diffuse green fluorescence, which is indicative of ECM production. The yellow fluorescence indicates colocalization of the cells and eDNA (red and green fluorescence combined). Higher magnification through the z stacks of calcofluor white-stained biofilms (blue fluorescence) indicated that propidium iodide-stained eDNA (red fluorescence) colocalized with hyphae (Fig. 1c), which were dispersed throughout the biofilm structure as part of the ECM (Fig. 1d). Controlling for nonspecific binding of PI to ECM was not possible; therefore, to confirm that the staining observed was eDNA, we next aimed to quantify the eDNA content throughout the biofilm. Significant release of eDNA was observed in a time-dependent manner (P < 0.0001) (Fig. 1e) for biofilms formed in RPMI medium. At the earliest time point (8 h), eDNA release was below detectable limits, whereas 4 h later 24 ng/ml eDNA was detected. In comparison to 8-h biofilms, a significant increase of ECM-associated eDNA was observed within biofilms at 24 h (279 ng/ml; P < 0.01), 48 h (473 ng/ml; P < 0.001), and 72 h (864 ng/ml; P < 0.001). In comparison to biofilms formed in RPMI medium, eDNA release from 48-h biofilms formed in YPD (155 ng/ml) and YNB (110 ng/ml) was significantly lower (P < 0.001) (Fig. 1f).
Fig 1.
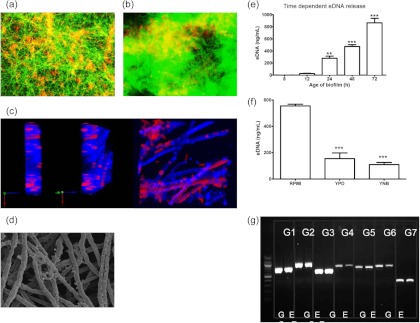
Aspergillus fumigatus biofilms contain extracellular DNA. (a and b) Fluorescence micrographs of A. fumigatus (Af-GFP strain) biofilms grown for 24 and 48 h on Thermanox coverslips in a 24-well plate at 37°C. Biofilms were stained with the DNA-binding dye propidium iodide (PI). Color-saturated images of live Af-GFP-expressing cells (green) and PI-stained eDNA (diffuse red) or dead cells (punctate red) are shown. Yellow indicates the presence of both cells and eDNA. Magnification, ×200. (c) CLSM images of 24-h A. fumigatus biofilms on Thermanox coverslips at 37°C. The different planar images show PI-stained eDNA (diffuse red), which diffusely covers and intersperses with calcofluor white (blue)-stained hyphae. (d) High-magnification SEM image of 24-h A. fumigatus biofilm showing the presence of hyphae covered and interspersed with ECM. Magnification, ×2,000. The images shown are representative of three independent experiments. (e and f) Af293 biofilms grown in RPMI medium for 8, 12, 24, 48, or 72 h (e) or in RPMI medium, YPD, or YNB for 48 h (f) were gently washed with EDTA to remove the ECM, an MFA was used to produce a standard curve, and eDNA within the ECM was quantified. Experiments were performed in triplicate on 3 separate occasions. Data were analyzed by ANOVA, and Bonferroni's correction for multiple comparisons was applied (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (g) RAPD PCR was carried out on extracted genomic (G) and extracellular (E) DNA samples, targeting seven different genes (G1, ANXC4; G2, BGT1; G3, CAT1; G4, LIP; G5, MAT1-2; G6, SODB; and G7, ZRF2), and the PCR products were electrophoresed in an agarose gel. Positive bands for seven genes indicate that the eDNA is similar in sequence to genomic DNA.
To investigate whether eDNA released from maturing biofilms was different from genomic DNA, we undertook RAPD PCR analysis using defined primer sets comprising 7 conserved genes from throughout the A. fumigatus genome. We hypothesized that if the genetic material of the endogenous genomic DNA differed from extracellular DNA, then primers designed for multilocus sequence typing (MLST) analysis would generate differently sized PCR products. Gel electrophoresis showed equivalent bands for all 7 genes amplified for both genomic DNA and eDNA (Fig. 1g). This suggests that eDNA is a result of release from dead or dying cells within the maturing biofilm rather than being synthesized uniquely. Whether eDNA plays a direct functional role within the biofilm remained to be determined.
Extracellular DNA provides structural integrity to Aspergillus fumigatus biofilms.
Since as the biofilm matures eDNA release increases proportionally to the level of ECM, eDNA may play a specific role within the biofilm. Studies of P. aeruginosa have shown that eDNA plays a role in biofilm development (18), indicating that this may also be the case for A. fumigatus. To test this hypothesis, we investigated whether the addition of DNase would affect mature (24-h) biofilms. CLSM imaging showed that the dense structure of mature A. fumigatus biofilms (Fig. 2a) was extensively disaggregated by 0.25 mg/ml of DNase, leaving adherent germlings and a scattered number of hyphae (Fig. 2b). Quantitative analysis of biofilm depth showed that the depth was also reduced significantly, by approximately 57% (P < 0.01) (Fig. 2c).
Fig 2.
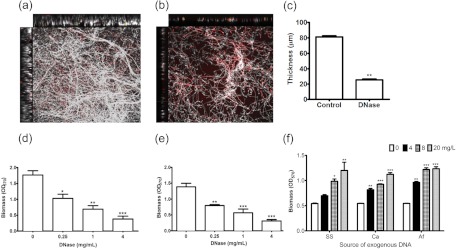
Extracellular DNA plays a role in biofilm structural integrity. (a and b) CLSM analysis. Mature 24-h biofilms were treated with vehicle control (a) or 0.25 mg/ml of DNase (b) for 24 h, and the resultant biofilms were assessed by CLSM imaging of the x-y, x-z, and y-z dimensions. Biofilms were stained with calcofluor white and propidium iodide. Artificially colored images of cells (white) and PI-stained eDNA (red) or dead cells (punctate red) are shown. (c) Biofilm depths with and without DNase were analyzed using Volocity software. The images shown are representative of three independent experiments. (d to f) Biomass analysis. Mature biofilms (d) and conidia (e) were treated with DNase (0, 0.25, 1, and 4 mg/ml), and the resultant biomasses were assessed using a crystal violet assay. (f) Salmon sperm (SS), C. albicans (Ca), and A. fumigatus (Af) genomic DNAs were added to conidia (0, 4, 8, and 20 mg/liter), and biofilms formed over 24 h at 37°C. The resultant biomasses were assessed using a crystal violet assay. Data were analyzed by ANOVA with Bonferroni's correction for multiple comparisons (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Next, we used biomass analysis to investigate how DNase would affect different phases of biofilm growth and structure, as has been described for studies of C. albicans (31). First, to control for the direct effect of DNase on cells, we demonstrated that no direct antimicrobial effect was observed at the concentrations tested (0.25, 2, and 4 mg/ml) (data not shown). We demonstrated that the structural integrity of the preformed biofilm was shown to be markedly affected by the addition of DNase, in a concentration-dependent manner. Compared to the control biofilm, 0.25 mg/ml of DNase significantly reduced the resultant biomass from an optical density at 570 nm (OD570) of 1.76 to 1.03 (P < 0.05), 1 mg/ml of DNase reduced it to 0.69 (P < 0.01), and the maximal effect was observed with 4 mg/ml of DNase (OD570 of 0.38) (P < 0.001) (Fig. 2d). In contrast, treatment with heat-inactivated DNase (0.25 mg/ml), RNase (0.25 mg/ml), and proteinase K (0.25 mg/ml) did not significantly reduce the biofilm biomass compared to the Tris-EDTA (TE)-buffered control (P > 0.05) (data not presented). To test the effect on biofilm development, conidia were treated as described above, and significant reductions of biofilm biomass were also observed in a concentration-dependent manner at DNase concentrations of 0.25, 1, and 4 mg/ml (P < 0.0001 by ANOVA with Bonferroni's multiple-comparison test) (Fig. 2e). Compared to the biofilm development control, 0.25 mg/ml of DNase reduced the resultant biomass from an OD570 of 1.34 to 0.79 (P < 0.01), 1 mg/ml of DNase reduced it to 0.57 (P < 0.001), and the maximal effect was observed with 4 mg/ml DNase (0.31) (P < 0.001).
Given that DNase treatment led to architectural instability, we hypothesized that adding exogenous DNA during initial adhesion would positively promote biofilm formation, as has been shown to be the case for C. albicans biofilms (17). Addition of exogenous DNA from salmon sperm, C. albicans, or A. fumigatus significantly increased the biofilm biomass at 4, 8, and 20 mg/liter (P < 0.0001 by ANOVA with Bonferroni's multiple-comparison test) (Fig. 2f). The proportional increase in biofilm biomass was 29%, 82% (P < 0.05), and 122% (P < 0.01), respectively, when 4, 8, and 20 mg/liter of exogenous salmon sperm DNA was added. However, when C. albicans and A. fumigatus exogenous genomic DNAs were added at 4, 8, and 20 mg/liter, the biofilm biomass increases were more notable, significantly increasing by 49% and 77% (P < 0.01), 70% and 124% (P < 0.001), and 106% and 126% (P < 0.001), respectively.
Collectively, these data demonstrate the presence of eDNA within dense biofilm populations and show that it plays a functional role in maintaining structural and architectural integrity. We demonstrated that eDNA is most likely genomic DNA released from A. fumigatus during biofilm growth, but whether or not this is an active process remains to be determined.
Extracellular DNA release is associated with autolysis.
Autolysis is often cited as a key mechanism for the release of eDNA in bacterial biofilms (26), and evidence exists for eukaryotes from studies of Aspergillus nidulans showing that autolysis is regulated by chitinase activity (42–44). It was also recently shown that autolysis is associated with chitinases in A. fumigatus biofilms (39). We hypothesize that autolysis leads to the release of eDNA in A. fumigatus biofilms.
First, we examined chitinase expression by using a biochemical assay. It was shown that as the biofilm matured, the levels of chitinase activity significantly increased (P < 0.01). No activity was observed for the 8-h phase, but at 12 and 24 h, values of 0.80 and 1.26 U/ml were detected. Addition of azetazolamide was shown to significantly reduce the activity of the 24-h biofilm, to 0.13 U/ml (P < 0.01) (Fig. 3a), indicating that chitinase activity is associated with biofilm maturation.
Fig 3.
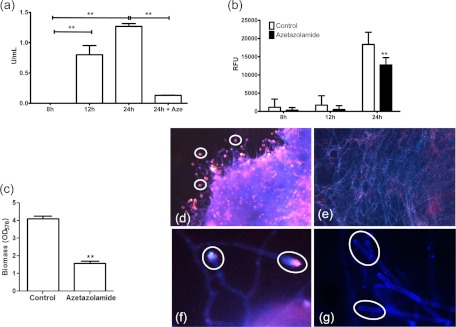
Autolysis is associated with extracellular DNA release. (a) Chitinase activity was assessed using a commercially available fluorometric assay for 8-, 12-, and 24-h biofilms, and also in the presence of azetazolamide (24 h + Aze). (b) eDNA release was assessed in the presence of azetazolamide for 8, 12, and 24 h, using the SYBR green MFA. RFU, relative fluorescence units. (c) Conidia were incubated with azetazolamide for 24 h, and the resultant biomass was assessed using a crystal violet assay. Data were analyzed by ANOVA with Bonferroni's correction for multiple comparisons (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (d to g) Fluorescence micrographs of A. fumigatus biofilm (24 h) controls (d and f) or biofilms grown in the presence of azetazolamide (e and g) on Thermanox coverslips. These biofilms were stained with calcofluor white and propidium iodide and imaged using Colorview software. Magnification, ×200 (d and e) or ×400 (f and g).
To test this hypothesis, we again used the chitinase inhibitor azetazolamide to test whether we could influence eDNA release. It was shown that both 8- and 12-h biofilms were not significantly affected in eDNA release, but in 24-h biofilms, a significant reduction of eDNA release was shown (P > 0.01) (Fig. 3b). In parallel, we investigated whether biofilm development was affected by azetazolamide, and here it was shown that the resultant biomass was significantly impeded (P < 0.01) (Fig. 3c). These data were confirmed by fluorescence microscopy, which showed that biofilms formed in the presence of azetazolamide were substantially compromised in their ECM and presence of eDNA. Figure 3d shows that untreated biofilms had more eDNA localization (diffuse red/pink) within the ECM than those treated with azetazolamide (Fig. 3e), which were largely devoid of red/pink staining. Moreover, evaluation of the same biofilms at a higher magnification demonstrated that the eDNA (red/pink) appeared to be colocalized at the ends of the hyphal tips (Fig. 3f), and this was not present in biofilms treated with azetazolamide (Fig. 3g).
Finally, we undertook a quantitative transcriptional analysis of 5 different type III chitinases, using real-time quantitative PCR (qPCR) as previously described (39). These data showed that all transcripts (class III [chit1, -2, -4, and -5]) except for chit3 were upregulated in a phase-dependent manner (8 < 12 < 24 h). Both chit2 and chit5 showed significant upregulation at both 12 h (P < 0.01) and 24 h (P < 0.001), whereas chit1 and chit4 were significantly upregulated only in 24-h biofilms (P < 0.01). Expression levels compared to those at 8 h ranged from a 2.6- to 3.43-fold increase at 12 h and a 3.6- to 7.25-fold increase at 24 h (Table 1).
Table 1.
Differential expression of class III chitinase genes during biofilm maturation
| Chitinase gene | Fold change in expression at 12 h relative to that at 8 h (mean ± SD) | P value | Fold change in expression at 24 h relative to that at 8 h (mean ± SD) | P value |
|---|---|---|---|---|
| chit1 | 2.85 ± 0.71 | >0.05 | 7.24 ± 1.79 | <0.01 |
| chit2 | 2.60 ± 0.60 | <0.01 | 6.10 ± 0.52 | <0.001 |
| chit3 | 3.43 ± 3.98 | >0.05 | 7.25 ± 6.70 | >0.05 |
| chit4 | 2.28 ± 0.10 | >0.05 | 3.61 ± 1.10 | <0.01 |
| chit5 | 2.76 ± 0.78 | <0.01 | 6.24 ± 0.39 | <0.001 |
Collectively, these data suggest that chitinases are an important factor in eDNA release from A. fumigatus biofilms, presumably through autolysis. Whether this process is regulated specifically or is a serendipitous effect of autolysis used to provide protection to the community of cells is unknown at this time.
Extracellular DNA release confers antifungal resistance upon Aspergillus fumigatus biofilms.
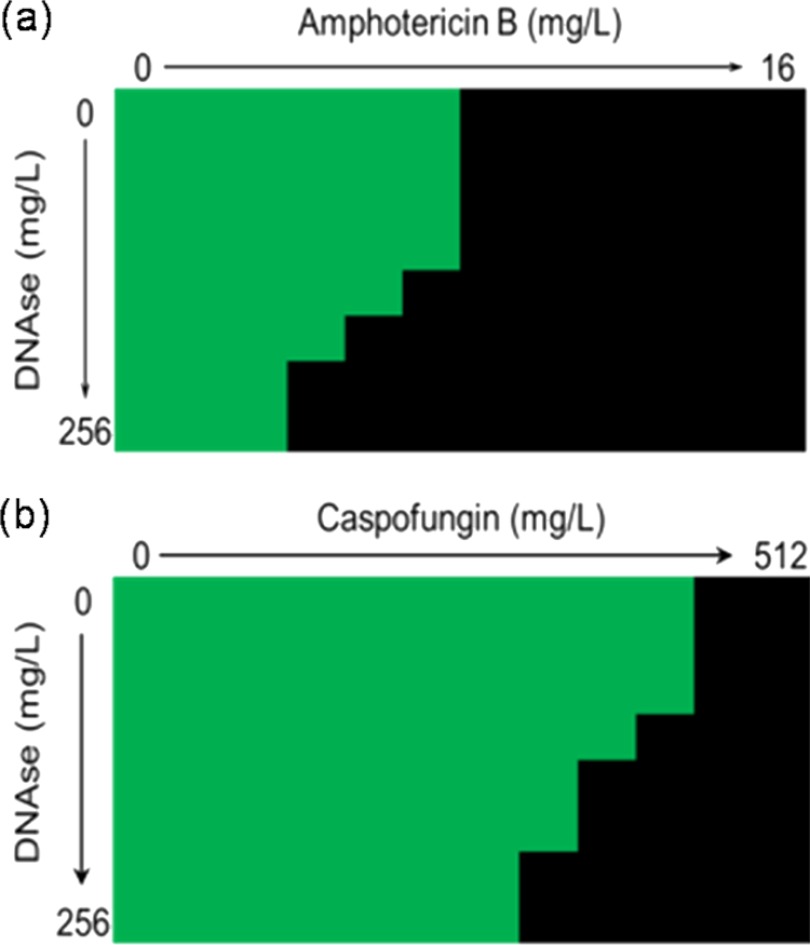
Numerous studies have shown the integral relationship between biofilm ECM and antifungal resistance (10). Most recently, it was shown that beta-glucans synthesized through the glucan synthase Fks1p are a key component of C. albicans biofilm ECM and actively confer resistance (14). Given that eDNA is an important component of the ECM and that a recent study of C. albicans has shown that biofilm sensitivity to antifungal agents is improved through the addition of DNase (31), we hypothesized that treatment of A. fumigatus biofilms with DNase in combination with different classes of antifungal agents would improve the activity. Antifungal testing using a checkerboard format on 24-h biofilms showed that the sMIC50 values for both amphotericin B- and caspofungin-treated cells decreased with increases in the DNase concentration (Fig. 4a and b). No significant change in the MICs of azoles in the presence of DNase was obtained (data not shown). Based on the checkerboard data, a fixed concentration of 128 mg/liter was used for further experimental testing of different biofilm phases. It was shown that the susceptibility of 8- and 12-h biofilms was relatively unaffected by antifungal and DNase treatment. However, the efficacy of both amphotericin B and caspofungin was improved as the biofilms matured. For amphotericin B, it was shown that the mean MIC changed from 0.125 to 0.0156 mg/liter (8-fold change) for 24-h biofilms and from 1 to 0.125 mg/liter (8-fold change) for 48-h biofilms (Table 2). For caspofungin, it was shown that the mean MIC changed from 64 to 16 mg/liter (4-fold change) for 24-h biofilms and from >256 to 64 mg/liter (>8-fold change) for 48-h biofilms (Table 3).
Fig 4.
Extracellular DNA contributes to antifungal resistance. A checkerboard assay was designed to evaluate the combined effects of DNase and antifungal agents on the sMIC50. A. fumigatus Af293 biofilms were grown for 24 h at 37°C prior to treatment. DNase was serially double diluted in RPMI medium down each column (0 to 256 mg/liter), and amphotericin B (0 to 16 mg/liter) (a) or caspofungin (0 to 512 mg/liter) (b) was then serially double diluted across each row of a 96-well microtiter plate to create a concentration gradient. The biofilms were treated for 24 h prior to their viability being assessed using an XTT reduction assay. The green areas indicate metabolically active growth of A. fumigatus, and the black areas indicate the inhibition of metabolism (<50% of the control).
Table 2.
sMICs of amphotericin B in the presence and absence of DNasea
| Strain | sMIC (mg/liter) at 8 h |
FC | sMIC (mg/liter) at 12 h |
FC | sMIC (mg/liter) at 24 h |
FC | sMIC (mg/liter) at 48 h |
FC | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −DNase | +DNase | −DNase | +DNase | −DNase | +DNase | −DNase | +DNase | |||||
| Af293 | 0.0625 | 0.0625 | 1 | 0.0625 | 0.0625 | 1 | 0.125 | 0.03125 | 4 | 1 | 0.125 | 8 |
| YHCF1 | 0.015 | 0.015 | 1 | 0.031 | 0.015 | 2 | 0.125 | 0.01563 | 8 | 1 | 0.125 | 8 |
| YHCF2 | 0.125 | 0.125 | 1 | 0.125 | 0.031 | 2 | 0.125 | 0.01563 | 8 | 2 | 0.125 | 16 |
| YHCF3 | 0.031 | 0.015 | 2 | 0.0625 | 0.015 | 2 | 0.125 | 0.00781 | 16 | 1 | 0.0625 | 16 |
| YHCF4 | 0.031 | 0.031 | 1 | 1 | 1 | 1 | 1 | 0.25 | 4 | 4 | 0.25 | 16 |
| YHCF5 | 0.007 | 0.007 | 1 | 0.25 | 0.25 | 1 | 1 | 0.25 | 4 | 4 | 0.25 | 16 |
Biofilms were treated with amphotericin B in the presence of 128 mg/liter of DNase (+DNase) or in the absence of DNase (−DNase). FC, fold change in MIC relative to control.
Table 3.
sMICs of caspofungin in the presence and absence of DNasea
| Strain | sMIC (mg/liter) at 8 h |
FC | sMIC (mg/liter) at 12 h |
FC | sMIC (mg/liter) at 24 h |
FC | sMIC (mg/liter) at 48 h |
FC | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −DNase | +DNase | −DNase | +DNase | −DNase | +DNase | −DNase | +DNase | |||||
| Af293 | 0.015 | 0.015 | 1 | 2 | 2 | 1 | 128 | 32 | 4 | >256 | 64 | >8 |
| YHCF1 | 0.062 | 0.03125 | 2 | 4 | 4 | 1 | 128 | 16 | 8 | >256 | 64 | >8 |
| YHCF2 | 0.062 | 0.062 | 1 | 8 | 4 | 2 | 64 | 8 | 8 | >256 | 64 | >8 |
| YHCF3 | 0.015 | 0.015 | 1 | 4 | 2 | 2 | 64 | 8 | 8 | 128 | 8 | 16 |
| YHCF4 | 0.062 | 0.03125 | 2 | 4 | 2 | 2 | 64 | 16 | 4 | >256 | 32 | >8 |
| YHCF5 | 0.015 | 0.015 | 1 | 2 | 2 | 1 | 64 | 16 | 4 | >256 | 32 | >8 |
Biofilms were treated with caspofungin in the presence of 128 mg/liter of DNase (+DNase) or in the absence of DNase (−DNase). FC, fold change in MIC relative to control.
Next, we assessed the MICs of biofilms formed in the presence of azetazolamide. We hypothesized that inhibition of chitinase activity would affect autolysis and the subsequent release of eDNA. We reasoned that biofilms would therefore become more sensitive to antifungal challenge. Analysis of amphotericin B treatment of biofilms compared to those also exposed to azetazolamide would show increased sensitivity. Indeed, this was the case, as the amphotericin B-treated biofilm sMIC50s ranged from 0.0625 to 0.125 mg/liter, compared to 0.125 to 1 mg/liter (1- to 8-fold change) for biofilms also treated with the chitinase inhibitor (Table 4). For azetazolamide combination studies, dose-dependent effects were observed, as the effects were minimal at the lower concentrations (data not shown). We therefore used a fixed concentration (128 mg/liter) in order to observe defined changes in sensitivity.
Table 4.
sMICs of amphotericin B in the presence and absence of azetazolamidea
| Strain | sMIC (mg/liter) |
FC | |
|---|---|---|---|
| Control | +Azetazolamide | ||
| Af293 | 0.125 | 0.03125 | 4 |
| YHCF1 | 0.125 | 0.125 | 1 |
| YHCF2 | 0.125 | 0.0625 | 2 |
| YHCF3 | 0.125 | 0.03125 | 4 |
| YHCF4 | 1 | 0.125 | 8 |
| YHCF5 | 1 | 0.25 | 4 |
Biofilms were pretreated with azetazolamide (256 mg/liter) or not pretreated with azetazolamide. FC, fold change in MIC relative to control.
To test whether the levels of antifungal resistance correlated with eDNA release, Spearman's rho correlation coefficient was determined. Statistically significant correlations were observed between the level of eDNA accumulation in biofilms and the biomass (R = 0.91364; P = 0.00003), the amphotericin B MIC (R = 0.71729; P = 0.00864), and the caspofungin MIC (R = 0.82664; P = 0.00092). Collectively, these data show that eDNA plays an important role in mature biofilm resistance.
DISCUSSION
Our results establish a novel serendipitous role for eDNA release in conferring antifungal resistance to A. fumigatus biofilms. A. fumigatus infections are typically difficult to manage with antifungal treatment and are associated with unacceptably high rates of mortality. It has been hypothesized that treatment failure may be related to the presence of innate resistance mechanisms associated with multicellular biofilms (10, 16, 41). Here we show for the first time that eDNA is a key structural component of maturing biofilms, possibly released through autolysis and imparting protective effects against antifungal treatment, as evidenced through the synergistic effects of DNase with either polyenes or echinocandins. Given that eDNA has been shown to be an important component of biofilm ECM in both bacteria and fungi, the data suggest that this may be a conserved and possibly active microbial biofilm process, but this has yet to be demonstrated.
The presence and function of biofilm-associated eDNA have been well documented for a variety of bacterial species (9, 18, 21), yet for fungal biofilms this has been limited to Candida spp. (17, 29). Recent investigations of C. albicans have confirmed that despite the use of different growth media, eDNA is present in mature biofilms, and based on digestion with DNase and the addition of exogenous DNA at different stages of biofilm development, it was shown that eDNA contributes to the maintenance and stability of mature biofilms (17). This is in agreement with our observations, as we have shown the detection of eDNA in different media, as well as its phase-dependent release. We also demonstrated the critical function of eDNA as a biofilm ECM structural component providing architectural stability to A. fumigatus biofilms. In spite of this, it is not known how eDNA is spatially organized within the ECM. Given its diffuse characteristics, it seems most likely that it is bound to ECM polymers. It has been shown that β-1,3-glucans, which are components of C. albicans biofilms (45), are able to complex with specific nucleotides (46). ECM of A. fumigatus contains α-1,3-glucans (11), so it is feasible that these interact with and trap eDNA, but at present this is not known and is under investigation. Given that there are subtle differences in ECM composition between in vivo and in vitro A. fumigatus biofilms (12), there is a question of whether the ECM of in vivo biofilms contains eDNA and, if so, how it is spatially organized within the ECM. Given that it is detected in different media, albeit in significantly different quantities, it may have a role within in vivo biofilms.
We next set out to determine how exactly eDNA becomes a key structural component of the biofilm ECM for A. fumigatus. Although not well understood, mechanisms of eDNA release in eukaryotic cells include active secretion, necrosis, and apoptosis (47), with the possibility of more than one pathway being involved (48). Bacterial autolysis is a commonly described mechanism of eDNA release to promote biofilm formation (49, 50). For fungi such as A. nidulans, the molecular basis of autolysis has been investigated, and it has been shown that the chitinase ChiB plays a pivotal role in fungal autolysis in A. nidulans under conditions of carbon starvation (51, 52). Fungal chitinases are secreted enzymes that have been shown to play a role in the digestion of exogenous chitin or utilization of fungal chitin for energy during autolysis. We hypothesized that eDNA is released as a consequence of autolysis, thus conferring structural support for the development and maintenance of the complex biofilm architecture. Both biochemical quantification of chitinase activity and targeted real-time qPCR analysis of the five specific chitinase genes showed a growth phase-dependent upregulation of chitinases. Moreover, biochemical inhibition of chitinase activity significantly affected eDNA release. It was also reported in an A. fumigatus biofilm study using microarray analysis that class IV chitinase mRNA was upregulated ∼5-fold in 48-h biofilms (53), whereas Gibbons and colleagues did not identify this expression, but they explained this difference by the potential production of pseurotin A, a potent inhibitor of chitinases (54).
Given that chitinases are markers of autolysis, this process is a possible mechanism for eDNA release. Within the biofilm, there is a buildup of toxic secondary metabolites, and nutrients may be limited by diffusion through the ECM. This may provide the environmental stresses that induce the expression of these cell wall remodeling enzymes in order to liberate nutrients from the cellular community. Our data indicate that exogenous DNA leads to an increased biomass, so in addition to providing structural support, it may also act as a nutrient source. This indicates that eDNA release may have a multifunctional role for A. fumigatus. The primary role ascribed to eDNA has been the regulation of biofilm formation and structure (18). However, while investigations proceed to elucidate other functions (55), eDNA has been involved, for instance, in biofilm antimicrobial resistance (24, 25).
To further investigate alternative functional roles of eDNA, we aimed to evaluate its impact on antifungal treatment. It has been shown that the ECM is a pivotal barrier for antifungal activity, as it is known to bind specific classes of antifungal agents and to limit penetration of others into C. albicans (56, 57). Moreover, it has been shown to have a functional role in A. fumigatus (2, 11). Given that eDNA is a key structural component of the ECM, we hypothesized that destabilizing it through the simultaneous addition of DNase would improve the penetration and sensitivity of antifungal agents, as has been shown for amphotericin B against preformed C. albicans biofilms (31). While DNase was shown to have no antifungal effect per se, its use in a checkerboard assay combined with antifungal agents increased the sensitivity of 24-h biofilms to polyenes and echinocandins. These data suggest that DNase treatment may offer an attractive chemotherapeutic strategy for the management of A. fumigatus infections. Interestingly, we observed no effect for azoles in these assays, as may be predicted from the C. albicans literature. We suspect that this may be because C. albicans biofilms consist of a combination of yeast cells and hyphae, thus giving yeast cells the opportunity to divide and proliferate. However, for A. fumigatus, mature biofilms are entirely hyphal, showing no further signs of proliferation. Therefore, we would predict that targeting of the ergosterol biosynthesis pathway would be futile, irrespective of ECM and biofilm stability. This is in agreement with our previous studies showing that inhibition of defined fungal biofilm resistance mechanisms, such as efflux pumps and heat shock proteins, has negligible effects against mature A. fumigatus biofilms (16, 41).
Recently, the combined use of drugs (58) or drugs with other agents, such as enzymes (59), has received considerable attention. ECM-degrading enzymes present broad-spectrum activity that is unlikely to induce antimicrobial resistance. The combined use of ECM-degrading agents and antifungals was previously tested in vitro. It was shown that C. albicans biofilm cell susceptibility to fluconazole and amphotericin B was increased by β-1,3-glucanase (45), whereas A. fumigatus biofilm cell susceptibility to polyenes was not changed by α-1,3-glucanase (11). Clinically, however, the DNase Pulmozyme is currently used as a therapeutic agent for cystic fibrosis treatment as an adjunct to antibiotic treatment (60). However, limitations to these chemotherapeutic approaches include the expense and increased risk of dispersing cells to distal sites of colonization (59).
To summarize, this is the first report to show the presence of eDNA in A. fumigatus and that it affects antifungal resistance. These observations may allow us to improve the treatment of different forms of aspergillosis by using agents, such as nucleases, allowing disaggregation of filamentous masses and improving the penetration and efficacy of existing antifungal agents.
ACKNOWLEDGMENTS
We thank Margaret Mullin (Integrated Microscopy Facility, University of Glasgow) for assistance with SEM processing and imaging. We also thank Jean Paul Latge and Anne Beauvais for their helpful and critical appraisals of the manuscript.
Footnotes
Published ahead of print 11 January 2013
REFERENCES
- 1. Muller FM, Seidler M, Beauvais A. 2011. Aspergillus fumigatus biofilms in the clinical setting. Med. Mycol. 49(Suppl 1): S96–S100 [DOI] [PubMed] [Google Scholar]
- 2. Ramage G, Rajendran R, Gutierrez-Correa M, Jones B, Williams C. 2011. Aspergillus biofilms: clinical and industrial significance. FEMS Microbiol. Lett. 324: 89–97 [DOI] [PubMed] [Google Scholar]
- 3. Escande W, Fayad G, Modine T, Verbrugge E, Koussa M, Senneville E, Leroy O. 2011. Culture of a prosthetic valve excised for streptococcal endocarditis positive for Aspergillus fumigatus 20 years after a previous fumigatus endocarditis. Ann. Thorac. Surg. 91: e92–e93 [DOI] [PubMed] [Google Scholar]
- 4. Jeloka TK, Shrividya S, Wagholikar G. 2011. Catheter outflow obstruction due to an aspergilloma. Perit. Dial. Int. 31: 211–212 [DOI] [PubMed] [Google Scholar]
- 5. Langer P, Kassim RA, Macari GS, Saleh KJ. 2003. Aspergillus infection after total knee arthroplasty. Am. J. Orthop. 32: 402–404 [PubMed] [Google Scholar]
- 6. Rosenblatt WB, Pollock A. 1997. Aspergillus flavus cultured from a saline-filled implant. Plast. Reconstr. Surg. 99: 1470–1472 [DOI] [PubMed] [Google Scholar]
- 7. Young RC, Bennett JE, Vogel CL, Carbone PP, DeVita VT. 1970. Aspergillosis. The spectrum of the disease in 98 patients. Medicine (Baltimore) 49: 147–173 [DOI] [PubMed] [Google Scholar]
- 8. Jayshree RS, Shafiulla M, George J, David JK, Bapsy PP, Chakrabarti A. 2006. Microscopic, cultural and molecular evidence of disseminated invasive aspergillosis involving the lungs and the gastrointestinal tract. J. Med. Microbiol. 55: 961–964 [DOI] [PubMed] [Google Scholar]
- 9. Flemming HC, Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8: 623–633 [DOI] [PubMed] [Google Scholar]
- 10. Ramage G, Rajendran R, Sherry L, Williams C. 2012. Fungal biofilm resistance. Int. J. Microbiol. 2012: 528521 doi:10.1155/2012/528521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beauvais A, Schmidt C, Guadagnini S, Roux P, Perret E, Henry C, Paris S, Mallet A, Prevost MC, Latge JP. 2007. An extracellular matrix glues together the aerial-grown hyphae of Aspergillus fumigatus. Cell. Microbiol. 9: 1588–1600 [DOI] [PubMed] [Google Scholar]
- 12. Loussert C, Schmitt C, Prevost MC, Balloy V, Fadel E, Philippe B, Kauffmann-Lacroix C, Latge JP, Beauvais A. 2010. In vivo biofilm composition of Aspergillus fumigatus. Cell. Microbiol. 12: 405–410 [DOI] [PubMed] [Google Scholar]
- 13. Seidler MJ, Salvenmoser S, Muller FM. 2008. Aspergillus fumigatus forms biofilms with reduced antifungal drug susceptibility on bronchial epithelial cells. Antimicrob. Agents Chemother. 52: 4130–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nett JE, Crawford K, Marchillo K, Andes DR. 2010. Role of Fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob. Agents Chemother. 54: 3505–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mowat E, Lang S, Williams C, McCulloch E, Jones B, Ramage G. 2008. Phase-dependent antifungal activity against Aspergillus fumigatus developing multicellular filamentous biofilms. J. Antimicrob. Chemother. 62: 1281–1284 [DOI] [PubMed] [Google Scholar]
- 16. Rajendran R, Mowat E, McCulloch E, Lappin DF, Jones B, Lang S, Majithiya JB, Warn P, Williams C, Ramage G. 2011. Azole resistance of Aspergillus fumigatus biofilms is partly associated with efflux pump activity. Antimicrob. Agents Chemother. 55: 2092–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martins M, Uppuluri P, Thomas DP, Cleary IA, Henriques M, Lopez-Ribot JL, Oliveira R. 2010. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia 169: 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295: 1487. [DOI] [PubMed] [Google Scholar]
- 19. Mulcahy H, Charron-Mazenod L, Lewenza S. 2010. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ. Microbiol. 12: 1621–1629 [DOI] [PubMed] [Google Scholar]
- 20. Molin S, Tolker-Nielsen T. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14: 255–261 [DOI] [PubMed] [Google Scholar]
- 21. Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59: 1114–1128 [DOI] [PubMed] [Google Scholar]
- 22. Berne C, Kysela DT, Brun YV. 2010. A bacterial extracellular DNA inhibits settling of motile progeny cells within a biofilm. Mol. Microbiol. 77: 815–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Izano EA, Amarante MA, Kher WB, Kaplan JB. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mulcahy H, Charron-Mazenod L, Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 4: e1000213 doi:10.1371/journal.ppat.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tetz GV, Artemenko NK, Tetz VV. 2009. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob. Agents Chemother. 53: 1204–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153: 2083–2092 [DOI] [PubMed] [Google Scholar]
- 27. Spoering AL, Gilmore MS. 2006. Quorum sensing and DNA release in bacterial biofilms. Curr. Opin. Microbiol. 9: 133–137 [DOI] [PubMed] [Google Scholar]
- 28. Kadurugamuwa JL, Beveridge TJ. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177: 3998–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Al-Fattani MA, Douglas LJ. 2006. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J. Med. Microbiol. 55: 999–1008 [DOI] [PubMed] [Google Scholar]
- 30. Paramonova E, Krom BP, van der Mei HC, Busscher HJ, Sharma PK. 2009. Hyphal content determines the compression strength of Candida albicans biofilms. Microbiology 155: 1997–2003 [DOI] [PubMed] [Google Scholar]
- 31. Martins M, Henriques M, Lopez-Ribot JL, Oliveira R. 2012. Addition of DNase improves the in vitro activity of antifungal drugs against Candida albicans biofilms. Mycoses 55: 80–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wasylnka JA, Moore MM. 2002. Uptake of Aspergillus fumigatus conidia by phagocytic and nonphagocytic cells in vitro: quantitation using strains expressing green fluorescent protein. Infect. Immun. 70: 3156–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mowat E, Butcher J, Lang S, Williams C, Ramage G. 2007. Development of a simple model for studying the effects of antifungal agents on multicellular communities of Aspergillus fumigatus. J. Med. Microbiol. 56: 1205–1212 [DOI] [PubMed] [Google Scholar]
- 34. Erlandsen SL, Kristich CJ, Dunny GM, Wells CL. 2004. High-resolution visualization of the microbial glycocalyx with low-voltage scanning electron microscopy: dependence on cationic dyes. J. Histochem. Cytochem. 52: 1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Beauvais A, Loussert C, Prevost MC, Verstrepen K, Latge JP. 2009. Characterization of a biofilm-like extracellular matrix in FLO1-expressing Saccharomyces cerevisiae cells. FEMS Yeast Res. 9: 411–419 [DOI] [PubMed] [Google Scholar]
- 36. Schuttelkopf AW, Gros L, Blair DE, Frearson JA, van Aalten DM, Gilbert IH. 2010. Acetazolamide-based fungal chitinase inhibitors. Bioorg. Med. Chem. 18: 8334–8340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leggate J, Allain R, Isaac L, Blais BW. 2006. Microplate fluorescence assay for the quantification of double stranded DNA using SYBR Green I dye. Biotechnol. Lett. 28: 1587–1594 [DOI] [PubMed] [Google Scholar]
- 38. Bain JM, Tavanti A, Davidson AD, Jacobsen MD, Shaw D, Gow NA, Odds FC. 2007. Multilocus sequence typing of the pathogenic fungus Aspergillus fumigatus. J. Clin. Microbiol. 45: 1469–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alcazar-Fuoli L, Clavaud C, Lamarre C, Aimanianda V, Seidl-Seiboth V, Mellado E, Latge JP. 2011. Functional analysis of the fungal/plant class chitinase family in Aspergillus fumigatus. Fungal Genet. Biol. 48: 418–429 [DOI] [PubMed] [Google Scholar]
- 40. Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution antifungal susceptibility testing of yeast; approved standard, 3rd ed. CLSI document M27-A3 CLSI, Wayne, PA [Google Scholar]
- 41. Robbins N, Uppuluri P, Nett J, Rajendran R, Ramage G, Lopez-Ribot JL, Andes D, Cowen LE. 2011. Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 7: e1002257 doi:10.1371/journal.ppat.1002257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emri T, Molnar Z, Szilagyi M, Pocsi I. 2008. Regulation of autolysis in Aspergillus nidulans. Appl. Biochem. Biotechnol. 151: 211–220 [DOI] [PubMed] [Google Scholar]
- 43. Molnar Z, Emri T, Zavaczki E, Pusztahelyi T, Pocsi I. 2006. Effects of mutations in the GanB/RgsA G protein mediated signalling on the autolysis of Aspergillus nidulans. J. Basic Microbiol. 46: 495–503 [DOI] [PubMed] [Google Scholar]
- 44. Shin KS, Kwon NJ, Kim YH, Park HS, Kwon GS, Yu JH. 2009. Differential roles of the ChiB chitinase in autolysis and cell death of Aspergillus nidulans. Eukaryot. Cell 8: 738–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nett J, Lincoln L, Marchillo K, Massey R, Holoyda K, Hoff B, VanHandel M, Andes D. 2007. Putative role of beta-1,3 glucans in Candida albicans biofilm resistance. Antimicrob. Agents Chemother. 51: 510–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mochizuki S, Sakurai K. 2009. A novel polysaccharide/polynucleotide complex and its application to bio-functional DNA delivery system. Polym. J. 2009: 343–353 [Google Scholar]
- 47. Tamkovich S, Vlassov V, Laktionov P. 2008. Circulating DNA in the blood and its application in medical diagnosis. Mol. Biol. 42: 9–19 [Google Scholar]
- 48. Fink SL, Cookson BT. 2005. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect. Immun. 73: 1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lappann M, Claus H, van Alen T, Harmsen M, Elias J, Molin S, Vogel U. 2010. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol. Microbiol. 75: 1355–1371 [DOI] [PubMed] [Google Scholar]
- 50. Thomas VC, Thurlow LR, Boyle D, Hancock LE. 2008. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 190: 5690–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pocsi I, Leiter E, Kwon NJ, Shin KS, Kwon GS, Pusztahelyi T, Emri T, Abuknesha RA, Price RG, Yu JH. 2009. Asexual sporulation signalling regulates autolysis of Aspergillus nidulans via modulating the chitinase ChiB production. J. Appl. Microbiol. 107: 514–523 [DOI] [PubMed] [Google Scholar]
- 52. Szilagyi M, Kwon NJ, Dorogi C, Pocsi I, Yu JH, Emri T. 2010. The extracellular beta-1,3-endoglucanase EngA is involved in autolysis of Aspergillus nidulans. J. Appl. Microbiol. 109: 1498–1508 [DOI] [PubMed] [Google Scholar]
- 53. Bruns S, Seidler M, Albrecht D, Salvenmoser S, Remme N, Hertweck C, Brakhage AA, Kniemeyer O, Muller FM. 2010. Functional genomic profiling of Aspergillus fumigatus biofilm reveals enhanced production of the mycotoxin gliotoxin. Proteomics 10: 3097–3107 [DOI] [PubMed] [Google Scholar]
- 54. Gibbons JG, Beauvais A, Beau R, McGary KL, Latge JP, Rokas A. 2012. Global transcriptome changes underlying colony growth in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot. Cell 11: 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grande R, Di Giulio M, Bessa LJ, Di Campli E, Baffoni M, Guarnieri S, Cellini L. 2011. Extracellular DNA in Helicobacter pylori biofilm: a backstairs rumour. J. Appl. Microbiol. 110: 490–498 [DOI] [PubMed] [Google Scholar]
- 56. Al-Fattani MA, Douglas LJ. 2004. Penetration of Candida biofilms by antifungal agents. Antimicrob. Agents Chemother. 48: 3291–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nett JE, Sanchez H, Cain MT, Andes DR. 2010. Genetic basis of Candida biofilm resistance due to drug-sequestering matrix glucan. J. Infect. Dis. 202: 171–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Espinel-Ingroff A. 2009. Novel antifungal agents, targets or therapeutic strategies for the treatment of invasive fungal diseases: a review of the literature (2005–2009). Rev. Iberoam. Micol. 26: 15–22 [DOI] [PubMed] [Google Scholar]
- 59. Kaplan JB. 2009. Therapeutic potential of biofilm-dispersing enzymes. Int. J. Artif. Organs 32: 545–554 [DOI] [PubMed] [Google Scholar]
- 60. Frederiksen B, Pressler T, Hansen A, Koch C, Hoiby N. 2006. Effect of aerosolized rhDNase (Pulmozyme) on pulmonary colonization in patients with cystic fibrosis. Acta Paediatr. 95: 1070–1074 [DOI] [PubMed] [Google Scholar]