Abstract
Oligotrichs are ciliates of great abundance, but their molecular systematics are rarely studied. In this study, nine species representing three genera (Strombidium, Novistrombidium, and Omegastrombidium) of marine oligotrich ciliates were collected from coastal waters of China. The small subunit (SSU) rRNA gene of two species and the internal transcribed spacers and 5.8S region (ITS1-5.8S-ITS2) for all nine species were sequenced for the first time. Phylogenetic trees using both the SSU rRNA gene and ITS1-5.8S-ITS2 region sequences were generated. In addition, the secondary structures of ITS2 RNA transcripts of 11 taxa representing four genera (Novistrombidium, Strombidium, Omegastrombidium, and Laboea) were investigated. The phylogenetic analyses show that (i) the family Strombidiidae is polyphyletic, (ii) the genus Novistrombidium is probably paraphyletic, containing at least two subclades, which is consistent with recent cladistic analyses based on morphological data, and (iii) the tail-less genus Laboea is separate from other genera of Strombidiidae, clustering instead with the tontoniids. Comparisons of the secondary structure of ITS2 regions also show that Laboea is clearly different from other strombidiids. These findings cast doubt on the monophyly of the family Strombidiidae.
INTRODUCTION
Most members of the ciliate subclass Oligotrichia are thought to be cosmopolitan (1). Because of their high abundance and growth rate, oligotrichs are an important component of marine planktonic ciliate communities and play a critical role in the trophic flux and nutrient cycling of the pelagial realm (2). To date only c. 60% of oligotrichs, or about 120 species, have been described or redescribed using modern methods to reveal details of the infraciliature and other morphological features of taxonomic importance (1, 3–6). Consequently, many issues concerning the systematics of members within the group remain unresolved.
In recent years, molecular methods have been applied to investigate phylogenetic relationships among oligotrichs, mostly based on small subunit (SSU) rRNA gene sequence data (5–9). However, it is increasingly recognized that any single gene has limitations for elucidating evolutionary relationships among ciliates (10). Furthermore, there appears to be more genetic variation among oligotrich and choreotrich genera than among other spirotrich ciliate groups, casting doubt on the likelihood that SSU rRNA gene sequence data alone can reflect this variation (11). Additional molecular markers that are increasingly used for investigating phylogenetic relationships among ciliates include the internal transcribed spacer 2 (ITS2) secondary structure and ITS1-5.8S-ITS2 region sequence data (12, 13). In the present study, we combine data from all three sources (i.e., SSU rRNA gene sequences, ITS1-5.8S-ITS2 region sequences, and ITS2 secondary structure) in order to infer evolutionary relationships within the Oligotrichia. Furthermore, in order to better resolve the phylogeny of the group, we increase the number of sampled taxa by adding molecular data for nine populations, representing seven species and three genera of oligotrichs.
MATERIALS AND METHODS
Ciliate collection and identification.
Samples were collected using 20-μm mesh plankton nets. Collection data for each species are given in Table 1. Culturing and morphological examination of these species were performed using the methods of previous studies (3, 14). Species identification was based on the literature (4, 15, 16). Terminology and systematics follow Lynn (17).
Table 1.
Oligotrichs sampled for this study
| Species | Sample location | Date (mo/yr) |
|---|---|---|
| Novistrombidium sinicum pop1 | Mangrove wetland, Shenzhen (22°32′N, 114°01′E; southern China) | 01/2008 |
| Omegastrombidium cf. elegans | Daya Bay, Guangdong (22°42′N, 114°32′E), southern China | 01/2008 |
| Strombidium stylifer | Mangrove wetland, Shenzhen | 04/2008 |
| Strombidium basimorphum | Mangrove wetland, Shenzhen | 04/2008 |
| Novistrombidium testaceum | Mangrove wetland, Shenzhen | 04/2008 |
| Novistrombidium orientale | Daya Bay, Guangdong | 03/2008 |
| Strombidium conicum | Daya Bay, Guangdong | 04/2008 |
| Novistrombidium sinicum pop2 | Daya Bay, Guangdong | 04/2008 |
| Strombidium cf. conicum | Daya Bay, Guangdong | 12/2008 |
DNA extraction, PCR amplification, cloning, and sequencing.
Genomic DNA was extracted as described by Liu et al. (16). In brief, 10 to 15 cells were starved overnight in sterile seawater at room temperature to minimize the contents of food vacuoles and eliminate contaminants (13). DNA was extracted using a RdExtract-N-Amp tissue PCR kit (Sigma, St. Louis, MO) according to the manufacturer's protocol, with the slight modification that only 1/10 of the volume suggested for each reagent solution was used.
Primers used for amplifying ITS1-5.8S-ITS2 region sequences were 5.8SF, 5′-GTA GGT GAA CCT GCG GAA GGA TCA TTA-3′, and 5.8SR, 5′-TAC TGA TAT GCT TAA GTT CAG CGG-3′. PCR conditions followed Yi and Song (13). Additionally, the universal eukaryotic forward primer Euk A (5′-AACCTGGTTGATCCTGCCAGT-3′) and reverse primer Euk B (5′-TGATCCTTCTGCAGGTTCACCTAC-3′) were used to amplify the SSU rRNA gene (18), using the PCR protocol described by Liu et al. (19).
The purified PCR product was inserted into the pUCm-T vector (Shanghai Sangon Biological Engineering & Technical Service Company, Shanghai, China) and transformed into E. coli DH5α cells. Sequencing was carried out on an ABI 3730 sequencer (Applied Biosystems).
Phylogenetic analyses.
Other sequences used in this study were obtained from the NCBI GenBank database (accession numbers are given in Fig. 1 and 2A). Sequences were aligned using Clustal W implemented in BioEdit 7.0.0 (20), and the unique regions for oligotrichs were exported using Seaview 4 (21). Ends were trimmed, and the ambiguously aligned sites were refined, yielding an alignment of 1,532 (versus 2,005 in the original alignment) sites for the SSU rRNA gene, 563 (versus 766 in the original alignment) sites for the ITS1-5.8S-ITS2 region, and 2,190 (versus 2,615 in the original alignment) sites for the concatenated SSU rRNA and ITS-5.8S gene sequences, respectively. For the phylogenetic analyses, the program Modeltest (22) selected GTR plus I (0.4092) plus G (0.4771) for the SSU rRNA gene, GTR plus I (0.0673) plus G (0.5883) for the ITS1-5.8S-ITS2 region, and GTR plus I (0.3208) plus G (0.5055) for the concatenated sequences as the best models under the AIC criterion, which were then used to construct maximum likelihood (ML) trees. Similarly, MrModeltest, version 2 (23), chose the same best-fitting model with the identical values for Bayesian inference (BI) analyses. ML trees were constructed with the PhyML program, version 2.4.4 (24). The reliability of internal branches was assessed using a nonparametric bootstrap method with 1,000 replicates. The BI analysis was performed with MrBayes 3.1.2 (25). Four simultaneous Markov chain Monte Carlo (MCMC) chains were run for 1,000,000 (2,000,000 for concatenated sequences) generations with a sample frequency of 100. The first 2,500 (5,000 for concatenated sequences) generations were discarded as burn-in. Posterior probabilities were calculated by applying the majority rule consensus. Protocruzia adherens of the subclass Protocruziidia was selected as the outgroup taxon in the analyses.
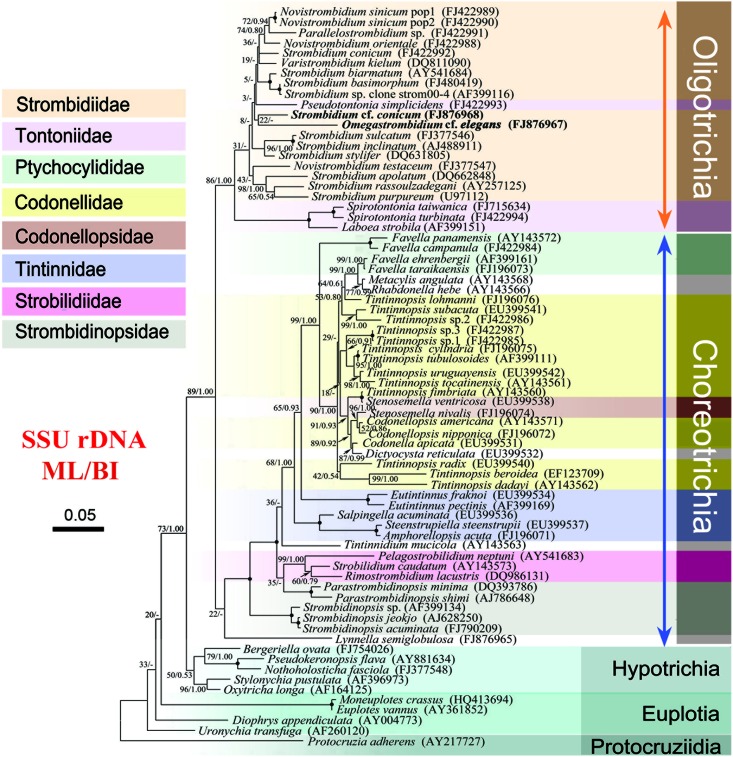
Fig 1.
Phylogenetic tree based on 72 SSU rDNA sequences inferred by maximum-likelihood (ML) and Bayesian inference (BI) analyses. Numbers near the nodes of branches represent the bootstrap value for the ML analysis and the posterior probability value of the BI analysis, respectively. Dashes (-) reflect disagreement between the two topologies. The scale bar corresponds to 5 substitutions per 100 nucleotide positions. Newly deposited sequences are in boldface. The codes in parentheses following the species name are the GenBank sequence accession numbers.
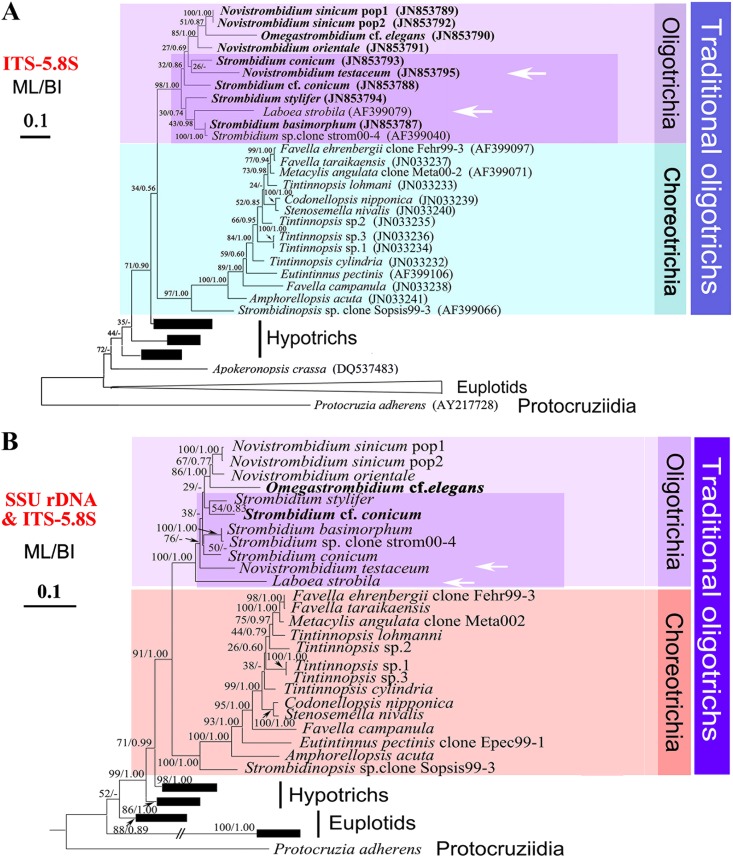
Fig 2.
Phylogenetic trees based on ITS1-5.8S-ITS2 region sequences (A) and SSU rRNA gene and ITS-5.8S region sequences (B) inferred by maximum likelihood (ML) and Bayesian inference (BI) analyses. Species newly sequenced in this study are in boldface. The codes following the names are the GenBank sequence accession numbers. Numbers on branches represent the bootstrap values from ML analysis and posterior probability of Bayesian analysis, respectively. Dashes (-) indicate disagreement between the ML and BI methods. The scale bar corresponds to 10 substitutions per 100 nucleotide positions.
ITS2 secondary structure prediction.
The ITS2 sequences were submitted to the mfold website (http://mfold.rna.albany.edu/?q=mfold) for secondary structure prediction with default settings (26). Structures were edited for esthetic purposes with RnaViz 2.0 (27) under the model for ciliates (28).
Topology testing.
In order to test the monophyly of the family Strombidiidae and of the genera Novistrombidium and Strombidium, the approximately unbiased (AU) test was used (29). Steps were as described in reference 12. Eight constrained ML analyses were carried out on the SSU rRNA alignment, as follows: (i) 18 strombidiids, (ii) 18 strombidiids and Pseudotontonia simplicidens, (iii) 18 strombidiids and Laboea strobila, (iv) 18 strombidiids, P. simplicidens, and L. strobila, (v) four species of Novistrombidium, (vi) Laboea and Spirotontonia, (vii) Pseudotontonia, Laboea, and Spirotontonia, and (viii) 11 species of Strombidium. The resulting constrained topologies were then compared to the unconstrained ML topologies. The internal relationships within each constrained group and the relationships among the remaining taxa were unspecified.
Nucleotide sequence accession numbers.
The SSU rRNA gene of two species (Strombidium cf. conicum and Omegastrombidium cf. elegans) and the ITS1-5.8S-ITS2 region for all nine species were sequenced for the first time. The sequences were deposited in the NCBI GenBank database with accession numbers, lengths, and GC contents as listed in Table 2.
Table 2.
List of species for which both the ITS1-5.8S-ITS2 region and SSU-rDNA gene were newly sequenceda or obtained from GenBank
| Species | ITS1-5.8S-ITS2 |
SSU-rRNA gene |
||||
|---|---|---|---|---|---|---|
| GenBank accession no. | Sequence length (bp) | GC content (%) | GenBank accession no. | Sequence length (bp) | GC content (%) | |
| Strombidium conicum | JN853793 | 546 | 48.53 | FJ422992 | 1775 | 47.61 |
| S. cf. conicum | JN853788 | 542 | 46.68 | FJ876968 | 1776 | 47.30 |
| Omegastrombidium cf. elegans | JN853790 | 542 | 44.10 | FJ876967 | 1772 | 43.74 |
| S. stylifer | JN853794 | 541 | 49.15 | DQ631805 | 1774 | 48.03 |
| S. basimorphum | JN853787 | 543 | 47.88 | FJ480419 | 1774 | 48.08 |
| Novistrombidium orientale | JN853791 | 536 | 48.51 | FJ422988 | 1772 | 47.35 |
| N. sinicum pop1 | JN853789 | 534 | 50.37 | FJ422989 | 1773 | 48.34 |
| N. sinicum pop2 | JN853792 | 534 | 50.56 | FJ422990 | 1773 | 48.11 |
| N. testaceum | JN853795 | 538 | 47.21 | FJ377547 | 1770 | 48.36 |
Accession numbers for new sequences are in boldface.
RESULTS
There is only a 1-bp difference in the ITS1-5.8S-ITS2 region sequences between the two populations of N. sinicum. The SSU rRNA gene and the ITS1-5.8S-ITS2 region sequences of the 11 strombidiids used in the phylogenetic analyses share similarities of 68.4% to 99.6% (86.0% to 99.6% without Strombidium sp.) and 73.2% to 99.8%, respectively.
Secondary structures of the order Strombidiida.
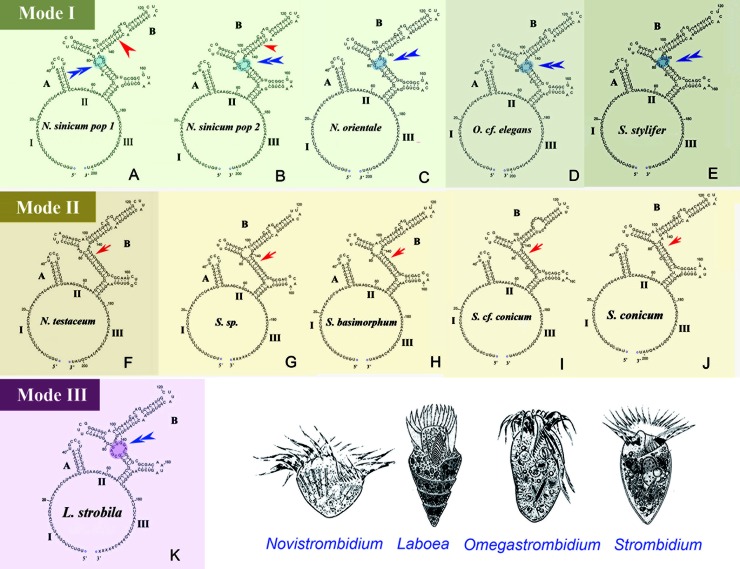
Within the order Strombidiida, ITS1-5.8S-ITS2 region sequence data were available for 11 lineages representing four genera (Novistrombidium, Omegastrombidium, Strombidium, and Laboea). The predicted secondary structure of the ITS2 region generally consists of a large loop separated by two helices. Considering the primary sequences, as well as the secondary structures, of their ITS2 region, we found that (i) although four genera were represented, the 11 structures were divided into three modes (Novistrombidium sinicum population 1 [pop1] and pop2, Novistrombidium orientale, Omegastrombidium cf. elegans, and Strombidium stylifer [Fig. 3A to E]; Novistrombidium testaceum, Strombidium sp., Strombidium basimorphum, Strombidium cf. conicum, and Strombidium conicum [Fig. 3F to J]; and Laboea strobila [Fig. 3K]), (ii) the only substituted nucleotide between two isolates of N. sinicum was located at the 136th site of the ITS2 region (see Fig. S1 in the supplemental material), and (iii) the unique nucleotide contributing to the classification of these three modes, inferred from diagrams in Figure 3, was the 146th base (see Fig. S1).
Fig 3.
Secondary structures of the internal transcribed spacer 2 (ITS2) RNA transcript of representative species of four oligotrich genera (Novistrombidium, Omegastrombidium, Strombidium, and Laboea). The diagrams illustrate the two helices, labeled A and B, present in the class Spirotrichea (28). The three parts of the largest loop are labeled I, II, and III, respectively. The red arrowheads (in panels A and B) mark the only nucleotide variation between two populations of N. sinicum. Red arrows or blue double arrowheads indicate the different structures in helix B (elliptical shadowed region) among these three categories.
Phylogenetic trees.
Trees were constructed using different algorithms and with different gene sequences. The topologies were identical at higher taxonomic levels, whereas the relationships among certain strombidiid genera were inconsistent.
Trees based on SSU rRNA gene sequences with more taxa selected showed the monophyly of both subclasses, Oligotrichia and Choreotrichia (Fig. 1). Laboea strobila clustered with two species of the tontoniid genus Spirotontonia with high support values (100% ML and 1.00 BI) and was the sister clade to the strombidiid assemblage.
Trees constructed using the ITS1-5.8S-ITS2 region (Fig. 2A) showed that the subclass Oligotrichia was monophyletic with strong support (98% ML and 1.00 BI) and was sister to the subclass Choreotrichia, which was also monophyletic with strong support values (97% ML and 1.00 BI). There were two clades within the Oligotrichia, (i) Laboea-Strombidium (except S. conicum) (30% ML and 0.74 BI) and (ii) Novistrombidium-Omegastrombidium-Strombidium conicum (27% ML and 0.69 BI). Within the second clade, there were two groups; one comprised the two populations of N. sinicum, O. cf. elegans and N. orientale, and the other comprised N. testaceum and S. conicum.
In order to mitigate the effect of base mutation among isolates, SSU ribosomal DNA (rDNA) sequences were, as far as possible, obtained from the same clones as those used to construct the ITS1-5.8S-ITS2 region tree and then combined with their corresponding ITS-5.8S region sequences. The resulting concatenated tree (SSU rDNA and ITS-5.8S region) shared a similar topology to that of the ITS1-5.8S-ITS2 tree (Fig. 2B). For example, in both trees, (i) both Oligotrichia and Choreotrichia were monophyletic with high support and (ii) both Strombidium and Novistrombidium were paraphyletic. However, the distribution patterns of the component species were different. In the ITS1-5.8S-ITS2 tree, three of four Novistrombidium sequences formed a paraphyletic clade nesting with Omegastrombidium species (85% ML and 1.00 BI), while N. testaceum clustered with S. conicum. In the concatenated tree, N. testaceum appeared as the second deepest branch of Oligotrichia and the other three Novistrombidium sequences formed a monophyletic clade. The inconsistency also applied to Strombidium. In the ITS1-5.8S-ITS2 tree, five species of Strombidium scattered into 4 clades, while they clustered into two groups in the concatenated tree. The most obvious difference is the position of Laboea. In the concatenated tree (Fig. 2B), L. strobila occupied a basal position within the oligotrichs (100% ML and 1.00 BI), whereas in the ITS1-5.8S-ITS2 tree (Fig. 2A), L. strobila grouped with two Strombidium species (43% ML and 0.98 BI).
Topology testing.
The AU test rejected the possibility that Laboea belongs to the family Strombidiidae (Table 3, constraint group 8, P = 0.002), but the possibility that all species of Strombidiidae except Laboea form a monophyletic group was not refuted (constraint group 3, P = 0.970). Moreover, even though species of Strombidium and Novistrombidium did not form monophyletic clades (Fig. 1), the possibility of monophyly of the genera Strombidium and Novistrombidium was not rejected (Table 3, constraint group 4, P = 0.759, and constraint group 5, P = 0.404). Furthermore, the topology was robust for the tontoniid Spirotontonia and Pseudotontonia together with Laboea (Table 3, constraint group 1, P = 0.970, and constraint group 2, P = 0.970).
Table 3.
Approximately unbiased test results
| Constraint group | Topology constraint | −ln likelihood | AU value (P)a |
|---|---|---|---|
| 1 | Laboea + Spirotontonia | 15,809.24,178 | 0.970 |
| 2 | Pseudotontonia + Laboea + Spirotontonia | 15,809.24,178 | 0.970 |
| 3 | 18 strombidiids | 15,809.24,178 | 0.970 |
| 4 | Strombidium monophyly | 15,813.87,249 | 0.759 |
| 5 | Novistrombidium monophyly | 15,820.11,228 | 0.404 |
| 6 | 18 strombidiids + Pseudotontonia | 15,832.74,004 | 0.039 |
| 7 | 18 strombidiids + Pseudotontonia + Laboea | 15,940.07,464 | 4e−004 |
| 8 | 18 strombidiids + Laboea | 20,311.37,195 | 0.002 |
AU values (P) below 0.05 are in boldface.
DISCUSSION
The genus Laboea.
In recently published SSU rRNA gene trees of the Oligotrichia, the tail-less genus Laboea is more closely related to the tailed tontoniid genera Spirotontonia and Pseudotontonia than to the tail-less taxa of the family Strombidiidae (6, 8). This relationship was also recovered with maximum support (100% ML and 1.00 BI) in the present analyses based on SSU rRNA gene sequence data (Fig. 1). Moreover, the AU test rejected all constrained trees which supposed the monophyly of Strombidiidae with the inclusion of the genus Laboea (P < 0.01), and the constrained tree which supposed Laboea with Spirotontonia was not rejected (P = 0.970). Besides the tree topology, there are another three pieces of evidence supporting the closer relationship between Laboea and tontoniids, as follows. (i) The sequence identities of Laboea with three tontoniids (92.3% to 94.8%) were higher than those of Laboea with strombidiids (88.3% to 90.4%). (ii) Laboea has more common unique characters with tontoniids than with strombidiids both in molecular information (see Fig. S2 in the supplemental material) and in morphological features. The morphology of Laboea is very similar to that of the tontoniid Spirotontonia, e.g., the irregular cone-shaped body with sinistrally spiraled girdle kinety, giving it a screw-like appearance, and the multiple macronuclear nodules (3) (see Fig. S3 in the supplemental material). (iii) The secondary structure of the ITS2 region shows that Laboea clearly differs from other strombidiids, such as Novistrombidium and Strombidium, by the presence of a loop in helix B composed of 10 nucleotides (versus 8 in Novistrombidium sinicum and N. orientale and no loop in Strombidium) (Fig. 3). These findings were examples of a disagreement between molecular information and morphology. The “tail” may be not a good family-level diagnostic feature for the separation of Tontoniidae from other oligotrich families (21), especially with the consideration of Laboea.
Is the family Strombidiidae paraphyletic?
As a species-rich group of oligotrichs, the family Strombidiidae is characterized by having a bipartite oral ciliature with anterior and ventral membranelles, a strongly reduced somatic ciliature, and a unique stomatogenetic process which takes place within a transient tube (3, 17). Within the family, the somatic ciliature, which consists of only one to several kineties, exhibits high diversity of arrangement and has been always regarded as an important generic character. Ten genera are currently assigned in the family Strombidiidae, i.e., Strombidium, Spirostrombidium, Parallelostrombidium, Novistrombidium, Omegastrombidium, Apostrombidium, Varistrombidium, Opisthostrombidium, Foissneridium, and Williophrya (3, 4, 30) (see Fig. S3 in the supplemental material). Recently, Agatha (3) separated strombidiids with a contractile tail from other family members and placed them in a new family, Tontoniidae, containing four genera: Tontonia, Paratontonia, Spirotontonia, and Pseudotontonia.
As mentioned above, members of the family Strombidiidae differ from the Tontoniidae in lacking a conspicuous, elongate, contractile tail (17). Previous studies have reported the monophyly of the family Strombidiidae (9, 11, 31). However, recent studies based on SSU rRNA gene sequence data revealed that the tail-less genus Laboea clusters with the Tontoniidae rather the Strombidiidae (8, 32), which renders the family Strombidiidae paraphyletic.
In the present study, the ITS1-5.8S-ITS2 region tree and the concatenated tree were constrained by the limited number of taxa. With the absence of tontoniids, it is hard to test whether the family Strombidiidae is monophyletic. Therefore, we constructed an expanded SSU rDNA tree with 11 additional oligotrichid and 26 additional choreotrichid sequences (Fig. 1). Although the bootstrap values are too low to define relationships among genera in Strombidiidae (Fig. 1), it is clear that the family Strombidiidae is not monophyletic if the genus Laboea is considered (Fig. 2 and Table 3).
Besides the intricate assignment of Laboea, it is noteworthy that the diversity of strombidiids is probably underestimated, with three new strombidiid genera, i.e., Williophrya Liu, 2011, Foissneridium Agatha, 2011, and Opisthostrombidium Agatha, 2011, being recently established (4, 30). Furthermore, there is a lack of molecular data, particularly multigene information, for many strombidiid species (especially the ITS of Spirotontonia and Pseudotontonia) and some genera are not represented in any form of molecular information. Consequently, more data are needed to determine the monophyly of the family Strombidiidae.
The known taxa in Strombidium belong to a nonmonophyletic assemblage.
The species-rich genus Strombidium was the basic and earliest-established taxon in the family Strombidiidae. To date, about 65 species have been assigned in this genus (1), while only about 10 nominal and 9 undetermined species have their molecular information in the NCBI database. However, even from this limited information, species of Strombidium have exhibited enormous diversities in both sequence identity and tree topology. The identities of the SSU rRNA gene sequences among Strombidium species ranged from 69.9% to 96.3% (89.5% to 96.3% without Strombidium sp. and S. biarmatum). The sequences of Strombidium sp. and S. biarmatum shared similarity of 95.5%, but they are both notably different from other Strombidium species (similarities with others ranged from 69.9% to 75.9%). In SSU rRNA gene trees (Fig. 1), the Strombidium species were separated into four clades, as follows: (i) S. conicum; (ii) S. biarmatum, S. basimorphum, and Strombidium sp.; (iii) S. apolatum, S. rassoulzadegani, and S. purpureum; and (iv) S. sulcatum, S. inclinatum, and S. stylifer. Intriguingly, the separation of congeners was supported by morphological differences. For example, S. conicum revealed itself in a separate branch by possessing a unique hemitheca with longitudinal lines rather than polygonal platelets (33). In addition, S. biarmatum and S. basimorphum justified a distinct clade by being the only two congeners with extra extrusomes attached to the anterior portion of cells (4, 31). Our findings, together with previous molecularly based results (4), further support that Strombidium should be split into several morphologically and ontogenetically defined genera (31). Although the possibility of the monophyly of Strombidium was not rejected (Table 3, constraint group 4, P = 0.759), it might still be possible that the intrageneric morphological differences are not genuinely reflected in SSU rRNA genes.
The genus Novistrombidium is paraphyletic.
Previous reports based on SSU rRNA gene sequence data have suggested that the genus Novistrombidium is paraphyletic when N. sinicum and N. orientale are included (4, 6, 8, 15, 16). The results of the present multigene analyses are consistent with this finding (Fig. 1 and 2). Data inferred from the secondary structure of the ITS2 region corroborate the paraphyly of Novistrombidium, with N. sinicum and N. orientale having a mode I structure (with seven loops in helix B) similar to that of S. stylifer and Omegastrombidium, whereas N. testaceum has a mode II structure (with six loops in helix B) similar to the majority of Strombidium species (Fig. 3).
It should be noted in the SSU rDNA tree that N. sinicum and N. orientale reside in the same clade as Parallelostrombidium. This topology could be explained by their shared characteristics with Parallelostrombidium, including (i) the broadly ellipsoidal cell shape, (ii) extrusomes equidistantly arranged along the girdle kinety, (iii) the presence of thigmotactic membranelles, (iv) an ovoid macronucleus, and (v) the localization of the anterior end of the ventral kinety below the right portion of the girdle kinety (15). Furthermore, the respective sequence identities of Parallelostrombidium with N. sinicum and N. testaceum are 97% and 96.2%, slightly higher than those between N. testaceum and two other congeners (95.1% and 95.7%). In contrast, N. testaceum, the type species of Novistrombidium, has a sausage-shaped macronucleus, extrusomes grouped in bundles, and localization of the ventral kinety extending longitudinally through the gap in the girdle kinety, which resulted in its separation from the other two Novistrombidium species (34). Nevertheless, the monophyly of the genus Novistrombidium could not be rejected by the AU test (Table 3, constraint group 5, P = 0.404), and this discrepancy between phylogenetic topology and AU test result may stem from undersampling.
Conclusions from the multigene phylogenetic analysis.
When the ITS1-5.8S-ITS2 region was first applied to the study of oligotrichs and choreotrichs, it was suggested that the ITS and 5.8S regions could provide adequate polymorphism data to assess genetic variation at the genus/population level within these groups (11). In the meantime, SSU rDNA sequences have been widely used to infer evolutionary relationships among spirotrichs, and phylogenetic trees based on such data are generally concordant with many morphological hypotheses, albeit with some discrepancies (17, 35). The value of using a single gene marker in order to elucidate evolutionary relationships among ciliates has been questioned (10). Therefore, we have used multigenes, i.e., SSU rRNA gene and ITS-5.8S region sequences, to increase the robustness of our analyses of phylogenetic relationships among oligotrichs. In the present study, the overall mean distances in oligotrichs are 0.104 in the SSU rRNA gene and 0.159 in the ITS-5.8S region, while in choreotrichs, they are 0.067 in the SSU rRNA gene and 0.090 in the ITS-5.8S region, confirming that oligotrichs are more genetically variable than choreotrichs in both the SSU rRNA gene and the ITS-5.8S region (11). Our findings support the removal of Laboea from the Strombidiidae to the Tontoniidae, thus rendering the family Strombidiidae monophyletic. Furthermore, the monophyly of Novistrombidium was doubted in the topologies of trees and morphological features. However, we are currently unable to resolve a number of phylogenetic relationships due to (i) differences between the two genes in length and variation rate, (ii) the lack of available ITS-5.8S region sequence data for oligotrichs, and (iii) undersampling of certain key taxa, such as Varistrombidium, Parallelostrombidium, and Omegastrombidium. Therefore, further studies are required to increase the resolution of the oligotrich systematics.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Natural Science of Foundation of China (project numbers 31201696, 31030059, and 31222050).
We thank Feng Gao (OUC), as well as three anonymous reviewers, for their help and advice with the early versions of this paper. Many thanks are also given to Jie Huang, Zhenzhen Yi, Qianqian Zhang (OUC), and Liang Zhou, Laboratory of Protozoology, South China Normal University, for their help with the laboratory work.
Footnotes
Published ahead of print 11 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00270-12.
REFERENCES
- 1. Agatha S. 2011. Global diversity of aloricate Oligotrichea (Protista, Ciliophora, Spirotricha) in marine and brackish sea water. PLoS One 6: e22466 doi:10.1371/journal.pone.0022466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pierce RW, Turner JT. 1992. Ecology of planktonic ciliates in marine food webs. Rev. Aquat. Sci. 6: 139–181 [Google Scholar]
- 3. Agatha S. 2004. Evolution of ciliary patterns in the Oligotrichida (Ciliophora, Spirotricha) and its taxonomic implications. Zoology (Jena) 107: 153–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu W, Yi Z, Warren A, Al-Rasheid KAS, Al-Farraj SA, Lin X, Song W. 2011. Taxonomy, morphology and molecular systematics of a new oligotrich ciliate, Williophrya maedai gen. nov., sp. nov., with redescriptions of Strombidium basimorphum and Pseudotontonia simplicidens (Protozoa, Ciliophora, Oligotrichia). Syst. Biodivers. 9: 247–258 [Google Scholar]
- 5. Modeo L, Petroni G, Rosati G, Montagnes DJS. 2003. A multidisciplinary approach to describe protists: redescriptions of Novistrombidium testaceum Anigstein 1914 and Strombidium inclinatum Montagnes, Taylor, and Lynn 1990 (Ciliophora, Oligotrichia). J. Eukaryot. Microbiol. 50: 175–189 [DOI] [PubMed] [Google Scholar]
- 6. Zhang Q, Yi Z, Xu D, Al-Rasheid KAS, Gong J, Song W. 2010. Molecular phylogeny of oligotrich genera Omegastrombidium and Novistrombidium (Protozoa, Ciliophora) for the systematical relationships within family Strombidiidae. Chin. J. Oceanol. Limnol. 28: 769–777 [Google Scholar]
- 7. Agatha S, Strüder-Kypke MC. 2007. Phylogeny of the order Choreotrichida (Ciliophora, Spirotricha, Oligotrichea) as inferred from morphology, ultrastructure, ontogenesis, and SSrRNA gene sequences. Eur. J. Protistol. 43: 37–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao S, Gong J, Lynn D, Lin X, Song W. 2009. An updated phylogeny of oligotrich and choreotrich ciliates (Protozoa, Ciliophora, Spirotrichea) with representative taxa collected from Chinese coastal waters. Syst. Biodivers. 7: 235–242 [Google Scholar]
- 9. Strüder-Kypke MC, Lynn DH. 2003. Sequence analyses of the small subunit rRNA gene confirm the paraphyly of oligotrich ciliates sensu lato and support the monophyly of the subclasses Oligotrichia and Choreotrichia (Ciliophora, Spirotrichea). J. Zool. 260: 87–97 [Google Scholar]
- 10. Dunthorn M, Foissner W, Katz LA. 2011. Expanding character sampling for ciliate phylogenetic inference using mitochondrial SSU-rDNA as a molecular marker. Protist 162: 85–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Snoeyenbos-West OL, Salcedo T, McManus GB, Katz LA. 2002. Insights into the diversity of choreotrich and oligotrich ciliates (Class: Spirotrichea) based on genealogical analyses of multiple loci. Int. J. Syst. Evol. Microbiol. 52: 1901–1913 [DOI] [PubMed] [Google Scholar]
- 12. Gao F, Katz LA, Song W. 2012. Insights into the phylogenetic and taxonomy of philasterid ciliates (Protozoa, Ciliophora, Scuticociliatia) based on analyses of multiple molecular markers. Mol. Phylogen. Evol. 64: 308–317 [DOI] [PubMed] [Google Scholar]
- 13. Yi Z, Song W. 2011. Evolution of the order Urostylida (Protozoa, Ciliophora): new hypotheses based on multi-gene information and identification of localized incongruence. PLoS One 6: e17471 doi:10.1371/journal.pone.0017471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McManus GB, Xu D, Costas BA, Katz LA. 2010. Genetic identities of cryptic species in the Strombidium stylifer/apolatum/oculatum cluster, including a description of Strombidium rassoulzadegani n. sp. J. Eukaryot. Microbiol. 57: 369–378 [DOI] [PubMed] [Google Scholar]
- 15. Liu W, Xu D, Lin X, Li J, Gong J, Al-Rasheid KA, Song W. 2009. Novistrombidium sinicum n. sp. and Novistrombidium orientale n. sp. (Protozoa: Ciliophora): two new oligotrich ciliates from a mangrove wetland, South China. J. Eukaryot. Microbiol. 56: 459–465 [DOI] [PubMed] [Google Scholar]
- 16. Liu W, Yi Z, Lin X, Al-Rasheid KA. 2011. Morphologic and molecular data suggest that Lynnella semiglobulosa n. g., n. sp. represents a new family within the subclass Choreotrichia (Ciliophora, Spirotrichea). J. Eukaryot. Microbiol. 58: 43–49 [DOI] [PubMed] [Google Scholar]
- 17. Lynn DH. 2008. The ciliated Protozoa. Characterization, Classification, and Guide to the Literature. Springer, New York, NY [Google Scholar]
- 18. Medlin L, Elwood HJ, Stickel S, Sogin ML. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71: 491–499 [DOI] [PubMed] [Google Scholar]
- 19. Liu W, Yi Z, Lin X, Warren A, Song W. 2012. Phylogeny of three choreotrich genera (Protozoa, Ciliophora, Spirotrichea), with morphological, morphogenetic and molecular investigations on three strobilidiid species. Zool. Scr. 41: 417–434 [Google Scholar]
- 20. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. (Oxf.) 41: 95–98 [Google Scholar]
- 21. Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27: 221–224 [DOI] [PubMed] [Google Scholar]
- 22. Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818 [DOI] [PubMed] [Google Scholar]
- 23. Nylander JA. 2004. MrModeltest version 2. Program Distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden [Google Scholar]
- 24. Guindon S, Gascuel O. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- 25. Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- 26. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De Rijk PD, De Wachter RD. 1997. RnaViz, a program for the visualisation of RNA secondary structure. Nucleic Acids Res. 25: 4679–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coleman AW. 2005. Paramecium aurelia revisited. J. Eukaryot. Microbiol. 52: 68–77 [DOI] [PubMed] [Google Scholar]
- 29. Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51: 492–508 [DOI] [PubMed] [Google Scholar]
- 30. Agatha S. 2011. Updated hypothesis on the evolution of oligotrichid ciliates (Ciliophora, Spirotricha, Oligotrichida) based on somatic ciliary patterns and ontogenetic data. Eur. J. Protistol. 47: 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Agatha S, Strüder-Kypke MC, Beran A, Lynn DH. 2005. Pelagostrobilidium neptuni (Montagnes and Taylor, 1994) and Strombidium biarmatum nov. spec. (Ciliophora, Oligotrichea): phylogenetic position inferred from morphology, ontogenesis, and gene sequence data. Eur. J. Protistol. 41: 65–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu D, Sun P, Shin MK, Kim YO. 2012. Species boundaries in tintinnid ciliates: a case study—morphometric variability, molecular characterization, and temporal distribution of Helicostomella species (Ciliophora, Tintinnina). J. Eukaryot. Microbiol. 59: 351–358 [DOI] [PubMed] [Google Scholar]
- 33. Maeda M, Carey PG. 1985. An illustrated guide to the species of the family Strombidiidae (Oligotrichida, Ciliophora), free-swimming protozoa common in the aquatic environment. Bull. Ocean Res. Inst. 19: 1–65 [Google Scholar]
- 34. Agatha S. 2003. Morphology and ontogenesis of Novistrombidium apsheronicum nov. comb. and Strombidium arenicola (Protozoa, Ciliophora): a comparative light microscopical and SEM study. Eur. J. Protistol. 39: 245–266 [Google Scholar]
- 35. Foissner W, Moon-van der Staay SY, van der Staay GWM, Hackstein JHP, Krautgartner Berger W-DH. 2004. Reconciling classical and molecular phylogenies in the stichotrichines (Ciliophora, Spirotrichea), including new sequences from some rare species. Eur. J. Protistol. 40: 265–281 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.