Abstract
Candida albicans is associated with humans, as both a harmless commensal organism and a pathogen. Adaption to human body temperature is extremely important for its growth and morphogenesis. Saccharomyces cerevisiae Esa1, a member of the MYST family HATs (histone acetyltransferases) and the catalytic subunit of the NuA4 complex, and its homologues in other eukaryotes have been shown to be essential for cell growth. To investigate the functional roles of two MYST family HATs, Esa1 and Sas2 in C. albicans, we deleted ESA1 and SAS2 in the C. albicans genome and performed cell growth analyses. Our results demonstrated that C. albicans Esa1 is not essential for general growth but is essential for filamentous growth. The esa1/esa1 mutant cells exhibited sensitivity to thermal, genotoxic, and oxidative stresses but tolerance to cold, osmotic, and cell wall stresses. In contrast, the sas2/sas2 mutant adapted to growth at higher temperatures and promoted filament formation at lower temperatures, resembling the phenotype of a C. albicans strain overexpressing ESA1. Cells with deletions of both ESA1 and SAS2 were inviable, reflecting the functional redundancy in cell growth. C. albicans Esa1 and Sas2 have distinct and synergistic effects on histone acetylation at H4K5, H4K12, and H4K16. Esa1 contributes mainly to acetylation of H4K5 and H4K12, whereas Sas2 contributes to acetylation of H4K16. Our findings suggest that C. albicans Esa1 and Sas2 play opposite roles in cell growth and morphogenesis and contribute coordinately to histone acetylation and gene regulation.
INTRODUCTION
The MYST proteins, which represent the largest family of HATs (histone acetyltransferases) (1) and are named for the founding members MOZ (2), Ybf2/Sas3 (3), Sas2 (4), and TIP60 (5), are conserved from yeasts to humans and are involved in diverse biological functions, including gene regulation, DNA repair, cell cycle regulation, and development (6). Esa1 (Essential SAS family acetyltransferase, also called KAT5), the human TIP60 orthologue in Saccharomyces cerevisiae, is essential for cell growth. Inactivation of the Esa1 HAT activity renders the yeast temperature sensitive (37°C) and leads to abnormal cell morphology (7, 8). Moreover, Esa1 is required for transcriptional silencing at the rDNA locus (9). As the catalytic subunit of the NuA4 HAT complex, Esa1 is involved in acetylation of four conserved lysines of the histone H4 N-terminal tail (lysines 5, 8, 12, and 16) (10). Furthermore, Esa1 is involved in acetylation of nonhistone substrates and contributes to the regulation of metabolism and stress response (11). Recently, Esa1 has been shown to function as a lysine acetyltransferase (KAT) required for autophagy via acetylation of the autophagy signaling component Atg3 (12).
Another MYST family member in yeast, Sas2 (Something About Silencing, also called KAT8), is the catalytic subunit of the SAS HAT complex (Sas2p-Sas4p-Sas5p) and can acetylate both free histones and nucleosomes and is involved in regulation of transcriptional silencing (13, 14). Sas2 is mainly required for acetylation of histone H4 at lysine 16 (H4K16) and plays an important role in maintaining the boundary of telomeric heterochromatin by opposing effects of Sir2 on deacetylation of H4K16Ac. Sas2 and Sir2 function in a concerted manner with transcriptional regulation in yeast, by acetylating and deacetylating H4K16 via a mechanism that may be common to all eukaryotes (15, 16). The human MYST family member MOF (Males Absent on the First), exists in at least two distinct complexes, MOF-MSL and MOF-MSL1v1, which have indistinguishable activities on H4K16 but differ dramatically in acetylating the nonhistone substrate p53 (17).
The MYST proteins have well-established roles on histone acetylation; however, their substrate spectrum is broader, and new nonhistone protein substrates are continuously being discovered (18). Other typical substrates of the MYST enzymes are components of the multisubunit MYST complexes, including the MYST proteins themselves (6). Discovering novel substrates of MYST proteins is pivotal for understanding the diverse functions of these essential acetyltransferase in nuclear processes, signaling, stress response, and metabolism.
Candida albicans is a common fungal pathogen. It colonizes skin and mucosal surfaces of the majority of healthy individuals in the human population but can cause various forms of candidiasis, ranging from superficial mucosal infections to life-threatening systemic diseases in immunocompromised patients (19, 20). The ability of C. albicans to undergo morphological transition to survive in different human niches contributes to its pathogenicity and adaptability. So far, the biological functions of the MYST proteins in C. albicans have not been studied, although the components of the NuA4 and SAS complexes have been predicted and sequenced. We previously reported that Yng2, a component of the Piccolo NuA4, plays an important role in morphogenesis of C. albicans (21). In this study, we report the characterization of two MYST proteins in C. albicans, namely, C. albicans Esa1 (CaEsa1), the catalytic subunit of the NuA4 complex, and C. albicans Sas2 (CaSas2), the catalytic subunit of the SAS complex. We demonstrate that CaEsa1 and CaSas2 have distinct and redundant effects on histone H4 acetylation and cell growth. In addition, we describe their functional roles in morphogenesis, stress response, and other cellular processes of C. albicans.
MATERIALS AND METHODS
Strains and culture conditions.
The C. albicans strains used in this study are listed in Table 1. Yeast strains were routinely grown on YPD medium (1% yeast extract, 2% peptone, and 2% glucose). Synthetic complete media with different carbon sources were used for the growth assay. YPD plus 10% bovine serum was used for hyphal induction. YPS medium (1% yeast extract, 2% peptone, and 2% sucrose) with 1% agar was used for the colony morphology assay under embedded conditions (22).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| SC5314 | Wild type | 24 |
| CAI4 | ura3::λ imm434/ura3::λ imm434 | 24 |
| BWP17 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 23 |
| CWX1 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG ESA1/esa1::HIS1 | This study |
| CWX2 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG esa1::HIS1/esa1::hisG-URA3-hisG | This study |
| CWX3 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG esa1::HIS1/esa1::hisG | This study |
| CWX4 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG SAS2/sas2::HIS1 | This study |
| CWX5 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG sas2::HIS1/sas2::hisG-URA3-hisG | This study |
| CWX6 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG sas2::HIS1/sas2::hisG | This study |
| CWX7 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG esa1::ARG4 Ptet-ESA1::URA3 | This study |
| CWX8 | ura3::λ imm434/ura3::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG sas2::HIS1/sas2::hisG-ura3-hisG esa1::ARG4 Ptet-ESA1::URA3 | This study |
| Plasmids | ||
| pCUB6 | Replacement of S. cerevisiae URA3 by C. albicans URA3 in pNKY50 | 24 |
| pESA1-KO | 0.53- and 0.6-kb KO fragments of C. albicans ESA1 and hisG-URA3-hisG in pBluescript SK(+) | This study |
| pSAS2-KO | 0.55- and 0.5-kb KO fragments of C. albicans SAS2 and hisG-URA3-hisG in pBluescript SK(+) | This study |
| pBES116 | C. albicans URA3 vector for integration at ADE2 locus | 53 |
| pBES116-ESA1 | 2.18-kb CaESA1 in pBES116 for ESA1 expression under control of ESA1 promoter | This study |
| pBA1 | C. albicans ADH1 promoter in BES116 | 25 |
| pBA1-WX1 (pBA1-ESA1) | 1.56-kb full-length CaESA1 in pBA1 for ESA1 expression under control of ADH1 promoter | This study |
| pBA1-WX2 | 1.2 kb of CaESA1 with deletion of chromo domain in pBA1 | This study |
| pBA1-WX3 | 0.9 kb of CaESA1 with deletion of HAT domain in pBA1 | This study |
| pBA1-WX4 | 1.3 kb of CaESA1 with deletion of insertion sequence in pBA1 | This study |
| pBA1-WX5 | 1.45-kb full-length ScESA1 in pBA1 | This study |
| pBA1-WX6 | ScESA1 G315E mutation in pBA1 | This study |
| pBA1-WX7 | ScESA1 L327S mutation in pBA1 | This study |
| pBA1-WX8 | 1.45-kb full-length human TIP60 cDNA in pBA1 | This study |
| pBA1-WX9 | Hybrid of CaEsa1 chromo domain and hTIP60 MYST domain | This study |
| pBA1-WX10 | Hybrid of hTIP60 chromo domain and CaEsa1 MYST domain | This study |
| p671 | C. albicans ACT1 promoter, 13×myc-FLAG1, HIS1 | 25 |
| p671-ARG4 | HIS1-to-ARG4 substitution in p671 | This study |
| pCaUME6-3 | Supplies ADH1p-cartTA-GAL4AD fragment | 54 |
| pCPC39 | Supplies tet-on promoter and URA3 marker | This study |
| pCPC42 | Supplies tet-off promoter and URA3 marker | This study |
| pCPC97 | Infusion of pUC18 and fragment containing CaESA1 (bp −521 to +1979) | This study |
| pCPC98 (pPtet-ESA1) | Infusion of pCPC97 and pCPC42 for doxycycline-controlled ESA1 expression | This study |
Plasmids and strain constructions.
SC5314 genomic DNA was used as the template for PCR amplification of all C. albicans genes. All constructs were verified by DNA sequencing. The plasmids used in this study are listed in Table 1. The primers used for PCR amplification are listed in Table S1 of the supplemental material.
For construction of the esa1/esa1 mutant, the first copy of ESA1 was deleted by PCR-based homologous recombination (23). C. albicans HIS1 was amplified from plasmids pGEM-HIS1 and substituted for ESA1 in C. albicans parent strain BWP17. The second copy of ESA1 was disrupted by the “URA-blaster” method (24). PstI-digested pESA1-KO was transformed into the ESA1/esa1 mutant to generate the esa1/esa1 mutant (CWX2) (see Fig. S1A in the supplemental material). The same strategy was used for construction of the sas2/sas2 mutant. The first copy of SAS2 was deleted by the PCR-based method, and the second copy of SAS2 was disrupted by the URA-blaster method. A PstI-linearized pSAS2 knockout (pSAS2-KO) was introduced into the SAS2/sas2 mutant to produce the sas2/sas2 mutant (CWX5) (see Fig. S1B). All constructs were verified by PCR analysis (see Fig. S1C and D). To generate the sas2 esa1 double mutant, the first copy of ESA1 in the sas2/sas2 mutant was deleted by replacing ESA1 with ARG4 by using a PCR-based knockout strategy, and the second copy of ESA1 was replaced by a PCR fragment amplified from pCPC98 (pPtet-ESA1), in which ESA1 expression is under doxycycline control (CWX8).
pESA1-KO for knockout of ESA1, pSAS2-KO for knockout of SAS2, and pCPC98 (pPtet-ESA1) for doxycycline-controlled C. albicans ESA1 expression are described in the supplemental material. pBA1-WX1 (pBA1-ESA1) to pBA1-WX10 were constructed for the ectopic expression of genes under the control of the ADH1 promoter (see the supplemental material).
Western blot analysis.
Protein extraction and immunoblotting were performed as described previously (25). C. albicans cells were grown in YPD and harvested and resuspended in 0.5 ml of lysis buffer (10 mM Tris-HCl [pH 8.0], 250 mM NaCl, 0.1% NP-40, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 2 mM benzamidine, 0.5 g/ml leupeptin, 1.4 g/ml pepstatin, 2.4 μg/ml chymostatin, and 17 μg/ml aprotinin). Proteins were separated by 7.5% SDS-PAGE, transferred to a polyvinylidenedifluoride membrane and then subjected to Western blotting. Histone H4 acetylation levels were analyzed by using peroxidase-conjugated antibodies that specifically recognized unacetylated H4 (Sigma), acetylated H4 (H4Ac), acetylated H4 at lysine 5 (H4K5Ac), lysine 12 (H4K12Ac), and lysine 16 (H4K16Ac) (Abcam).
Quantitative reverse transcription-PCR (RT-PCR) analysis.
RNA extraction was performed as described by Lane et al. (26). Total RNA was reverse transcribed to cDNA with the ReverTra Ace qPCR RT kit from Toyobo. Quantitative PCR was performed on a StepOne qPCR instrument (Applied Biosystems) with a SYBR green master mix containing Fast Start Taq DNA polymerase (Roche) and SYBR green nucleic acid stain (Roche). Transcription levels of genes in different samples were normalized against the levels of ACT1. The data were measured in three independent experiments.
Stress response assay.
The susceptibilities of the mutants to hydroxyurea (HU), methyl methanesulfonate (MMS), camptothecin (CPT), and H2O2 were tested by using cells grown in YPD medium at room temperature (22°C) to exponential phase. Cells were serially diluted 10-fold, and 5 μl was spotted onto a YPD plate with or without 10 mM hydroxyurea, 0.01% methyl methanesulfonate, 15 μg/ml camptothecin, or 2 mM hydrogen peroxide and then incubated at room temperature for 3 days. For analysis of resistance of the mutants to NaCl (5 M), Congo red (100 μg/ml), caspofungin (0.32 μg/ml), nonfermentable carbon sources (2% glucose, 2% maltose, 2% ethanol, or 2% glycerol), and thermal stress (42°C or 45°C), cells grown in YPD medium at room temperature to exponential phase (optical density at 600 nm [OD600], 0.6 to 0.8) were serially diluted in YPD, YP, or synthetic complete medium (SC) plates containing the appropriate supplements and examined after incubation for 56 h at room temperature or the temperatures specified.
Growth rate analysis.
For analysis of the cell growth rate above 10°C, the doubling time was determined by measuring OD600 values of growing cells. Overnight cultures were reinoculated into fresh liquid YPD and grown at the specified temperature, and OD600 values were measured during the log phase at various time points and used for calculation of generation times. The data were measured in three independent experiments. The growth curves, with doubling times in minutes, were drawn using the Origin 6.1 software (MicroCal) or sigmaplot10 for data statistical analysis. For analysis of the cell growth rate at 4°C, the doubling time was determined by a method used for a yeast replicative life span assay (27). Strains were patched onto fresh solid medium and grown for 2 days at 25°C. Cells were arrayed onto YPD plates by using a micromanipulator, allowed to grow for 3 h at 25°C, and then incubated at 4°C. Virgin daughter cells were isolated as buds from mother cells and subjected to life span analysis. The doubling time was calculated between generations. Each experiment consisted of more than 10 mother cells and was independently repeated at least twice.
RESULTS
Candida albicans Esa1 plays a role in cell growth in sensing temperature.
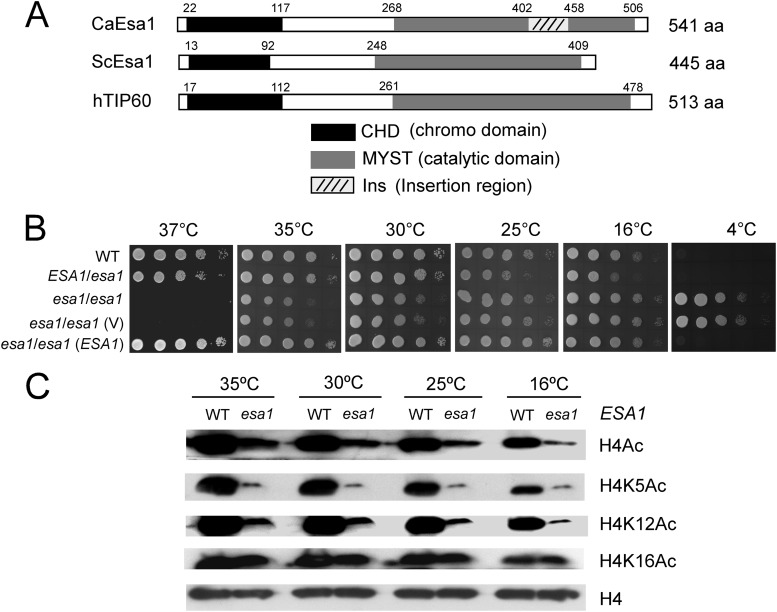
To identify the MYST family members of C. albicans, we searched the C. albicans genome database (http://www.candidagenome.org/) and found a protein designated Esa1 (orf19.5416), which shares the highest similarity with S. cerevisiae Esa1 (ScEsa1), and a protein designated Sas2 (orf19.2087), which shares the highest similarity with S. cerevisiae Sas2 (ScSas2). Compared with ScEsa1, C. albicans Esa1 (CaEsa1) contains a MYST domain at the C terminus with 84% identity and a chromo domain (CHD) at the N terminus with 61% identity. However, there is an insertion sequence between residues 402 to 458 in the MYST domain of CaEsa1 (Fig. 1A).
Fig 1.
Candida albicans open reading frame 19.5416 encodes a conserved MYST protein, Esa1, that plays a role in cell growth. (A) Schematic depiction of the MYST domains and chromo domains in C. albicans Esa1, S. cerevisiae Esa1, and human TIP60. (B) Growth of esa1/esa1 mutant strains on YPD plates at different temperatures. Strains are wild type (WT; BWP17/pBES116), ESA1/esa1 (CWX1/pBES116), esa1/esa1 (CWX2), esa1/esa1 (V) (CWX3/pBES116), and esa1/esa1(ESA1) (CWX3/pBES116-ESA1). The cells were spotted onto YPD plates and incubated at 37°C and 35°C for 2 days, at 30°C and 25°C for 3 days, or at 16°C for 5 days and at 4°C for 2 weeks. (C) Western analyses of histone H4 acetylation levels in the wild type and the esa1/esa1 mutant. Cells were grown in liquid YPD at 35°C, 30°C, 25°C, or 16°C and harvested for Western analysis with antibodies that specifically recognized acetylated H4 (H4Ac), H4 acetylated at lysine residues 5 (H4K5Ac), 12 (H4K12Ac), 16 (H4K16Ac) (all from Abcam), and histone H4 (Sigma).
Esa1 and its homologues have been shown to be essential in all eukaryotes examined, including S. cerevisiae (7, 8), Aspergillus nidulans (28), Schizosaccharomyces pombe (29), Drosophila melanogaster (30), and Mus musculus (31). To examine whether C. albicans ESA1 is also essential for cell growth, we knocked out the ESA1 gene in C. albicans by using a combined PCR-based and “URA-blaster” method. Surprisingly, the mutant cells with deletion of both copies of ESA1 were still viable (see Fig. S1A and C in the supplemental material), indicating that CaEsa1 is not essential for general growth. Furthermore, we found that the esa1/esa1 mutant strains could grow on YPD plates in the cold room (4°C) with a doubling time of 486 min, in contrast to the wild-type cells, which entered into the quiescent stage (phase) and stopped growing in the cold room (4°C). On the other hand, the esa1/esa1 mutant cells stopped growing at the high temperature (37°C) (Fig. 1B). These unusual phenomena could be attributed to the ESA1 deletion as reintroducing ESA1 back into its own locus under the control of the ESA1 endogenous promoter and rescuing the wild-type phenotype. The ESA1/esa1 heterozygote showed a similar phenotype to the revertant (Fig. 1B). These results indicated that deletion of C. albicans ESA1 caused reduced cell growth at high temperatures (>25°C) but increased growth at low temperatures (<25°C), suggesting that CaEsa1 is required for cell survival at high temperatures (37°C) and for cell quiescence at cold temperatures (4°C).
S. cerevisiae Esa1 is a HAT required for H4 and H2A acetylation in vivo (7, 10, 32, 33). To test if CaEsa1 exhibited HAT activity, we performed Western blot analysis that detected acetylation levels of histone H4 in cells lacking ESA1. At all temperatures examined, the esa1/esa1 mutant cells exhibited a significantly decreased overall level of H4 acetylation. In particular, the acetylation levels of H4K5 and H4K12 were dramatically reduced, but that of H4K16 was not (Fig. 1C), suggesting that CaEsa1 is required for H4 acetylation at H4K5 and H4K12 independent of the growth temperature.
C. albicans Esa1 is a functional homologue of ScEsa1 and hTIP60.
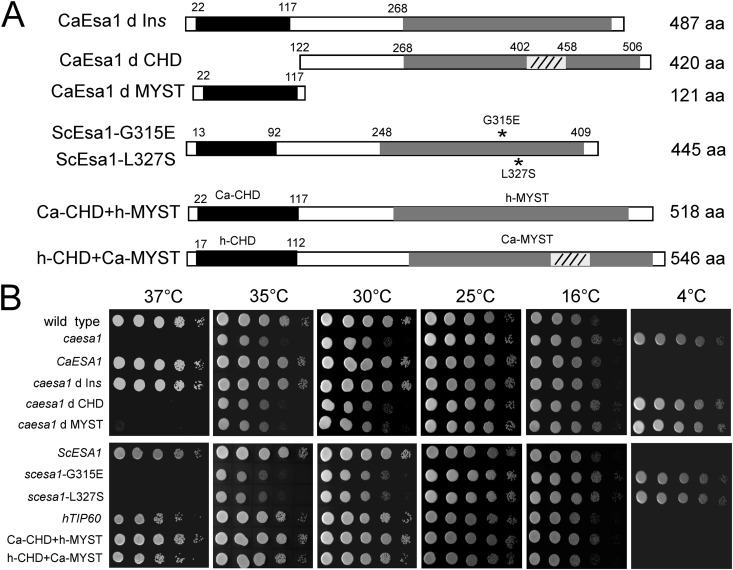
The MYST domain of CaEsa1 shares a high sequence identity with that of ScEsa1 (84% identity) and human TIP60 (hTIP60; 79% identity), and its N-terminal chromo domain shares relatively low sequence identity with that of ScEsa1 (61% identity) and hTIP60 (52% identity). In addition, CaEsa1 contains an insertion sequence (residues 402 to 458) in the MYST domain. To determine whether CaEsa1 is a functional homologue (orthologue) of ScEsa1 or hTIP60, we examined the abilities of ScEsa1 and hTIP60 to complement the growth defect in the Caesa1 mutant cells. We constructed a series of ectopic expression vectors for functional complementary assays (Fig. 2A; Table 1). ScESA1 and hTIP60 were introduced into C. albicans and expressed under the control of the ADH1 promoter. At 37°C, the ectopically expressed ScESA1 and hTIP60 suppressed the lethal phenotype of the Caesa1 mutant (Fig. 2B, left panel); at 4°C, they suppressed the cold-resistant phenotype of the Caesa1 mutant (Fig. 2B, right panel). These results together showed that both ScESA1 and hTIP60 can rescue the growth defect of the Caesa1 mutant cells at high temperature and suppress the abnormal growth at low temperature (Fig. 2B), indicating that CaEsa1 is a functional homologue of ScEsa1 and hTIP60.
Fig 2.
Expression of both S. cerevisiae ESA1 and human TIP60 can complement the growth defect of the esa1/esa1 mutant in C. albicans. (A) Strategy for ectopic expression. CaEsa1, wild-type C. albicans Esa1; CaEsa1 d Ins, deletion of the insertion region in CaEsa1; CaEsa1 d CHD, deletion of the chromo domain in CaEsa1; CaEsa1 d MYST, deletion of the MYST domain in CaEsa1; ScESA1, wild-type S. cerevisiae Esa1; ScEsa1-G315E, G315E substitution in S. cerevisiae Esa1; ScEsa1-L327S, L327S substitution in S. cerevisiae Esa1; hTIP60, wild-type human TIP60; Ca-CHD+h-MYST, hybrid of C. albicans Esa1 chromo domain and human TIP60 MYST domain; h-CHD+Ca-MYST, hybrid of human TIP60 chromo domain and C. albicans Esa1 MYST domain. (B) Growth of strains on YPD plates at different temperatures. The exponentially growing cells were spotted onto YPD plates and incubated at the temperatures indicated. Ectopic expression vectors pBA-WX1 to pBA-WX10 were introduced into C. albicans esa1/esa1 mutant (CWX3) cells for the complementation assay.
We further analyzed the contributions of the chromo domain, the MYST domain, and the insertion sequence of CaEsa1 to cell growth. As shown in Fig. 2B, deletion of the CHD or MYST domain failed to rescue the phenotype of the Caesa1 mutant, but deletion of the insertion sequence had no effect on cell growth, suggesting that both the CHD and MYST domain are required for CaEsa1 function. It has been shown that two S. cerevisiae variants containing mutant ScEsa1 (G315E and L327S) lacking HAT activity are temperature sensitive (37°C) (7, 8). As expected, our complementation assay results showed that neither the Scesa1 G315E mutant nor the Scesa1 L327S mutant could rescue the growth defect of the Caesa1 mutant (Fig. 2B, lower panels).
Unlike the MYST domain, the CHDs of ScEsa1, CaEsa1, and TIP60 are less conserved. The CHDs have been considered to mediate chromatin interactions in a variety of different protein contexts. The CHDs can also mediate interactions with histones, DNA, and RNA (34). Interaction of the TIP60 CHD with the trimethylated H3K9 (H3K9me3) links DNA damage detection to activation of Tip60 (35). Considering the low sequence similarity between the CHDs, we constructed two hybrid proteins to exchange the CHD and MYST domain of CaEsa1 with those of TIP60, and we tested their effects on cell growth. Both hybrid proteins suppressed the growth deficiency of the Caesa1 mutant (Fig. 2B, lower panels), albeit not to the same extent. The TIP60-MYST domain fully complemented the CaEsa1 MYST domain, whereas TIP60-CHD partially complemented the CaEsa1 CHD at 37°C, reflecting a functional diversity of the chromo domains which may add an additional layer to their functional regulation.
C. albicans Esa1 is essential for filamentous growth.
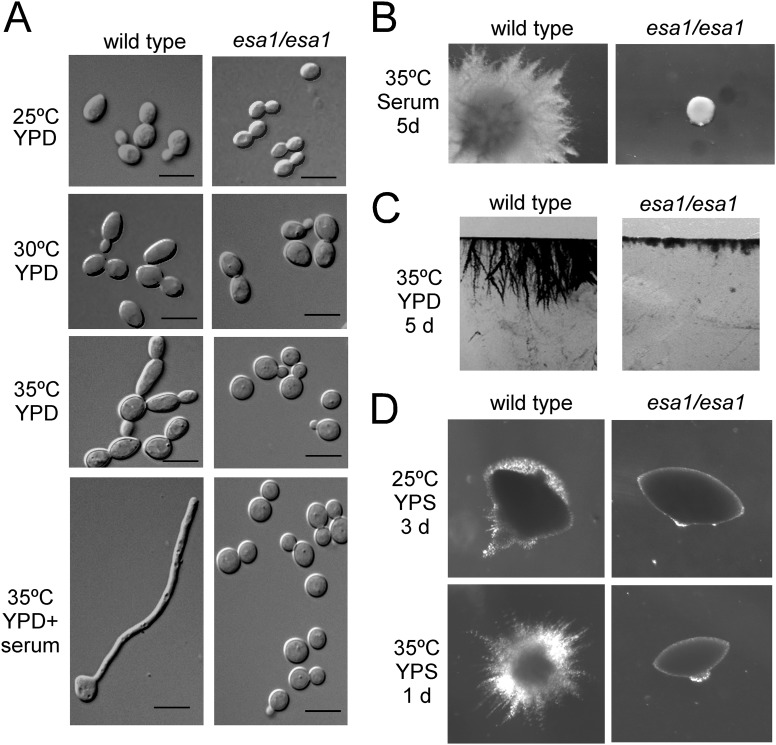
Esa1 is the catalytic subunit of the NuA4 HAT complex, which acetylates histones H4 and H2A. In S. cerevisiae, among the three subunits of the NuA4 core module (Piccolo NuA4), Yng2 is the only one not essential for cell growth. Our previous study showed that, like the yeast homologue, C. albicans Yng2 is not essential for cell growth but is involved in hyphal regulation via NuA4 HAT activity (21). Thus, we predicted that the catalytic subunit Esa1 plays a critical role in hyphal development. As expected, deletion of C. albicans ESA1 blocked filament formation under all hyphal induction conditions examined (Fig. 3). Since the esa1 mutant could not grow at 37°C, we lowered the temperature to 35°C. At 35°C, wild-type cells formed pseudohyphae in liquid YPD medium without serum and formed true hyphae in the presence of serum, whereas the esa1/esa1 mutant cells displayed the yeast form (Fig. 3A). In solid serum-containing medium, the wild-type strain produced florid filamentous colonies, while the esa1/esa1 mutant formed small smooth yeast colonies (Fig. 3B). On solid surfaces of YPD plates, the wild-type strain invaded into the agar and formed filaments below the surface, whereas the esa1/esa1 mutant failed in invasion and was defective in invasive growth (Fig. 3C). To analyze the role of Esa1 in filamentous growth under embedded conditions, we examined the phenotype of the esa1/esa1 mutant at high and low temperatures under microaerophilic conditions. After growth in YPS agar at 35°C for 1 day, the wild-type colonies generated long heterogeneous filaments, but the esa1/esa1 mutant produced smooth colonies (Fig. 3D). At 25°C, the wild-type colonies produced limited amounts of filaments after 3 days incubation, while the esa1/esa1 mutant formed smooth colonies (Fig. 3D). These observations indicate that Esa1 is essential for hyphal development and filamentous growth of C. albicans.
Fig 3.
C. albicans Esa1 is essential for hyphal development. (A) Cell morphology of wild-type (BWP17/pBES116) and esa1/esa1 mutant (CWX3/pBES116) cells under liquid conditions. The cells were cultured in liquid YPD medium at 25°C, 30°C, or 35°C for 6 h for yeast growth and in YPD plus 10% serum at 35°C for 3.5 h for hyphal growth. Bars, 5 μm. (B) Colony morphologies of the strains. The cells were plated on solid serum-containing medium and incubated at 35°C for 5 days. (C) Invasive growth on agar. The strains were patched on YPD plates and grown at 35°C for 5 days. After washing with water, the agar containing cells was cut into slices for photographing. (D) Filamentous growth under embedded conditions. The cells were plated with molten YPS agar and grown at 25°C for 3 days or 35°C for 1 day.
C. albicans Esa1 is involved in regulation of gene expression.
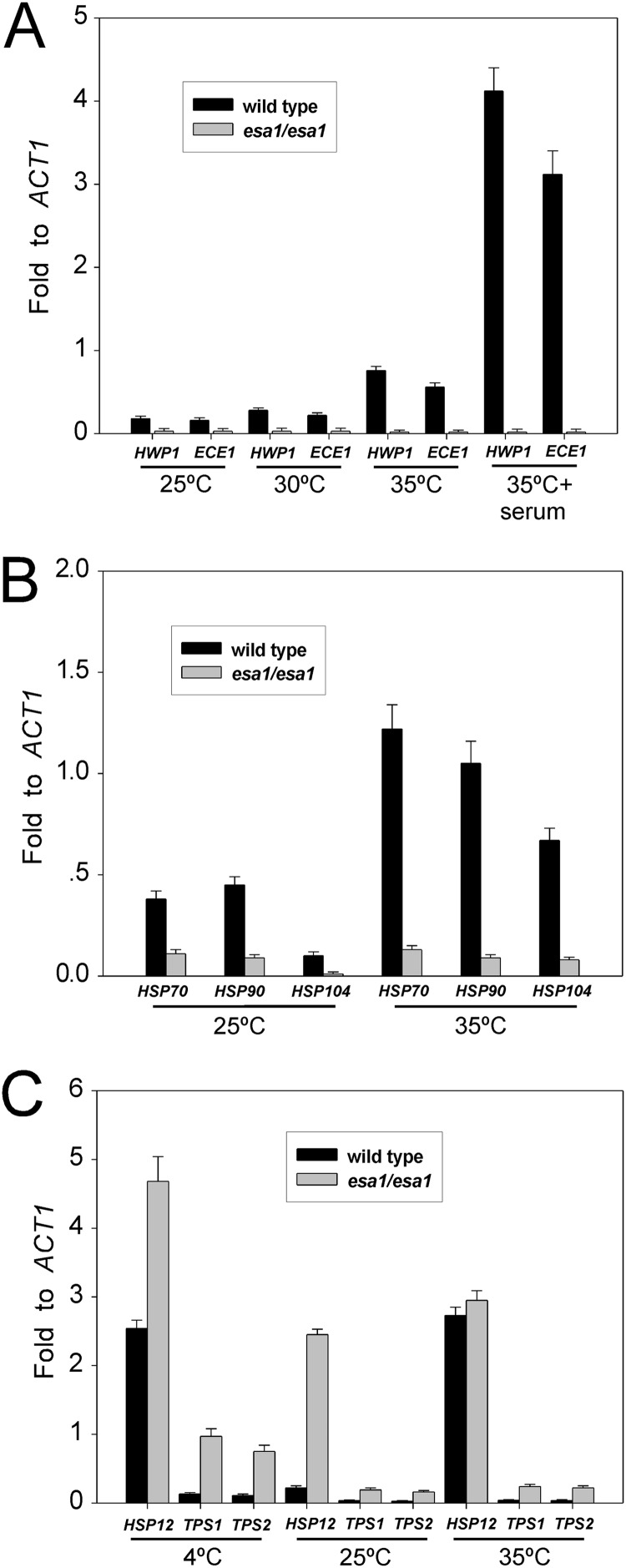
C. albicans Esa1, as a HAT, was predicted to have effects on gene regulation. HWP1 and ECE1 are the first few hypha-specific genes identified (36, 37). Consistent with the defect in hyphal morphogenesis, the esa1/esa1 mutant was also defective in the expression of hypha-specific genes. HWP1 and ECE1 were highly induced in the wild-type cells under hypha-inducing conditions (YPD plus 10% serum; 35°C), but they were undetectable under yeast growth conditions (YPD; 25°C) (Fig. 4A). The expression levels of HWP1 and ECE1 in the esa1/esa1 mutant were undetectable under hypha-inducing conditions (Fig. 4A). These results indicated that Esa1 is essential for hyphal development and the induction of hypha-specific genes.
Fig 4.
Effects of C. albicans Esa1 on gene regulation. (A) Quantitative RT-PCR analyses showed defective induction of hypha-specific genes HWP1 and ECE1 in esa1/esa1 mutants. Cells were grown in YPD plus 10% serum at 35°C for 3.5 h or grown in YPD at 25°C, 30°C, or 35°C for 6 h. (B) Expression levels of heat shock genes HSP90, HSP70, and HSP104 were inhibited in esa1/esa1 mutants. Cells were grown in YPD at 25°C and then heat shocked at 35°C for 0.5 hour. (C) Expression levels of stress response genes HSP12, TPS1, and TPS2 were derepressed in esa1/esa1 mutants. Cells were cultured in YPD at 25°C and then upshifted to 35°C or downshifted to 4°C for 0.5 hour, and total RNAs were extracted for qRT-PCR analyses.
To characterize the molecular basis of the Esa1 effects on C. albicans growth at different temperatures, we examined the expression levels of several heat shock response genes, including HSP90, HSP70 and HSP104 in the esa1/esa1 mutant cells. Heat shock proteins play key roles in protein biogenesis and have different functions in cell growth, morphogenesis, and pathogenesis of C. albicans (38–40). All three heat shock genes, HSP90, HSP70, and HSP104, were induced in response to heat shock stress (25 to 35°C) in the wild-type C. albicans cells but failed to be induced in the esa1/esa1 mutant cells (Fig. 4B). Requirement of Esa1 for induction of the heat shock genes may correlate its essential role in cell growth at a high temperature (37°C).
We further examined the expression levels of several stress response genes, including HSP12, TPS1, and TPS2 in the esa1/esa1 mutant cells. Hsp12, a small heat shock protein conserved among fungal species (41, 42), is induced during stationary-phase growth and under stress conditions, including heat shock, osmotic, and oxidative stress in C. albicans (42). In the wild-type cells, HSP12 was greatly induced in response to both heat shock stress (25°C to 35°C) and cold shock stress (25°C to 4°C) (Fig. 4C). Interestingly, the expression of HSP12 was 10-fold derepressed at 25°C in log-phase esa1/esa1 mutant cells. The derepression was enhanced by cold shock, but not by heat shock (Fig. 4C). Trehalose, an important stress protectant, is synthesized in a two-step process in fungi by trehalose-6-phosphate (Tre6P) synthase and Tre6P phosphatase, which are encoded by TPS1 and TPS2, respectively (43). C. albicans Tps1 and Tps2 are proteins that are enriched during and play roles in osmotic and thermal stresses (44–46). Under exponential growth conditions at 25°C, the expression levels of both TPS1 and TPS2 were derepressed in the esa1/esa1 mutant cells compared to the wild-type cells (Fig. 4C). Like HSP12, the TPS derepression was also enhanced by cold shock, but not by heat shock (Fig. 4C). Together, our data indicate that Esa1 has negative effects on transcription of HSP12, TPS1, and TPS2. Derepression of these genes under cold stress may largely account for the inhibitory role of Esa1 on cell growth at cold temperatures.
C. albicans Esa1 is required for stress responses.
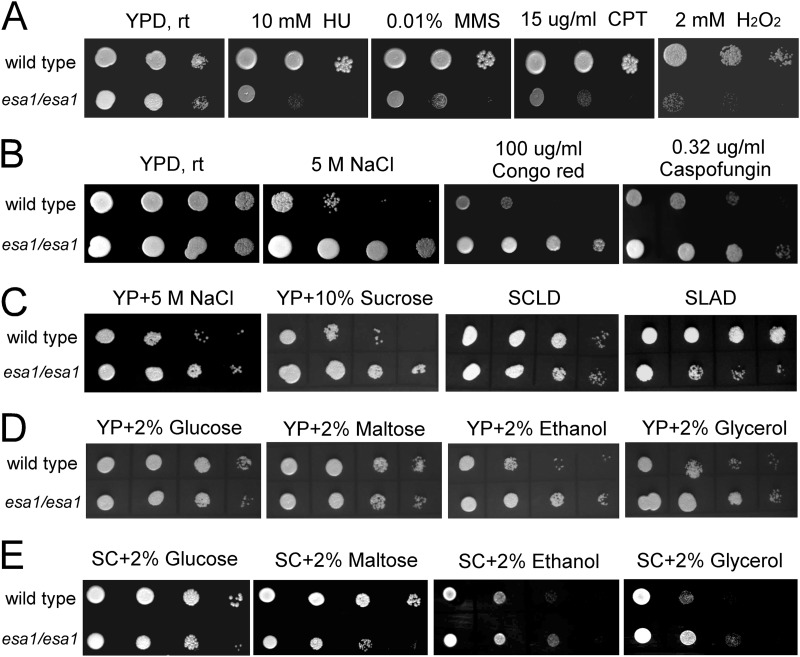
Adaption to the host environment is important for survival of C. albicans (47, 48). The esa1/esa1 mutant cells displayed sensitivity to DNA damage induced by HU, MMS, and CPT (Fig. 5A). This sensitivity was not due to the growth defect of the mutant, as we used the growth conditions were YPD medium and room temperature (22°C), under which both wild-type and esa1/esa1 mutant cells have similar growth rates. The viability of the esa1/esa1 mutant cells was at least 5-fold lower than that of the wild-type cells when spotted on the plates containing HU, MMS, or CPT (data not shown), suggesting that CaEsa1 is required for DNA damage repair in C. albicans. The esa1/esa1 mutant cells were also sensitive to peroxide (Fig. 5A), suggesting that Esa1 plays a critical role in protecting C. albicans from H2O2-induced cell death.
Fig 5.
C. albicans Esa1 is involved in multiple stress responses. (A) esa1/esa1 mutants are sensitive to genotoxic and oxidative stresses. Wild-type (BWP17/pBES116) and esa1/esa1 mutant (CWX3/pBES116) cells were serially diluted 10-fold, spotted onto YPD plates containing DNA-damaging agents (10 mM HU, 0.01% MMS, and 15 μg/ml CPT) or 2 mM H2O2, and incubated at room temperature (rt; 22°C) for 3 days. (B and C) esa1/esa1 mutants were resistant to osmotic and cell wall stresses. YPD plates containing 5 M NaCl, 100 μg/ml Congo red, or 0.32 μg/ml caspofungin (B), or YP with 5 M NaCl or 10% sucrose (C) were used for growth assays. SC plates containing 0.1% glucose (SCLD) and low ammonia (SLAD) were used for a nutrient-sensing assay. (D and E) YP plates (D) or SC plates (E) containing 2% glucose, maltose, ethanol, or glycerol were used for a carbon source-sensing assay.
In addition to temperature, genotoxic, and oxidative stresses, we extended our study to the impact of Esa1 on other stress responses. On solid medium containing 5 M NaCl (YPD plus NaCl or YP plus NaCl) or high concentration of sucrose (YP plus 10% sucrose), the esa1/esa1 mutant cells grew much faster than the wild-type cells at room temperature (22°C) (Fig. 5B and C), suggesting that inactivation of Esa1 increases the ability of C. albicans to resist osmotic stress. The esa1/esa1 mutant cells also exhibited cell wall stress-resistant phenotypes and grew faster in the presence of Congo red and the antifungal drug caspofungin, both of which interfere with glucan biosynthesis (Fig. 5B). Consistent with their resistance to cold stress, the esa1/esa1 mutant cells displayed osmotic and cell wall stress resistance.
We further sought to study the function of Esa1 in metabolic regulation. Compared to the wild-type cells, the esa1/esa1 mutant cells grew slowly in media containing a fermentable sugar, including glucose and maltose, but grew faster in media containing a nonfermentable carbon source, such as ethanol and glycerol (Fig. 5D). A similar phenomenon was observed in synthetic growth medium (SC plates with fermentable and nonfermentable carbon sources), excluding the effect of nutrients (Fig. 5E). The faster growth phenotype of the esa1/esa1 mutant in nonfermentative media was observed not only at room temperature (22°C) but also at 30°C and 16°C, but it was hardly observed at 35°C, due to a reduced growth rate of the esa1/esa1 mutant. Lack of Esa1 enhanced the ability of C. albicans to utilize nonfermentable carbon sources. In some synthetic growth media, including SCLD (SC plus 0.1% glucose) and SLAD (SD plus low ammonia), the esa1/esa1 mutant cells grew poorly, suggesting a defect in nutrient uptake (Fig. 5C). Taken together, these data indicate that Esa1 mediates multiple cellular processes and plays important roles in temperature adaptation, DNA damage repair, the oxidative stress response, osmotic stress tolerance, cell wall assembly, and metabolism regulation.
C. albicans Sas2 plays a role opposite to Esa1 in cell growth and morphogenesis.
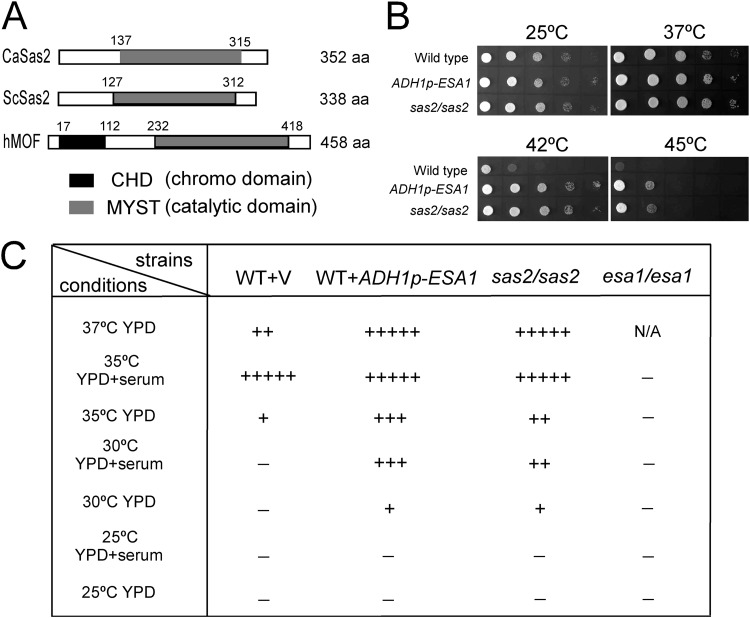
We suspected that the nonlethal phenotype of the esa1/esa1 mutant might be due to the existence of a functionally redundant MYST HAT in C. albicans. Thus, we searched the C. albicans genome database and found another putative MYST protein (orf19.2087) that was homologous to S. cerevisiae Sas2. Unlike most of the other MYST family proteins, such as human MOF, ScSas2 and CaSas2 contain only a MYST domain at the C terminus, with 83% sequence identity, but lack a chromo domain at the N terminus (Fig. 6A). To test the role of C. albicans SAS2 in cell growth, we deleted both copies of SAS2 in C. albicans. Deletion of C. albicans SAS2 did not cause a lethal phenotype, but the cells exhibited resistance to high temperature. The sas2/sas2 mutant cells grew much faster than the wild-type cells at high temperature, including 37°C, and especially at 42°C and 45°C (Fig. 6B). The ESA1-overexpressing strain displayed a phenotype similar to the sas2/sas2 mutant and grew faster at high temperature (Fig. 6B), in contrast to the sensitive phenotype of the esa1/esa1 mutant. These results indicate that CaSas2 and CaEsa1 play opposite roles in cell growth at high temperature.
Fig 6.
Candida albicans open reading frame 19.2087 encodes MYST protein Sas2, which plays a role opposite to that of Esa1 in cell growth and morphogenesis. (A) Schematic depiction of the MYST domain and chromo domain in C. albicans Sas2, S. cerevisiae Sas2, and human MOF. (B) sas2/sas2 mutant cells were tolerant to high temperature. The cells were spotted onto YPD plates and incubated at 25°C, 37°C, 42°C, or 45°C for 2 days. (C) Sas2 and Esa1 have opposite effects on filament formation. Wild-type (BWP17/pBA1), wild-type with ADH1p-ESA1 (BWP17/pBA1-ESA1), esa1/esa1 mutant (CWX3/pBA1), and sas2/sas2 mutant (CWX6/pBA1) cells were grown in liquid YPD medium with or without 10% serum at different temperatures for 3 h. −, yeast growth; +, filamentous growth. The level of cell elongation is indicated by the number of + symbols. N/A, not available (cells failed to grow).
To confirm the data, we analyzed the doubling time of the strains (see Fig. S2 in the supplemental material). Above room temperature (22°C), the esa1/esa1 mutant cells grew much slower than the wild-type cells; below room temperature, the esa1/esa1 mutant cells grew faster than the wild-type cells (see Fig. S2A). The sas2/sas2 mutant cells showed a slightly higher growth rate above 30°C but a slightly lower rate below 16°C. As expected, overexpression of ESA1 reversed the phenotype of the esa1/esa1 mutant and the phenotype resembled that of the sas2/sas2 mutant (see Fig. S2B).
CaSas2 has an inhibitory effect on hyphal growth. At high temperature (37°C), the wild-type cells could not sustain hyphal growth in YPD media without serum, whereas the sas2/sas2 mutant cells formed true hyphae (Fig. 6C). At an intermediate temperature (30°C), the wild-type cells could not produce hyphae in YPD media with or without serum, but the sas2/sas2 cells promoted filament formation even in the absence of serum (Fig. 6C). At a lower temperature (25°C), the sas2/sas2 cells exhibited a phenotype similar to wild-type cells and grew in yeast form (Fig. 6C). Overexpression of ESA1 resulted in a similar phenotype as the sas2/sas2 mutant in hyphal production, opposite to the phenotype of the esa1/esa1 mutant (Fig. 6C). Consistent with their opposite effects on cell growth, CaEsa1 and CaSas2 also have opposite effects on hyphal development.
Esa1 and Sas2 link histone H4 acetylation to cell growth.
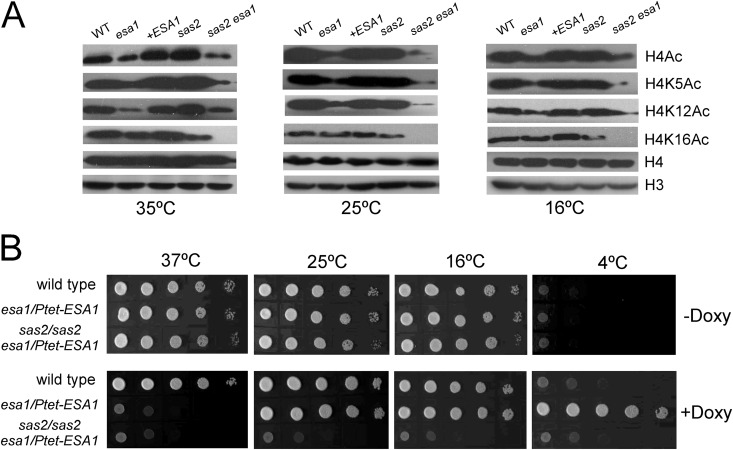
Deletion of C. albicans ESA1 caused dramatic reductions in acetylation at H4K5 and H4K12, but not at H4K16 (Fig. 1C and 7A), probably due to the existence of a redundant HAT for H4K16, similar to S. cerevisiae (15, 16). In S. cerevisiae, Sas2 is mainly required for H4K16 acetylation, and Sas2 links H4K16 acetylation to inhibition of the spread of subtelomeric heterochromatin through opposing Sir proteins binding to euchromatin (15, 16). In mammals, MOF also plays important roles in transcription activation by acetylating H4K16 (17). Thus, we predicted that CaSas2 also contributes to H4K16 acetylation. Indeed, deletion of C. albicans SAS2 reduced the acetylation level of H4K16 but had subtle effects on acetylation of H4K5 and H4K12 (Fig. 7A). Interestingly, at a higher temperature (35°C), CaSas2 seemed to have inhibitory effects on acetylation of H4K5 and H4K12. Deletion of SAS2 slightly increased the acetylation levels of H4K5 and H4K12 at higher temperature (35°C); however, the increase was not obvious at lower temperature (16°C) (Fig. 7A).
Fig 7.
C. albicans Esa1 and Sas2 have synergistic effects on histone acetylation and cell growth. (A) Western analyses of histone H4 acetylation levels in the wild type and mutants. (B) Double deletion of ESA1 and SAS2 showed synthetic lethality. The cells were precultured in YPD medium containing 50 μg/ml doxycycline for 24 h and then spotted onto YPD plates in the absence or presence of doxycycline and incubated at the indicated temperatures. Strains are wild type (BWP17/pBA1), wild type with ADH1p-ESA1 (BWP17/pBA1-ESA1), esa1/esa1 (CWX3/pBA1), sas2/sas2 (CWX6/pBA1), esa1/Ptet-ESA1 (CWX7), and sas2/sas2 esa1/Ptet-ESA1 (CWX8). All Ptet-ESA1 cells were treated with doxycycline to knock down the ESA1.
To determine whether the two MYST proteins have synergistic effects on H4 acetylation, we tried to delete both the ESA1 and SAS2 genes in C. albicans, but we failed to obtain the esa1/esa1 sas2/sas2 double mutant. Hence, we constructed a conditional knockdown esa1 sas2 double mutant by downregulating the expression of ESA1 in a tet-off system. At 25°C, the ESA1 mRNA levels in both esa1/Ptet-ESA1 and sas2/sas2 esa1/Ptet-ESA1 mutant cells decreased greatly after addition of doxycycline (see Fig. S3A in the supplemental material), indicating that the ESA1 expression can be specifically repressed by doxycycline. After treatment with doxycycline at 25°C, the acetylation level of H4 in the sas2/sas2 esa1/Ptet-ESA1 mutant was decreased. At 6 h, the acetylation levels of H4K5, H4K12, and H4K16 were all significantly decreased (see Fig. S3B). When we extending the treatment time to 24 h, ESA1 expression under tet promoter control was almost completely repressed, and the mutant cells stopped budding and displayed growth arrest. At this time point, H4K16 acetylation was completely abolished, and H4K5 and H4K12 acetylation was also decreased to very low levels at the temperatures examined (Fig. 7A). All of these results together indicate that the two MYST HATs Esa1 and Sas2 have redundant and synergistic effects on H4 acetylation.
To link H4 acetylation to cell growth, we tested the growth of the wild-type, esa1/Ptet-ESA1, and sas2/sas2 esa1/Ptet-ESA1 cells on solid medium containing doxycycline. The ESA1-downregulated cells were sensitive to high temperature (37°C) and resistant to cold temperature (4°C) but could grow at intermediate temperatures (Fig. 7B), i.e., showed a phenotype resembling the phenotype of the esa1/esa1 mutant cells. However, downregulating ESA1 in the sas2/sas2 esa1/Ptet-ESA1 mutant cells blocked cell growth at all temperatures (Fig. 7B). Inactivating both Esa1 and Sas2 caused a synthetic lethal phenotype, suggesting that Esa1 and Sas2 have synergistic effects on cell growth. Based on these results, we conclude that the two MYST HATs Esa1 and Sas2 play distinct and redundant roles in cell growth and morphogenesis of C. albicans.
DISCUSSION
Esa1 and Sas2 have distinct and redundant effects on H4 acetylation and cell growth.
In this study, we identified two MYST proteins, Esa1 and Sas2, in the human pathogen C. albicans. C. albicans Esa1 is a functional homologue (orthologue) of S. cerevisiae Esa1 and human TIP60 and is the catalytic subunit of the NuA4 HAT complex (49). C. albicans Sas2 is the orthologue of S. cerevisiae Sas2 and is the catalytic subunit of the SAS HAT complex (15, 16). Esa1 and Sas2 have opposite effects on cell growth. Loss of Esa1 changes gene expression in a way that supports growth at low temperature. In contrast, loss of Sas2 supports cell growth at high temperature. Deletion of ESA1 is not lethal, but deletion of both ESA1 and SAS2 causes lethality, indicating that Esa1 and Sas2 function redundantly in cell growth. Esa1 and Sas2 also play opposite roles in C. albicans morphogenesis, similar to the opposite roles of two histone deacetylases (HDACs), HOS2 and HDA1, in the regulation of hyphal formation (50). The synergistic effects of Esa1 and Sas2 on H4 acetylation at H4K5, H4K12, and H4K16 are correlated with their essential roles in cell growth. S. cerevisiae Esa1 has been proven a major HAT for acetylation of H4K5 and H4K12 and a secondary HAT for acetylation of H4K16 (10). The capability of S. cerevisiae Esa1 to acetylate H4K5, H4K12, and H4K16 and the synergy of C. albicans Esa1 and Sas2 on H4 acetylation link the acetylation of H4 at the K5, K12, and K16 sites to essential roles in cell growth.
Esa1 and Sas2 are involved in regulation of multiple cellular processes of C. albicans.
The MYST family of HATs is associated with diverse functions. C. albicans Esa1 and Sas2 target histone tails and are involved in transcriptional regulation of genes responsible for hyphal induction, heat shock, and stress resistance. The requirement of C. albicans Esa1 for the expression of hypha-specific genes is consistent with its essential role in C. albicans hyphal development. The activation of C. albicans Esa1 during cell growth at high temperatures correlates with the expression of heat shock genes. The repression of C. albicans Esa1 in response to stress is linked to the expression of stress response genes. Therefore, C. albicans Esa1 could function as a transcription coactivator as well as a transcription corepressor via modulation of histone acetylation.
C. albicans displays robust stress responses to promote survival in the host. Trehalose and glycerol are stress protectants, and accumulation of trehalose and glycerol is induced under stress conditions, including osmotic stress (43), high temperature (51), and cold stress (52). Loss of Esa1 caused cells to superaccumulate trehalose in stationary phase at all temperatures examined (see Fig. S4A in the supplemental material), which correlated well with its stress resistance and nonlethal phenotypes. Loss of Sas2 leads to accumulation of more trehalose at high temperature (42°C), which is associated with its heat-resistant phenotype (see Fig. S4C). Interestingly, the esa1/esa1 mutant cells did not accumulate more glycerol than the wild-type cells (see Fig. S4B), suggesting that the glycerol level may not correlate with survival of the esa1/esa1 mutant.
In the esa1/esa1 mutant, trehalose accumulation does not seem to be correlated with TPS gene expression in sensing temperature. TPS1 and TPS2 are expressed at the highest level at 4°C, yet the trehalose concentration is the highest at 35°C and the lowest at 4°C. The reason is that different states of the esa1/esa1 mutant cells were collected for measurement of TPS gene expression and the trehalose concentration. TPS gene expression was measured under cold shock or heat shock conditions, but the trehalose concentration was measured in cells from stationary phase. In stationary-phase cells, TPS gene expression in the esa1/esa1 mutant is hard to detect due to low level of TPS mRNA. On the other hand, superaccumulation of trehalose in the esa1/esa1 mutant is not increased by cold shock or heat shock (data not shown). Higher TPS gene expression and a higher trehalose concentration in the esa1/esa1 mutant may account for the inhibitory role of Esa1 on cell growth at cold temperatures but could not account for the decreased fitness of the mutant at high temperatures. A reasonable explanation might be that the downregulation of heat shock proteins plays a predominant role in the heat sensitivity of the esa1/esa1 mutant, which is supported by the essential role of Hsp90 for C. albicans viability. Indeed, downregulated HSP90 expression causes a lethal phenotype (40).
C. albicans Esa1 and Sas2 may target nonhistone substrates and function as KATs in the regulation of multiple cellular processes. S. cerevisiae Esa1 has been proven to be able to acetylate a variety of substrates, including Pck1, Yng2, and Atg3 (1, 11, 12), which are involved in multiple cellular processes. More substrates of the MYST proteins have been identified, including factors involved in transcription, heterochromatin formation, the cell cycle, DNA repair, and gluconeogenesis, and other subunits of MYST protein complexes (6). The mediation of several cellular processes, including temperature adaptation, stress response, cell cycle progress, metabolism, and morphogenesis, suggests that C. albicans Esa1 and Sas2 are involved in multiple regulatory processes via histone and/or nonhistone modification cross talk.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Chinese National Natural Science Foundation grants 30830003 and 31070074, Chinese MOST grant 2010CB912103, and Shanghai STCSM grant 10XD1404900.
Footnotes
Published ahead of print 25 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00275-12.
REFERENCES
- 1. Lin YY, Qi Y, Lu JY, Pan X, Yuan DS, Zhao Y, Bader JS, Boeke JD. 2008. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 22: 2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Borrow J, Stanton VP, Jr, Andresen JM, Becher R, Behm FG, Chaganti RS, Civin CI, Disteche C, Dube I, Frischauf AM, Horsman D, Mitelman F, Volinia S, Watmore AE, Housman DE. 1996. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat. Genet. 14: 33–41 [DOI] [PubMed] [Google Scholar]
- 3. Reifsnyder C, Lowell J, Clarke A, Pillus L. 1996. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat. Genet. 14: 42–49 [DOI] [PubMed] [Google Scholar]
- 4. Ehrenhofer-Murray AE, Rivier DH, Rine J. 1997. The role of Sas2, an acetyltransferase homologue of Saccharomyces cerevisiae, in silencing and ORC function. Genetics 145: 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. 1996. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology 216: 357–366 [DOI] [PubMed] [Google Scholar]
- 6. Sapountzi V, Cote J. 2011. MYST-family histone acetyltransferases: beyond chromatin. Cell Mol. Life Sci. 68: 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clarke AS, Lowell JE, Jacobson SJ, Pillus L. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19: 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith ER, Eisen A, Gu WG, Sattah M, Pannuti A, Zhou JX, Cook RG, Lucchesi JC, Allis CD. 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. U. S. A. 95: 3561–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clarke AS, Samal E, Pillus L. 2006. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol. Biol. Cell 17: 1744–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18: 5108–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin YY, Lu JY, Zhang J, Walter W, Dang W, Wan J, Tao SC, Qian J, Zhao Y, Boeke JD, Berger SL, Zhu H. 2009. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136: 1073–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J, Ma C, Sun Y, Zhang S, Feng W, Zhu L, Le Y, Gong X, Yan X, Hong B, Jiang FJ, Xie Z, Miao D, Deng H, Yu L. 2012. Function and molecular mechanism of acetylation in autophagy regulation. Science 336: 474–477 [DOI] [PubMed] [Google Scholar]
- 13. Osada S, Sutton A, Muster N, Brown CE, Yates JR, III, Sternglanz R, Workman JL. 2001. The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev. 15: 3155–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shia WJ, Osada S, Florens L, Swanson SK, Washburn MP, Workman JL. 2005. Characterization of the yeast trimeric-SAS acetyltransferase complex. J. Biol. Chem. 280: 11987–11994 [DOI] [PubMed] [Google Scholar]
- 15. Kimura A, Umehara T, Horikoshi M. 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32: 370–377 [DOI] [PubMed] [Google Scholar]
- 16. Suka N, Luo K, Grunstein M. 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32: 378–383 [DOI] [PubMed] [Google Scholar]
- 17. Li X, Wu L, Corsa CA, Kunkel S, Dou Y. 2009. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol. Cell 36: 290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840 [DOI] [PubMed] [Google Scholar]
- 19. Gudlaugsson O, Gillespie S, Lee K, Vande Berg J, Hu J, Messer S, Herwaldt L, Pfaller M, Diekema D. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 37: 1172–1177 [DOI] [PubMed] [Google Scholar]
- 20. Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20: 133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lu Y, Su C, Mao X, Raniga PP, Liu H, Chen J. 2008. Efg1-mediated recruitment of NuA4 to promoters is required for hypha-specific Swi/Snf binding and activation in Candida albicans. Mol. Biol. Cell 19: 4260–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brown DH, Jr, Giusani AD, Chen X, Kumamoto CA. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34: 651–662 [DOI] [PubMed] [Google Scholar]
- 23. Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181: 1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fonzi WA, Irwin MY. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134: 717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cao F, Lane S, Raniga PP, Lu Y, Zhou Z, Ramon K, Chen J, Liu H. 2006. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell 17: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lane S, Birse C, Zhou S, Matson R, Liu H. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276: 48988–48996 [DOI] [PubMed] [Google Scholar]
- 27. Fu XH, Meng FL, Hu Y, Zhou JQ. 2008. Candida albicans, a distinctive fungal model for cellular aging study. Aging Cell 7: 746–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soukup AA, Chiang YM, Bok JW, Reyes-Dominguez Y, Oakley BR, Wang CC, Strauss J, Keller NP. 2012. Overexpression of the Aspergillus nidulans histone 4 acetyltransferase EsaA increases activation of secondary metabolite production. Mol. Microbiol. 86: 314–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gomez EB, Nugent RL, Laria S, Forsburg SL. 2008. Schizosaccharomyces pombe histone acetyltransferase MstI (KAT5) is an essential protein required for damage response and chromosome segregation. Genetics 179: 757–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu X, Singh N, Donnelly C, Boimel P, Elefant F. 2007. The cloning and characterization of the histone acetyltransferase human homolog Dmel\TIP60 in Drosophila melanogaster: Dmel\TIP60 is essential for multicellular development. Genetics 175: 1229–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu Y, Fisher JB, Koprowski S, McAllister D, Kim MS, Lough J. 2009. Homozygous disruption of the Tip60 gene causes early embryonic lethality. Dev. Dyn. 238: 2912–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bird AW, Yu DY, Pray-Grant MG, Qiu QF, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415 [DOI] [PubMed] [Google Scholar]
- 33. Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, Buratowski S. 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20: 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brehm A, Tufteland KR, Aasland R, Becker PB. 2004. The many colours of chromodomains. Bioessays 26: 133–140 [DOI] [PubMed] [Google Scholar]
- 35. Sun YL, Jiang XF, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. 2009. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat. Cell Biol. 11: 1376–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Birse CE, Irwin MY, Fonzi WA, Sypherd PS. 1993. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect. Immun. 61: 3648–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Staab JF, Sundstrom P. 1998. Genetic organization and sequence analysis of the hypha-specific cell wall protein gene HWP1 of Candida albicans. Yeast 14: 681–686 [DOI] [PubMed] [Google Scholar]
- 38. La Valle R, Bromuro C, Ranucci L, Muller HM, Crisanti A, Cassone A. 1995. Molecular cloning and expression of a 70-kilodalton heat shock protein of Candida albicans. Infect. Immun. 63: 4039–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leach MD, Stead DA, Argo E, Brown AJ. 2011. Identification of sumoylation targets, combined with inactivation of SMT3, reveals the impact of sumoylation upon growth, morphology, and stress resistance in the pathogen Candida albicans. Mol. Biol. Cell 22: 687–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Swoboda RK, Bertram G, Budge S, Gooday GW, Gow NA, Brown AJ. 1995. Structure and regulation of the HSP90 gene from the pathogenic fungus Candida albicans. Infect. Immun. 63: 4506–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Praekelt UM, Meacock PA. 1990. HSP12, a new small heat shock gene of Saccharomyces cerevisiae: analysis of structure, regulation and function. Mol. Gen. Genet. 223: 97–106 [DOI] [PubMed] [Google Scholar]
- 42. Sheth CC, Mogensen EG, Fu MS, Blomfield IC, Muhlschlegel FA. 2008. Candida albicans HSP12 is co-regulated by physiological CO2 and pH. Fungal Genet. Biol. 45: 1075–1080 [DOI] [PubMed] [Google Scholar]
- 43. Hounsa CG, Brandt EV, Thevelein J, Hohmann S, Prior BA. 1998. Role of trehalose in survival of Saccharomyces cerevisiae under osmotic stress. Microbiology 144: 671–680 [DOI] [PubMed] [Google Scholar]
- 44. Van Dijck P, De Rop L, Szlufcik K, Van Ael E, Thevelein JM. 2002. Disruption of the Candida albicans TPS2 gene encoding trehalose-6-phosphate phosphatase decreases infectivity without affecting hypha formation. Infect. Immun. 70: 1772–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zaragoza O, Blazquez MA, Gancedo C. 1998. Disruption of the Candida albicans TPS1 gene encoding trehalose-6-phosphate synthase impairs formation of hyphae and decreases infectivity. J. Bacteriol. 180: 3809–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zaragoza O, de Virgilio C, Ponton J, Gancedo C. 2002. Disruption in Candida albicans of the TPS2 gene encoding trehalose-6-phosphate phosphatase affects cell integrity and decreases infectivity. Microbiology 148: 1281–1290 [DOI] [PubMed] [Google Scholar]
- 47. Brown AJ, Haynes K, Quinn J. 2009. Nitrosative and oxidative stress responses in fungal pathogenicity. Curr. Opin. Microbiol. 12: 384–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Enjalbert B, MacCallum DM, Odds FC, Brown AJ. 2007. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect. Immun. 75: 2143–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Squatrito M, Gorrini C, Amati B. 2006. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 16: 433–442 [DOI] [PubMed] [Google Scholar]
- 50. Zacchi LF, Schulz WL, Davis DA. 2010. HOS2 and HDA1 encode histone deacetylases with opposing roles in Candida albicans morphogenesis. PLoS One 5: e12171 doi:10.1371/journal.pone.0012171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Doehlemann G, Berndt P, Hahn M. 2006. Trehalose metabolism is important for heat stress tolerance and spore germination of Botrytis cinerea. Microbiology 152: 2625–2634 [DOI] [PubMed] [Google Scholar]
- 52. Schade B, Jansen G, Whiteway M, Entian KD, Thomas DY. 2004. Cold adaptation in budding yeast. Mol. Biol. Cell 15: 5492–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Feng Q, Summers E, Guo B, Fink G. 1999. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 181: 6339–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zeidler U, Lettner T, Lassnig C, Muller M, Lajko R, Hintner H, Breitenbach M, Bito A. 2009. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res. 9: 126–142 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.