Abstract
The genus Paracoccidioides includes the thermodimorphic species Paracoccidioides brasiliensis and P. lutzii, both of which are etiologic agents of paracoccidioidomycosis, a systemic mycosis that affects humans in Latin America. Despite the common occurrence of a sexual stage among closely related fungi, this has not been observed with Paracoccidioides species, which have thus been considered asexual. Molecular evolutionary studies revealed recombination events within isolated populations of the genus Paracoccidioides, suggesting the possible existence of a sexual cycle. Comparative genomic analysis of all dimorphic fungi and Saccharomyces cerevisiae demonstrated the presence of conserved genes involved in sexual reproduction, including those encoding mating regulators such as MAT, pheromone receptors, pheromone-processing enzymes, and mating signaling regulators. The expression of sex-related genes in the yeast and mycelial phases of both Paracoccidioides species was also detected by real-time PCR, with nearly all of these genes being expressed preferentially in the filamentous form of the pathogens. In addition, the expression of sex-related genes was responsive to the putative presence of pheromone in the supernatants obtained from previous cocultures of strains of two different mating types. In vitro crossing of isolates of different mating types, discriminated by phylogenetic analysis of the α-box (MAT1-1) and the high-mobility-group (HMG) domain (MAT1-2), led to the identification of the formation of young ascocarps with constricted coiled hyphae related to the initial stage of mating. These genomic and morphological analyses strongly support the existence of a sexual cycle in species of the genus Paracoccidioides.
INTRODUCTION
Fungi can reproduce asexually or sexually. This characteristic, called pleomorphism (1), is commonly observed in members of the host-associated dimorphic fungal pathogens of the family Ajellomycetaceae (2). These species can reproduce asexually by hyphal fragmentation or conidium production in specialized hyphae or by budding in the yeast form. Also, a sexual cycle may occur. Mating via pheromone exchange in a heterothallic bipolar system in the presence of mycelial cells of different mating types has been described for Ajellomyces dermatitidis (3), Ajellomyces capsulatus (4, 5), Ajellomyces parva, and Ajellomyces crescens (6).The initial step of mating in species of the Ajellomycetaceae family is triggered by hyphal constriction of cells of different mating types, coiling with one another several times to produce young ascocarps. Once hyphal aggregations become tighter, the cleistothecium is formed, with evanescence of the ascus, which harbors meiosporic spores (4). Sexual reproduction followed by meiosis and recombination plays an important role in the evolution of pathogenic fungi, by fitting populations and altering their virulence traits (7, 8).
Mating processes in filamentous ascomycetes are controlled by the presence of the MAT1-1 idiomorph, which encodes an alpha-domain transcription factor, while the MAT1-2 idiomorph encodes a high-mobility-group (HMG)-domain transcription factor that confers sexual identity to a haploid cell (9). These alleles or idiomorphs exhibit low comparative sequence similarity and are responsible for fungal sexuality and control of mating. Dimorphic pathogenic fungi, including Histoplasma capsulatum, Coccidioides immitis, Coccidioides posadasii, Paracoccidioides brasiliensis, and Paracoccidioides lutzii, exhibit a syntenic organization which has previously been characterized by comparative genomic analysis (10, 11). Besides the MAT locus, molecules such as pheromones, pheromone receptors, and pheromone-processing and response enzymes are involved in the sexual process, acting in opposite cell recognition through nuclear regulation of the mating process.
Paracoccidioides species are thermodimorphic fungal pathogens of humans; they are also adapted to other mammalian hosts, such as armadillos (12). Once infective propagules are inhaled by immunocompetent or immunodepressed humans, paracoccidioidomycosis can be induced (13). This systemic mycosis affects mainly rural workers in Latin America, frequently found in Brazil, Venezuela, and Colombia (14). Based on molecular phylogenetic tools, extensive genetic divergence has been detected among Paracoccidioides isolates. Applying the method of genealogical concordance for phylogenetic species recognition, two major species have been proposed: P. brasiliensis, with three phylogenetically cryptic species, i.e., S1, PS2, and PS3 (15); and P. lutzii, harboring “Pb01-like” strains, which have so far been collected mainly in Ecuador and the northern-central region of Brazil (16). Also, recombination events have been shown within these two Paracoccidioides species that could explain their highly genotypic and phenotypic diversity and which have prompted the hypothesis of a sexual cycle in these pathogens.
To test this hypothesis, the sexual genomic traits of the teleomorphic species A. capsulatus (anamorph Histoplasma capsulatum) and A. dermatitidis (anamorph Blastomyces dermatitidis) were searched in silico in Paracoccidioides species. The presence of a Paracoccidioides mating type locus, MAT1-1, which is a homologue of that in Aspergillus nidulans and other components of the pheromone response pathway, was previously reported from transcriptomic analyses of both morphological phases of P. lutzii strain Pb01 (17, 18). Torres et al. (19) identified the presence and expression of MAT1-1 and MAT1-2 in the P. brasiliensis phylogenetic species complex, suggesting a sexual cycle in this fungus.
In the present report, we provide additional information about (i) a comparative genomic analysis of mating regulator genes in all dimorphic fungi with available genome data that supports vertical transfer of the genomic trait for sexual reproduction through the Ajellomycetaceae family; (ii) the predominant expression of six sex-related genes in mycelial cells, including the MAT locus and genes for the pheromone receptors and components of the pheromone response cascade; (iii) the distribution of mating type idiomorphs in clinical and environmental strains; and (iv) strong evidence of direct activation of the mating pathway in cocultured strains of opposite mating types and the in vitro development of the initial steps of sexual reproduction, namely, the formation of young ascocarps in mated strains of P. brasiliensis and P. lutzii, with the possibility of homothallism in the Paracoccidioides genus.
MATERIALS AND METHODS
Maintenance of Paracoccidioides strains.
Environmental and clinical strains used in this study (see Table S1 in the supplemental material) were kept in the mycelial phase in semisolid potato dextrose agar (PDA; Acumedia) at 25°C and were subcultured every 21 days. Yeast cells were maintained in yeast-peptone-dextrose agar (YPD; 0.5% casein peptone, 0.5% yeast extract, 1.0% glucose) at 37°C and were subcultured every 15 days.
Quantitative expression of sex-related genes in yeast and mycelial forms.
To assess the levels of expression of sex-related genes (the α-box gene MAT1-1, the HMG gene MAT1-2, the pheromone receptor genes ste2 and ste3, the mitogen-activated protein kinase [MAPK] gene fus3, and the transcriptional factor gene ste12), we used real-time reverse transcription-PCR (RT-PCR). P. brasiliensis strains Pb18, Pb3, T15LN1, T1F1, and T9B1 and P. lutzii strains Pb01 and EE were cultured in yeast and mycelial forms in YPD for 14 days at 37°C and 25°C, respectively. Cells were harvested by centrifugation (5,000 × g for 10 min), and RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. To remove any genomic DNA contamination, RNA was treated with RNase-free DNase I (Promega), followed by ethanol precipitation. RNA samples were then used for real-time RT-PCR. Equal amounts of RNA (2 μg) were reverse transcribed (Superscript III; Invitrogen) using an oligo(dT)12–18 primer and then subjected to real-time PCR. Amplification assays were carried out with a model 7500 SDS Fast system (Applied Biosystems), using a 10-μl reaction volume containing 0.2 μM (each) primers (see Table S2 in the supplemental material), 5 μl of SYBR green PCR master mix (2×), and 0.02 μl template cDNA. After initial denaturation at 95°C for 20 s, amplifications were done for 40 cycles at 95°C for 3 s and 60°C for 20 s. All assays were carried out with experimental triplicates. In order to confirm the amplification specificity, the PCR products were subjected to a melting curve analysis. The comparative crossing threshold (CT) method, employing the constitutive P. brasiliensis α-tubulin gene for normalization, was used to evaluate the expression value (fold change) for each gene of interest. All primers used in real-time RT-PCR experiments were determined using the software Primer 3, which is available online (http://primer3.wi.mit.edu/).
Pheromone stimulation.
For pheromone stimulation, mycelia (about 500 mg wet weight) from strains of opposite mating types (T9B1 [MAT 1-1] and T15LN1 [MAT1-2]) were cocultivated in Erlenmeyer flasks containing 500 ml of YEM medium (0.1% MgSO4 · 7H2O, 0.15% KH2PO4, 0.1% NaNO3, 0.05% yeast extract). Fungi were incubated at 25°C for 21 days at an agitation rate of 150 rpm, after which the supernatant, representing conditioned medium (CM), was collected using a Sterifil aseptic system equipped with a 0.22-μm membrane (Millipore). In parallel, isolates T9B1 and T15LN1 were grown independently in 20 ml of YEM medium for 14 days, and mycelia were collected by centrifugation for 5 min at 2,300 × g and washed three times with 1× phosphate-buffered saline (PBS). Each mycelial sample was stimulated separately for 24 h at 25°C with agitation at 150 rpm in 20 ml of the previously collected CM. RNA extractions were performed with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. As a negative control, each isolate grown individually was stimulated with 20 ml YEM medium. All experiments were carried out in triplicate. The total RNA extracted was treated with RNase-free DNase I (Promega), precipitated, and used for real-time RT-PCR as described above.
DNA extraction and population analysis of MAT locus distribution in Paracoccidioides species.
DNA extractions were performed as described by Raeder and Broda (20), with modifications. Briefly, about 100 mg of either yeast cells or mycelium from each isolate was suspended in 500 μl of extraction buffer (25 mM EDTA, 250 mM NaCl, 0.5% [wt/vol] sodium dodecyl sulfate [SDS], 200 mM Tris-HCl, pH 8.5), to which 500 mg of 425- to 600-μm-diameter glass beads (Sigma-Aldrich) was added. The mixture was shaken vigorously for 15 min. A mixture of phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol/vol]; 500 μl) (BioAgency, Germany) was added to the solution, briefly mixed, and centrifuged for 60 min at 12,000 × g. The aqueous phase was treated with RNase A (final concentration, 100 μg/ml; Fisher Scientific) for 15 min at 37°C. An equal volume of chloroform was added and roughly mixed, and the solution was centrifuged for 10 min at 12,000 × g. The aqueous phase was precipitated with 0.5v (vol/vol) isopropanol by centrifugation for 20 min at 12,000 × g and 4°C. DNA was washed with 70% ethanol, dried at room temperature, and eluted with ultrapure water. The DNA integrity was confirmed by 0.8% agarose gel electrophoresis with ethidium bromide (0.5 μl/ml) staining.
For PCR amplification, 20 to 50 ng of genomic DNA from each isolate (see Table S1 in the supplemental material) was used for PCR amplification. Primers were designed (see Table S2) to target regions inside the open reading frames (ORFs) of MAT1-1 (α-box domain) and MAT1-2 (HMG domain) sequences taken from the P. brasiliensis genome (http://www.broadinstitute.org/annotation/genome/paracoccidioides_brasiliensis/MultiHome.html). PCR amplifications were done in a Veriti 96-well thermocycler (Applied Biosystems) and using PyroStart PCR master mix (Fermentas) as recommended by the manufacturers. The annealing temperature for both regions was 60°C. PCR samples were loaded into 0.8% agarose gels and analyzed by electrophoresis to identify the sexual idiomorph of each isolate.
Sequencing and phylogenetic analysis.
Sixteen amplicons from MAT1-1 strains and 11 amplicons from MAT1-2 strains were selected for sequencing. Sequences of the amplicons were determined by automatic capillary Sanger sequencing in a MegaBACE 500 machine, using a DYEnamic ET dye terminator kit (GE Healthcare, United Kingdom). Fragments were sequenced on both strands, and bases with phred scores of >30 were considered for alignments, increasing the quality of the sequence data. The MAT1-1 (GenBank accession numbers HQ687769 to HQ687784) and MAT1-2 (GenBank accession numbers HQ687785 to HQ687795) sequences were aligned using the PRANK algorithm (21) implemented in PRANKSTER software and manually corrected in order to avoid mispaired bases. Maximum parsimony analyses were carried out using PAUP, version 4.0b.10a (22). Unweighted parsimony was chosen, and character transformation states were considered disordered. Heuristic searches were performed by random stepwise sequence addition in order to obtain the initial tree; branch rearrangements were made by the tree bisection and reconnection (TBR) technique. One thousand bootstrap replicates were used to estimate confidence values for individual clades. Significant branches with bootstrap values of ≥50% were considered in the phylogeny (23). Phylogenetic trees were done with the help of FigTree v1.3.1 software (http://tree.bio.ed.ac.uk/software/figtree/).
Identification of sex-related genes in thermodimorphic fungal pathogens.
BLAST searches (24, 25) were carried out on P. lutzii, P. brasiliensis, H. capsulatum, B. dermatitidis, C. immitis, and C. posadasii genomes (http://www.broadinstitute.org/annotation/genome/dimorph_collab/MultiHome.html), using protein sequences from the aforementioned species as queries. tblastn searches were performed using an E value cutoff of 1e−13 to assign homology. The ORFs retrieved by BLAST searches were subjected to blastp searches of the Saccharomyces cerevisiae genome database (http://www.yeastgenome.org/), and protein domains were aligned and manually inspected.
Mating tests.
Mating assays were performed on pairwise MAT1-1 and MAT1-2 strains by slide culture and direct streaking of isolates on agar medium (see Table S3 in the supplemental material). The media selected for both mating methods were V8 juice agar (20% [vol/vol] Campbell's V8 juice, 0.3% CaCO3, and 1.5% agar), soil extract agar (SEA; 0.2% glucose, 0.1% yeast extract, 1.5% agar, and 50% [vol/vol] soil extract collected from armadillo burrows), and oatmeal tomato paste agar (OTA; 2% Elefante's tomato paste, 2% Dr. Oetker's oatmeal flakes, 0.3% CaCO3, and 1.5% agar). Crossing combinations were performed with isolates from the same phylogenetic species: P. brasiliensis (15) and P. lutzii (16). For slide culture mating tests, plugs of mycelia from strains of each mating type were planted onto an agar block pairwise in a side-by-side pattern. For mating assays performed in plates, mycelial fragments were planted about 1 cm apart. Plates were sealed with Parafilm (Pechiney Plastic Packaging) and kept at 25°C in the dark. Mating crossings were visualized by stereomicroscopy using a Stemi 2000C apparatus (Zeiss) and by light microscopy with lactophenol cotton blue staining monthly for up to 8 months. The presence or absence of sex-related structures was documented in a model PM-30 photomicrographic system (Olympus, Japan) by inspection of at least five random microscopic fields.
Confocal microscopy.
Slide cultures made as described above were also used for confocal microscopy experiments. The cocultured strains used were JAL × EE (P. lutzii) and T1F1 × T15LN1 (P. brasiliensis). After mating of the strains, the coverslips were removed, fixed, and permeabilized with 70% ethanol at 4°C overnight. The coverslips were then stained for 24 h with 1× SYBR green and for 1 h with 20 μg/ml calcofluor white. After washing, the coverslips were mounted with Prolong Gold antifade medium (Invitrogen), sealed, and imaged in a Leica SP5 laser scanning confocal microscope equipped with a 63×, 1.4-numerical-aperture (NA) objective. Confocal images were processed using ImageJ and Adobe Photoshop software, without any nonlinear processing.
RESULTS
Identification of sex-related genes.
The majority of components of the pheromone response pathway were inferred by comparative analysis of the genome sequences of the dimorphic fungi P. brasiliensis, P. lutzii, H. capsulatum, B. dermatitidis, C. immitis, and C. posadasii (see Table S4 in the supplemental material). The data revealed a conserved mating machinery that was vertically transferred from a common ancestor of these species. Both a- and α-pheromone receptors (Ste2 and Ste3, respectively) were identified in all dimorphic fungal genomes.
Pheromone molecules bind to G-protein-coupled receptors and stimulate the downstream MAP kinase cascade. The G protein is composed of three subunits, Gα, Gβ, and Gγ, encoded by the Gpa1, Ste4, and Ste18 genes, respectively. All three genes were identified in the analyzed genomes. In yeast, the binding of pheromone to its receptor releases Gβγ from its inhibitor, Gα, which in turn activates the initial steps of the mating process, i.e., polarization and shmooing, at the G1 arrest stage (26–28). In S. cerevisiae and C. albicans, Gβγ recruits the scaffold protein Ste5 to the plasma membrane, bringing together the protein kinases Ste20, Ste11, Ste7, and Fus3 and activating the MAPK pathway (29–31). In yeast, when Fus3p is activated by its dissociation from Ste5, it is translocated to the nucleus, activating downstream mating genes. Interestingly, the gene encoding the scaffold protein Ste5 was not detected in the dimorphic fungal genomes.
We extended the search to species in the subphylum Pezizomycotina; the Ste5 homologue was not found in any genome analyzed and was present exclusively in the Saccharomycotina. Fus3p that is released in the nucleus activates Ste12p, which is required for mating and filamentation-specific activation of several genes, including pheromone precursor genes (32). The latter genes were not identified by BLAST searches of the dimorphic fungal genomes. On the other hand, homologues of pheromone-processing enzymes were detected in all genomes analyzed.
In ascomycetes, the block structures of the pheromone genes are similar, but their mature pheromone peptides are highly divergent across species, controlling the sexual selection in fungi. The pheromone precursor protein α-factor is comprised of 2 to 5 identical repeat sequences flanked by Kex2 cleavage sites (KK or KR). Once processed by Kex2, the released peptides (9 to 13 amino acid residues) are matured by the proteases Kex1 and Ste13 in the Golgi internal membrane and then released to the extracellular space (33).
Like α-factor, a-factor is also assembled as a precursor polypeptide, but it is differentiated by the presence of a single-copy molecule. In fungi, the a-precursor protein ends in a Cys-aliphatic amino acid-aliphatic amino acid-X-COOH motif (the CAAX box), which is a target for lipid modification. The first step is cleavage downstream of the CAAX box cysteine by Rce1 and Ste24, the CAAX proteases. After this, two proteins (Ram1 and Ram2) catalyze the addition of lipid prenyl groups. Following prenylation, the carboxyl group of the cysteine is methylated by the Ste14 methyltransferase. All modifications in both α- and a-factor are necessary for binding to receptors, guaranteeing mating signaling (34, 35).
Population distribution of mating types in Paracoccidioides species.
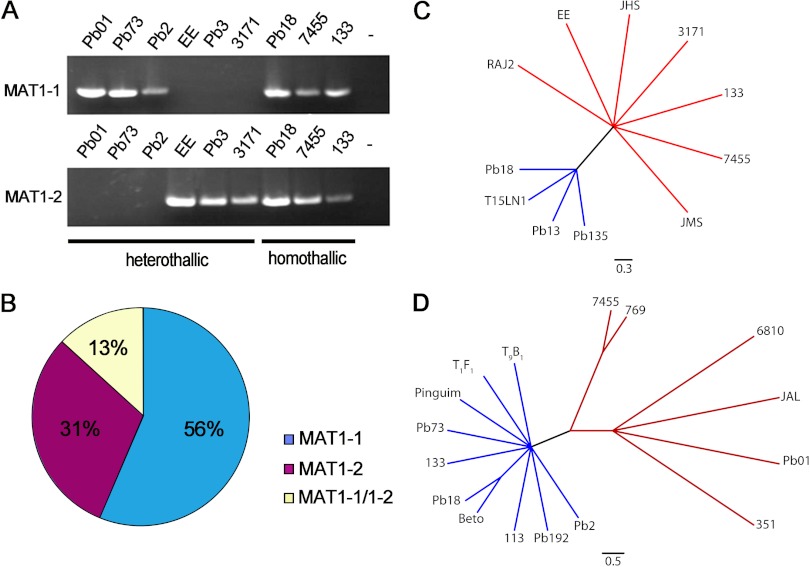
A total of 98 isolates representing the P. brasiliensis species complex (S1, PS2, and PS3) and P. lutzii were examined for the MAT locus, and they exhibited both homothallic and heterothallic profiles. Fifty-five isolates possessed the MAT1-1 locus and 30 isolates possessed the MAT1-2 locus, as demonstrated by single amplicons (Fig. 1A). Unexpectedly, 13 isolates presented both MAT1-1 and MAT1-2 amplified loci, suggesting that homothallism could exist in the Paracoccidioides genus. The prevalences of heterothallic MAT1-1 and MAT1-2 and homothallic MAT1-1/MAT1-2 populations were 56%, 31%, and 13%, respectively, which approximated a ratio of 4:2:1 (Fig. 1B). Amplicons from 16 MAT1-1 strains and 11 MAT1-2 strains were sequenced and submitted to phylogenetic analysis. The MAT locus cladogram clearly revealed the genetic divergence of the isolates belonging to the P. brasiliensis species complex and P. lutzii, in agreement with the new species proposal (16). The P. lutzii and P. brasiliensis standard strains Pb01 and Pb18, respectively, were clustered apart from each other, which helped in species recognition (Fig. 1C and D). These data also helped in the selection of isolates from the same phylogenetic cluster for mating assays. Twenty-one isolates of P. lutzii and 24 isolates of P. brasiliensis (S1 and PS2 group) were analyzed separately to verify whether the distribution of mating type ratios was equivalent inside each species of Paracoccidioides. Considering the two species apart, the ratio of heterothallic MAT1-1 (12 isolates of P. lutzii and 16 isolates of P. brasiliensis) and MAT1-2 (6 isolates of P. lutzii and 4 isolates of P. brasiliensis) strains and homothallic strains (3 isolates of P. lutzii and 4 isolates of P. brasiliensis) was 4:2:1 for P. lutzii and 4:1:1 for P. brasiliensis. Also, we evaluated 12 isolates, belonging to P. brasiliensis species S1 and PS2, recovered from armadillos in Botucatu City and surrounding neighborhoods located in the central-south region of São Paulo State, Brazil (11). The idiomorphic distribution of the MAT locus for these isolates was 4:2:1. The MAT locus data revealed a slight predominance of heterothallic MAT1-1 isolates, followed by MAT1-2 and putative homothallic MAT1-1/MAT1-2 isolates, in Paracoccidioides species, even between species or in isolates recovered from armadillos.
Fig 1.
MAT locus amplification, population distribution, and phylogenetic analysis of isolates belonging to the Paracoccidioides species complex. (A) Sexual identification in Paracoccidioides, evaluated by PCR amplification of MAT1-1 and MAT1-2 idiomorphs and revealing homothallic and heterothallic isolates. (B) Idiomorphic distribution of 98 isolates revealing that 56% harbored MAT1-1, 31% harbored MAT1-2, and 13% harbored both MAT1-1 and MAT1-2. Unrooted phylogenies generated by the maximum parsimony method are shown and allow discrimination between P. lutzii (red clades) and P. brasiliensis (blue clades), using both the MAT1-1 (C) and MAT1-2 (D) loci.
Expression of sex-related genes in yeast and mycelial cells of Paracoccidioides species.
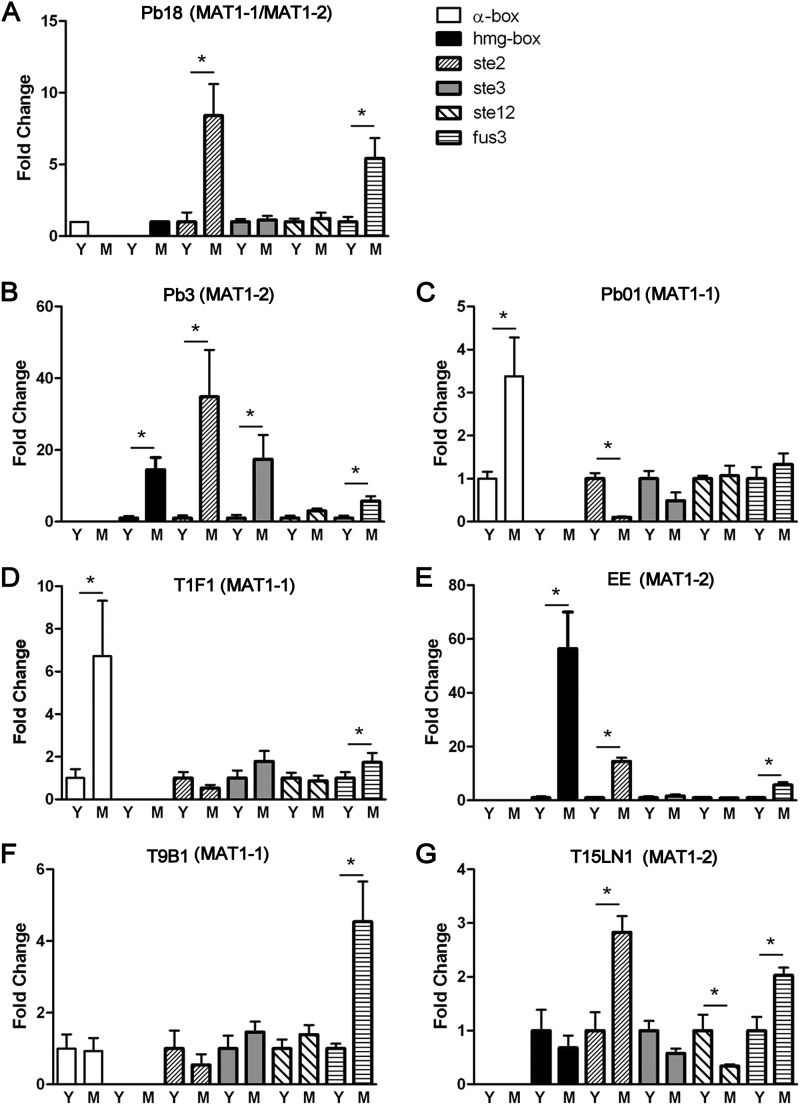
To assess the expression of genes involved in the mating process, the mRNA levels of MAT1-1 (α-box), MAT1-2 (HMG box), the pheromone receptor genes ste2 and ste3, the MAPK gene fus3, and the transcriptional factor gene ste12 were quantified by real-time RT-PCR analysis of yeast and mycelia of seven isolates of P. lutzii and P. brasiliensis. MAT1-1 and MAT1-2 genes were expressed exclusively, in a cell type-specific manner, in the heterothallic strains. Surprisingly, we detected differential expression of the MAT genes in the putative homothallic isolate Pb18: MAT1-1 was expressed only in the yeast phase, while MAT1-2 was expressed only in the mycelium phase. In general, all genes were expressed more exuberantly in the mycelium phase, in a species-independent fashion and irrespective of whether the isolate was homothallic or heterothallic (Fig. 2). The MAPK gene (fus3) was differentially expressed in six of seven analyzed isolates. The higher level of fus3 mRNA in the mycelial phase could be related to the saprophytic lifestyle required for the mating process in Paracoccidioides.
Fig 2.
Quantitative expression of the sex-related genes MAT1-1 (α-box), MAT1-2 (HMG), ste2 and ste3 (pheromone receptors), fus3 (MAPK), and ste12 (transcriptional factor) in yeast and mycelial phases of the homothallic strain Pb18 (A) and the heterothallic strains Pb3 (B), Pb01 (C), T1F1 (D), EE (E), T9B1 (F), and T15LN1 (G).
Evidence of pheromone induction in P. brasiliensis strains T9B1 and T15LN1.
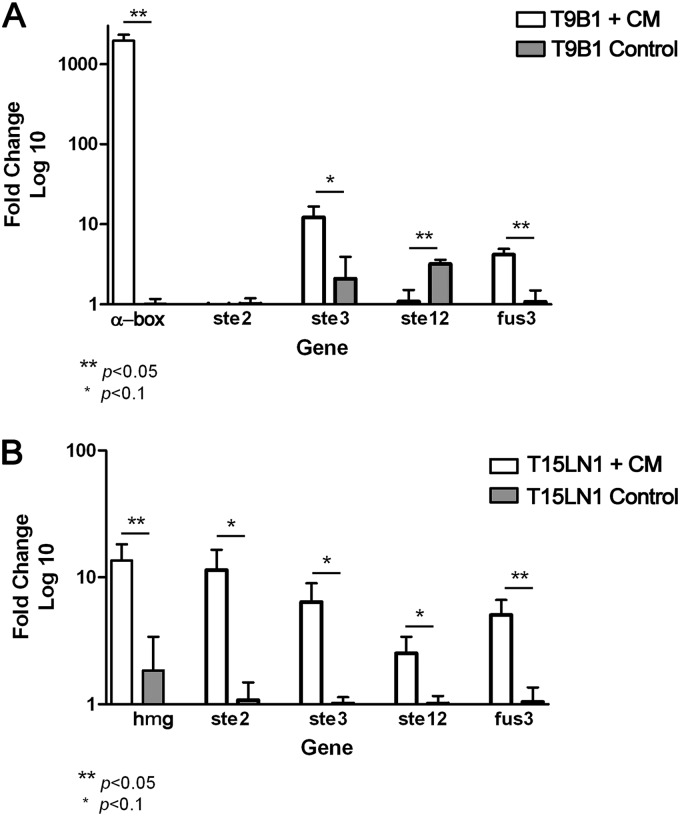
The cocultivation of haploid compatible a- and α-cells in minimal medium induces pheromone production in some ascomycetes, as already reported for S. cerevisiae (36) and for the dimorphic fungus H. capsulatum (37). We tested the effect of mating stimulation by the CM of cocultured T9B1 (MAT1-1) and T15LN1 (MAT1-2) strains. After 24 h of stimulation of mycelial cells, total mRNA was extracted and the sex-related genes were quantified by real-time RT-PCR (Fig. 3). All genes of the mating cascade were highly upregulated after CM stimulation, except for the pheromone receptor gene (ste3) in the T9B1 isolate, indicating an activation of the mating machinery by a- and α-pheromone molecules, probably due to their presence in the CM. The pheromone-induction test was carried out only on T9B1 and T15LN1 isolates of the P. brasiliensis S1 cryptic species, because these two strains were able to produce initial ascocarps in mating assays. More experiments are in progress to extend the results to other strains of P. brasiliensis (the PS2 cryptic species) and P. lutzii. Even so, the available data are extremely important in guiding further studies of mating in dimorphic fungi.
Fig 3.
Pheromone stimulation after coculture of strains of opposite mating types (T9B1 [MAT1-1] and T15LN1 [MAT1-2]) in Erlenmeyer flasks containing YEM medium (CM). mRNA levels of the sex-related genes MAT1-1 (α-box), MAT1-2 (HMG), ste2 and ste3 (pheromone receptors), fus3 (MAPK), and ste12 (transcriptional factor) were evaluated in isolates T9B1 (A) and T15LN1 (B), grown independently and stimulated for 24 h with the previously collected CM or with YEM medium (negative control).
Mating assays.
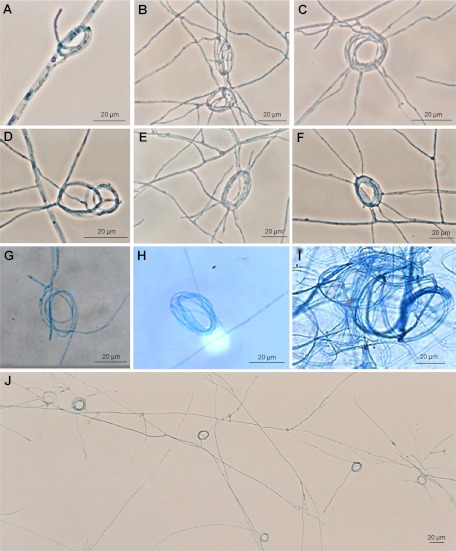
The coincubation of MAT1-1 and MAT1-2 strains of P. brasiliensis (S1) and P. lutzii revealed coiled hyphae, indicating the formation of young ascocarps (Fig. 4; see Table S3 in the supplemental material). These results were also obtained previously by laboratory crossings with their relatives H. capsulatum and B. dermatitidis (3, 4). Coiled hyphae were observed in the paired T1F1 × T15LN1 and T9B1 × T15LN1 P. brasiliensis isolates (Fig. 4A to F). In P. lutzii, the coiled hyphae were also observed for JAL × EE, 66 × EE, and PLRO1 × ED01 strains (Fig. 4G to J). Undifferentiated hyphal tips began to wind around and multiply encircle hyphae of the opposite mating type beginning 2 months after cocultivation (Fig. 4A and G). The hypha constriction tightened into numerous coils 4 to 6 months after cocultivation (Fig. 4B and F). In Fig. 4J, many coils are observed in one field for the 66 × EE P. lutzii crossing. The paired isolates, whether P. brasiliensis or P. lutzii, produced sexual structures, and no morphological alterations in young ascocarp formation were observed in the three different media (OTA, V8 juice agar, and SEA) used for mating experiments. Mating assays with P. brasiliensis isolates belonging to the PS2 phylogenetic species—Pb2 × Pb3 and Pb4 × Pb3—or with the putative homothallic isolate Pb18 did not produce any coiled hyphae, indicating an absence of sexual structure formation (see Table S3). No mature cleistothecium or ascus formation was observed, even 8 months after mating of cocultures. Mating of the standard laboratorial strains Pb18 and Pb01 also did not result in the production of sexual structures, likely because they were maintained in culture for a long time and lost their mating ability, as has clearly been demonstrated for H. capsulatum (5, 37, 38).
Fig 4.
Initial stages of sexual development in Paracoccidioides species. The sexual structures were obtained by mating experiments using V8, SEA, or OTA medium, and the samples were monitored for 6 months. The description of each panel gives the following information: isolates crossed, time of incubation (months), medium used, and magnification. (A) T9B1 × T15LN1, 2, SEA, ×400; (B) T9B1 × T15LN1, 4, SEA, ×400; (C) T9B1 × T15LN1, 4, SEA, ×400; (D) T9B1 × T15LN1, 6, SEA, ×400; (E) T1F1 × T15LN1, 4, SEA, ×400; (F) T1F1 × T15LN1, 6, SEA, ×400; (G) ED01 × PLRO1, 2, OTA, ×400; (H) ED01 × PLRO1, 4, OTA, ×400; (I) 66 × EE, 6, OTA, ×400; (J) 66 × EE, 4, V8, ×100.
To further evaluate the coiled hypha structure, we stained JAL × EE (P. lutzii) (Fig. 5A) and T1F1 × T15LN1 (P. brasiliensis) (Fig. 5B) matings with the cell wall dye calcofluor white and the nucleic acid stain SYBR green. In addition to coiled hyphae, we observed the formation of knob-like structures, which are dilated cells that form in the region where the two winding hyphae initially contact (4, 5). Moreover, we observed a single cell in a coiled hypha containing multiple nuclei, which was possibly a consequence of nuclear migration during mating.
Fig 5.
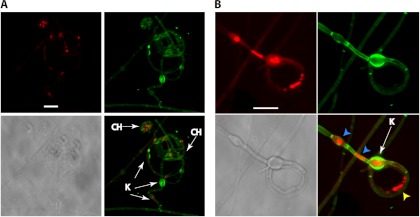
Confocal microscopy examination of Paracoccidioides species sexual structures. Samples from mating experiments using SEA medium and JAL × EE cocultures (P. lutzii) (A) or T1F1 × T15LN1 cocultures (P. brasiliensis) (B) were stained with calcofluor white, which labels cell wall chitin and is pseudocolored green, and SYBR green, which labels DNA in nuclei and is pseudocolored red. The arrows show coiled hyphae (CH) and three knob-like structures (K). In the coiled hyphae shown in detail in panel B, blue arrowheads point to single nuclei in adjacent cells and the yellow arrowhead points to a single cell in the sexual structure with four individual nuclei.
DISCUSSION
The first report of the presence and expression of mating type genes in Paracoccidioides was done by Fernandes et al. (18). The MAT1-1 transcript of P. lutzii strain Pb01 was identified during the EST sequencing project (17, 39). Recently, the chromosomal structures of the MAT1-1 and MAT1-2 loci of P. brasiliensis Pb3 and Pb18 and P. lutzii Pb01 were identified based on comparative genomic analyses with other dimorphic fungal pathogens and dermatophytes (11). The MAT locus of the Paracoccidioides genus represents a syntenic organization and locus expansion compared with the case in H. capsulatum, C. immitis, and C. posadasii (10, 11, 37). The presence of a single copy of each mating type gene (MAT1-1 and MAT1-2) in the Paracoccidioides genome (Pb18, Pb3, and Pb01) points to a potential bipolar mating system. The genomic identification of the mating components in Paracoccidioides and other dimorphic fungi led us to hypothesize that the genetic apparatus for sexual reproduction is conserved and vertically transferred among these fungi. The sexual cycle has been demonstrated for H. capsulatum and B. dermatitidis, and all orthologs were identified in their genomes compared to P. brasiliensis and P. lutzii (see Table S4 in the supplemental material).
The ratio of MAT1-1/MAT1-2 idiomorphs in bipolar fungal species is often 1:1 in sexual populations; this was observed for C. immitis, C. posadasii (40), Aspergillus fumigatus (41, 42), and, more recently, P. brasiliensis (19). In contrast, a high prevalence of MAT1-2 was observed in patients with disseminated histoplasmosis (43). Also, α-cells of the pathogenic basidiomycete Cryptococcus neoformans are 30 to 40 times more frequent than a-cells in both environmental and clinical samples (44). The high frequency of α-strains in clinical isolates has been correlated with their capacity to infect or colonize hosts (45). In the present study, there was a slight predominance of the MAT1-1 mating type, as well as the presence of both idiomorphs in a single genome, when P. lutzii and P. brasiliensis populations were considered together or individually (56% MAT1-1, 31% MAT1-2, and 13% MAT1-1/MAT1-2).
In this work, expression analysis of the mating components (MAT1-1, MAT1-2, ste2, ste3, fus3, and ste12) in yeast and mycelial cells of seven Paracoccidioides isolates revealed the preferential expression of these genes in the filamentous phase (Fig. 2). This study also investigated the ability of the supernatant (CM) of cocultured mycelia from the T9B1 (MAT1-1) and T15LN1 (MAT1-2) strains in YEM medium after 21 days of growth to stimulate pheromone production. CM stimulation in individual strains provoked an upregulation of the components of the mating/pheromone pathway, providing evidence that a- and α-cells of P. brasiliensis respond to CM stimulation (Fig. 3). These results agree with those reported for S. cerevisiae (46) and H. capsulatum (37). The present results are also consistent with the mating behavior of the Ajellomycetaceae species H. capsulatum and B. dermatitidis, since functional analysis of sex-related genes revealed an upregulation in the mycelium stage and the capability to respond to CM containing pheromone.
Homothallism is an intrinsic characteristic of fungi in which a single individual harbors both mating types and can undergo sexual reproduction. In Aspergillus species, the homothallic characteristic appears to be an ancestral state and heterothallism is acquired after speciation (47), as has also been reported for the closely related Cochliobolus and Stemphylium genera (48). The present study provides the first evidence of the possibility of homothallism in the Ajellomycetaceae family. Both MAT1-1 and MAT1-2 idiomorphs were present in a single individual, as confirmed by PCR analysis of 13 Paracoccidioides isolates (Fig. 1). Phylogenetic analysis revealed that the MAT1-1 and MAT1-2 loci from isolates Pb18 and 7455 clustered with their respective species, P. brasiliensis and P. lutzii (Fig. 1). Phylogenetic analysis of MAT1-1 positioned isolate 133 (which contains both MAT loci) within P. brasiliensis, while the MAT1-2 idiomorph clustered it together with P. lutzii, so this isolate shows genomic signatures of both Paracoccidioides species. Thus, consistent with a recent suggestion (16), the evidence supports isolate 133 as a hybrid or an ancestral state of Paracoccidioides. Our results suggest that the Paracoccidioides genus, or at least its common ancestor, could be homothallic. In the homothallic species A. nidulans (a filamentous fungus), both MAT1-1 and MAT1-2 are coexpressed under in vitro mating conditions (41). In contrast, gene expression analysis of the MAT genes in the P. brasiliensis Pb18 strain (putatively homothallic) revealed a differential pattern, with the α-box gene (MAT1-1) detected only in the yeast phase and the HMG gene (MAT1-2) detected only in the mycelial cells. Thus, what we determined to be “homothallic” isolates (MAT1-1/MAT1-2) could be hybrid strains. We listed isolate 133 as an example of strains where, based on sequence information, MAT1-1 and MAT1-2 are derived from P. brasiliensis and P. lutzii (47). Thus, these strains are not likely “true” homothallic strains but are fused products of interspecies or intraspecies matings between MAT1-1 and MAT1-2 strains. This is consistent with these species being heterothallic. These particular isolates are stuck in the hybrid state, probably due to an impaired ability to undergo meiosis, consistent with speciation. Strains of the basidiomycete C. neoformans serotype hybrid AD, which are intervarietal or interspecies hybrids, cannot undergo meiosis due to genetic divergence (49).
The ploidy of Paracoccidioides was assessed by different methods, leading to controversial results. PFGE combined with microfluorometry (50–53) revealed that genome sizes vary from 23 to 31 Mb, and microfluorometric data indicated twice the genome content (46 to 61 Mb), indicating that P. brasiliensis yeast cells could be diploid, haploid, or at least aneuploid (50, 51). Also, by means of flow cytometry combined with the haploid genome size previously measured by electrophoretic karyotyping, the ploidy was assessed in several P. brasiliensis isolates, revealing haploid or aneuploid nuclei (54). Another possibility for the amplification of both mating type genes is that these isolates represent mixed populations of two heterothallic strains. This consideration should not be discarded, but we demonstrated a differential expression of MAT1-1 and MAT1-2 for Pb18, recovered from single colonies, indicating a single genotype. Since the diploid and/or aneuploid state was observed for Pb18 isolates by Feitosa et al. (51) and Almeida et al. (54), “homothallism” in Paracoccidioides species must be investigated further.
Fungal sexual development is initiated by cell chemoattraction that is governed by pheromone induction. In filamentous and dimorphic fungi, the initial ascocarp is developed when hyphae of opposite mating types match and start to coil with each other (4). The present study offers the first evidence for initial in vitro formation of ascocarps under mating conditions for P. brasiliensis (S1 group) and also for P. lutzii (Fig. 4). Knob-like structures were found in regions of coiled hyphae in the mating assay (Fig. 5). Similar results were observed during ascogenesis of H. capsulatum and also Mucor circinelloides, suggesting that these structures are important for the maintenance of ascocarp development (4, 55). Moreover, the occurrence of a multinucleated single hypha cell in the mating interactions led us to suggest an exchange of genetic material between cells of opposite mating types (Fig. 5). Such observations are well described for A. nidulans, Schizophyllum commune, and Ustilago maydis during the initial steps of mating, suggesting that it is a consequence of nuclear migration (56–58). These results were obtained in mating experiments using V8, SEA, and OTA media, with samples being monitored for 6 months. The resemblance of the coiled hypha aggregations to immature ascocarps echoes previous observations in H. capsulatum, B. dermatitidis, Emmonsia spp., A. fumigatus, A. nidulans, and Trichophyton mentagrophytes, whose sexual cycles are known (3, 4, 59–62). Coiled hyphae were detected after 2 months of mating assays of isolates of opposite mating types for both species of Paracoccidioides; in fact, this is the first step of ascogenesis, as also reported for H. capsulatum and Aspergillus spp. (4, 60). However, no cleistothecium or ascus production was detected, even after 1 year of cocultivation. Further studies are being performed to find key environmental or physical conditions that could allow for cleistothecium and recombinant ascospore formation in Paracoccidioides, resulting in the description of a complete sexual stage. The balanced regulation of MAT locus expression could be an important factor for the progression of a sexual cycle, and specific environmental conditions might have to be present to allow sexual differentiation. Despite the detection of increased levels of transcripts of mating genes in hyphae, no correlation was observed between the expression levels of the sex-related genes and the ability to produce young ascocarps.
With the increase in fungal genome sequencing projects, many questions regarding the biology of mating in fungi are being answered. Components of the pheromone-dependent pathway involved in the mating system in ascomycetes are well characterized, as demonstrated for Saccharomyces cerevisiae (63), Neurospora crassa (6), and Aspergillus fumigatus (64). Combining structural and functional genomics and mating assays, we have shown that the sexual cycle of Paracoccidioides must occur during its saprophytic life cycle. In addition, we have identified all orthologs of the mating machinery in both C. immitis and C. posadasii (see Table S4 in the supplemental material), although no evidence of sexual structures has yet been uncovered. It is well known that the Coccidioides genus represents a cryptic speciation (65), and there is also evidence of a recombination process in this genus (66), supporting the existence of a sexual cycle. Furthermore, the ratio of MAT1-1/MAT1-2 idiomorphs is 1:1, providing molecular support for the presence of a sexual stage in the life cycle (40). The human pathogen A. fumigatus was long considered asexual. Genomic comparisons with S. cerevisiae and other ascomycetes and mRNA expression analyses of sex-related genes were revealed prior to the discovery of the sexual state of A. fumigatus (teleomorph = Neosartorya fumigata), thus showing the relevance of genomic data in defining the mating behavior of fungi (61, 64).
The other example is Trichophyton rubrum, which was also once considered asexual, since its mating machinery was discovered by comparative genomic analysis (11) and was recently confirmed by mating assays (67). Collectively, these data provide important information about the sexual behavior of fungi, especially for Paracoccidioides, and could be useful for further studies regarding the biology and speciation of this genus.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rosane Christine Hahn, Eduardo Honda, and Patrícia Cisalpino for supplying P. lutzii isolates and Erika Seki Kioshima for helping in improvement of the final artwork.
This work was supported by Fundação de Amparo à Pesquisa do Distrito Federal (FAP-DF) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Footnotes
Published ahead of print 2 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.05052-11.
REFERENCES
- 1. Sugiyama J. 1987. Pleomorphic fungi: the diversity and its taxonomic implications. Elsevier, Amsterdam, Netherlands [Google Scholar]
- 2. Untereiner WA, Scott JA, Naveau FA, Sigler L, Bachewich J, Angus A. 2004. The Ajellomycetaceae, a new family of vertebrate-associated Onygenales. Mycologia 96: 812– 821 [DOI] [PubMed] [Google Scholar]
- 3. McDonough ES, Lewis AL. 1967. Blastomyces dermatitidis: production of the sexual stage. Science 156: 528– 529 [DOI] [PubMed] [Google Scholar]
- 4. Kwon-Chung KJ. 1973. Studies on Emmonsiella capsulata. I. Heterothallism and development of the ascocarp. Mycologia 65: 109– 121 [PubMed] [Google Scholar]
- 5. Kwon-Chung KJ. 1975. Perfect state (Emmonsiella capsulata) of the fungus causing large-form African histoplasmosis. Mycologia 67: 980– 990 [PubMed] [Google Scholar]
- 6. Shiu PK, Glass NL. 2000. Cell and nuclear recognition mechanisms mediated by mating type in filamentous ascomycetes. Curr. Opin. Microbiol. 3: 183– 188 [DOI] [PubMed] [Google Scholar]
- 7. Heitman J. 2006. Sexual reproduction and the evolution of microbial pathogens. Curr. Biol. 16: R711– R725 [DOI] [PubMed] [Google Scholar]
- 8. Milgroom MG. 1996. Recombination and the multilocus structure of fungal populations. Annu. Rev. Phytopathol. 34: 457– 477 [DOI] [PubMed] [Google Scholar]
- 9. Coppin E, Debuchy R, Arnaise S, Picard M. 1997. Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61: 411– 428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fraser JA, Stajich JE, Tarcha EJ, Cole GT, Inglis DO, Sil A, Heitman J. 2007. Evolution of the mating type locus: insights gained from the dimorphic primary fungal pathogens Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot. Cell 6: 622– 629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li W, Metin B, White TC, Heitman J. 2010. Organization and evolutionary trajectory of the mating type (MAT) locus in dermatophyte and dimorphic fungal pathogens. Eukaryot. Cell 9: 46– 58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bagagli E, Bosco SM, Theodoro RC, Franco M. 2006. Phylogenetic and evolutionary aspects of Paracoccidioides brasiliensis reveal a long coexistence with animal hosts that explain several biological features of the pathogen. Infect. Genet. Evol. 6: 344– 351 [DOI] [PubMed] [Google Scholar]
- 13. Franco M, Lacaz CS, Restrepo A, Del Negro G. 1993. Paracoccidioidomycosis. CRC Press, Boca Raton, FL [Google Scholar]
- 14. San-Blas G, Nino-Vega G, Iturriaga T. 2002. Paracoccidioides brasiliensis and paracoccidioidomycosis: molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med. Mycol. 40: 225– 242 [DOI] [PubMed] [Google Scholar]
- 15. Matute DR, McEwen JG, Puccia R, Montes BA, San-Blas G, Bagagli E, Rauscher JT, Restrepo A, Morais F, Nino-Vega G, Taylor JW. 2006. Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol. Biol. Evol. 23: 65– 73 [DOI] [PubMed] [Google Scholar]
- 16. Teixeira MM, Theodoro RC, de Carvalho MJ, Fernandes L, Paes HC, Hahn RC, Mendoza L, Bagagli E, San-Blas G, Felipe MS. 2009. Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol. Phylogenet. Evol. 52: 273– 283 [DOI] [PubMed] [Google Scholar]
- 17. Felipe MS, Torres FA, Maranhao AQ, Silva-Pereira I, Pocas-Fonseca MJ, Campos EG, Moraes LM, Arraes FB, Carvalho MJ, Andrade RV, Nicola AM, Teixeira MM, Jesuino RS, Pereira M, Soares CM, Brigido MM. 2005. Functional genome of the human pathogenic fungus Paracoccidioides brasiliensis. FEMS Immunol. Med. Microbiol. 45: 369– 381 [DOI] [PubMed] [Google Scholar]
- 18. Fernandes L, Araujo MA, Amaral A, Reis VC, Martins NF, Felipe MS. 2005. Cell signaling pathways in Paracoccidioides brasiliensis—inferred from comparisons with other fungi. Genet. Mol. Res. 4: 216– 231 [PubMed] [Google Scholar]
- 19. Torres I, Garcia AM, Hernandez O, Gonzalez A, McEwen JG, Restrepo A, Arango M. 2010. Presence and expression of the mating type locus in Paracoccidioides brasiliensis isolates. Fungal Genet. Biol. 47: 373– 380 [DOI] [PubMed] [Google Scholar]
- 20. Raeder U, Broda P. 1985. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1: 3 [Google Scholar]
- 21. Loytynoja A, Goldman N. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci. U. S. A. 102: 10557– 10562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA [Google Scholar]
- 23. Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetics analysis. Syst. Biol. 42: 10 [Google Scholar]
- 24. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389– 3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gish W, States DJ. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3: 266– 272 [DOI] [PubMed] [Google Scholar]
- 26. Bender A, Sprague GF., Jr 1986. Yeast peptide pheromones, a-factor and alpha-factor, activate a common response mechanism in their target cells. Cell 47: 929– 937 [DOI] [PubMed] [Google Scholar]
- 27. Fujimura HA. 1989. The yeast G-protein homolog is involved in the mating pheromone signal transduction system. Mol. Cell. Biol. 9: 152– 158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reed SI. 1991. Pheromone signaling pathways in yeast. Curr. Opin. Genet. Dev. 1: 391– 396 [DOI] [PubMed] [Google Scholar]
- 29. Elion EA. 2001. The Ste5p scaffold. J. Cell Sci. 114: 3967– 3978 [DOI] [PubMed] [Google Scholar]
- 30. Liao H, Thorner J. 1980. Yeast mating pheromone alpha factor inhibits adenylate cyclase. Proc. Natl. Acad. Sci. U. S. A. 77: 1898– 1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singh P, Ghosh S, Datta A. 1997. A novel MAP-kinase kinase from Candida albicans. Gene 190: 99– 104 [DOI] [PubMed] [Google Scholar]
- 32. Wong Sak Hoi J, Dumas B. 2010. Ste12 and Ste12-like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryot. Cell 9: 480– 485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Achstetter T. 1989. Regulation of alpha-factor production in Saccharomyces cerevisiae: a-factor pheromone-induced expression of the MF alpha 1 and STE13 genes. Mol. Cell. Biol. 9: 4507– 4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boyartchuk VL, Rine J. 1998. Roles of prenyl protein proteases in maturation of Saccharomyces cerevisiae a-factor. Genetics 150: 95– 101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Powers S, Michaelis S, Broek D, Santa Anna S, Field J, Herskowitz I, Wigler M. 1986. RAM, a gene of yeast required for a functional modification of RAS proteins and for production of mating pheromone a-factor. Cell 47: 413– 422 [DOI] [PubMed] [Google Scholar]
- 36. Strazdis JR, MacKay VL. 1982. Reproducible and rapid methods for the isolation and assay of a-factor, a yeast mating hormone. J. Bacteriol. 151: 1153– 1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bubnick M, Smulian AG. 2007. The MAT1 locus of Histoplasma capsulatum is responsive in a mating type-specific manner. Eukaryot. Cell 6: 616– 621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kwon-Chung KJ, Weeks RJ, Larsh HW. 1974. Studies on Emmonsiella capsulata (Histoplasma capsulatum). II. Distribution of the two mating types in 13 endemic states of the United States. Am. J. Epidemiol. 99: 44– 49 [DOI] [PubMed] [Google Scholar]
- 39. Felipe MS, Andrade RV, Petrofeza SS, Maranhao AQ, Torres FA, Albuquerque P, Arraes FB, Arruda M, Azevedo MO, Baptista AJ, Bataus LA, Borges CL, Campos EG, Cruz MR, Daher BS, Dantas A, Ferreira MA, Ghil GV, Jesuino RS, Kyaw CM, Leitao L, Martins CR, Moraes LM, Neves EO, Nicola AM, Alves ES, Parente JA, Pereira M, Pocas-Fonseca MJ, Resende R, Ribeiro BM, Saldanha RR, Santos SC, Silva-Pereira I, Silva MA, Silveira E, Simoes IC, Soares RB, Souza DP, De-Souza MT, Andrade EV, Xavier MA, Veiga HP, Venancio EJ, Carvalho MJ, Oliveira AG, Inoue MK, Almeida NF, Walter ME, Soares CM, Brigido MM. 2003. Transcriptome characterization of the dimorphic and pathogenic fungus Paracoccidioides brasiliensis by EST analysis. Yeast 20: 263– 271 [DOI] [PubMed] [Google Scholar]
- 40. Mandel MA, Barker BM, Kroken S, Rounsley SD, Orbach MJ. 2007. Genomic and population analyses of the mating type loci in Coccidioides species reveal evidence for sexual reproduction and gene acquisition. Eukaryot. Cell 6: 1189– 1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Paoletti M, Rydholm C, Schwier EU, Anderson MJ, Szakacs G, Lutzoni F, Debeaupuis JP, Latge JP, Denning DW, Dyer PS. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 15: 1242– 1248 [DOI] [PubMed] [Google Scholar]
- 42. Rydholm C, Szakacs G, Lutzoni F. 2006. Low genetic variation and no detectable population structure in Aspergillus fumigatus compared to closely related Neosartorya species. Eukaryot. Cell 5: 650– 657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kwon-Chung KJ, Bartlett MS, Wheat LJ. 1984. Distribution of the two mating types among Histoplasma capsulatum isolates obtained from an urban histoplasmosis outbreak. Sabouraudia 22: 155– 157 [PubMed] [Google Scholar]
- 44. Kwon-Chung KJ, Bennett JE. 1978. Distribution of alpha and alpha mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108: 337– 340 [DOI] [PubMed] [Google Scholar]
- 45. Kwon-Chung KJ, Edman JC, Wickes BL. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60: 602– 605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sengupta P, Cochran BH. 1991. MAT alpha 1 can mediate gene activation by a-mating factor. Genes Dev. 5: 1924– 1934 [DOI] [PubMed] [Google Scholar]
- 47. Yun SH, Berbee ML, Yoder OC, Turgeon BG. 1999. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc. Natl. Acad. Sci. U. S. A. 96: 5592– 5597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Inderbitzin P, Harkness J, Turgeon BG, Berbee ML. 2005. Lateral transfer of mating system in Stemphylium. Proc. Natl. Acad. Sci. U. S. A. 102: 11390– 11395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J. 2007. alpha AD alpha hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 3: 1975– 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cano MI, Cisalpino PS, Galindo I, Ramirez JL, Mortara RA, da Silveira JF. 1998. Electrophoretic karyotypes and genome sizing of the pathogenic fungus Paracoccidioides brasiliensis. J. Clin. Microbiol. 36: 742– 747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Feitosa LS, Cisalpino PS, dos Santos MR, Mortara RA, Barros TF, Morais FV, Puccia R, da Silveira JF, de Camargo ZP. 2003. Chromosomal polymorphism, syntenic relationships, and ploidy in the pathogenic fungus Paracoccidioides brasiliensis. Fungal Genet. Biol. 39: 60– 69 [DOI] [PubMed] [Google Scholar]
- 52. Montoya AE, Alvarez AL, Moreno MN, Restrepo A, McEwen JG. 1999. Electrophoretic karyotype of environmental isolates of Paracoccidioides brasiliensis. Med. Mycol. 37: 219– 222 [PubMed] [Google Scholar]
- 53. Montoya AE, Moreno MN, Restrepo A, McEwen JG. 1997. Electrophoretic karyotype of clinical isolates of Paracoccidioides brasiliensis. Fungal Genet. Biol. 21: 223– 227 [DOI] [PubMed] [Google Scholar]
- 54. Almeida AJ, Matute DR, Carmona JA, Martins M, Torres I, McEwen JG, Restrepo A, Leao C, Ludovico P, Rodrigues F. 2007. Genome size and ploidy of Paracoccidioides brasiliensis reveals a haploid DNA content: flow cytometry and GP43 sequence analysis. Fungal Genet. Biol. 44: 25– 31 [DOI] [PubMed] [Google Scholar]
- 55. Li CH, Cervantes M, Springer DJ, Boekhout T, Ruiz-Vazquez RM, Torres-Martinez SR, Heitman J, Lee SC. 2011. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog. 7: e1002086 doi:10.1371/journal.ppat.1002086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Raudaskoski M, Kothe E. 2010. Basidiomycete mating type genes and pheromone signaling. Eukaryot. Cell 9:847– 859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Snetselaar KM. 1993. Microscopic observation of Ustilago maydis mating interactions. Exp. Mycol. 17: 10 [Google Scholar]
- 58. Sohn KT, Yoon KS. 2002. Ultrastructural study on the cleistothecium development in Aspergillus nidulans. Mycobiology 30:10 [Google Scholar]
- 59. Ajello L, Cheng SL. 1967. The perfect state of Trichophyton mentagrophytes. Sabouraudia 5:230– 234 [PubMed] [Google Scholar]
- 60. Grosse V, Krappmann S. 2008. The asexual pathogen Aspergillus fumigatus expresses functional determinants of Aspergillus nidulans sexual development. Eukaryot. Cell 7: 1724– 1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. O'Gorman CM, Fuller HT, Dyer PS. 2009. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457: 471– 474 [DOI] [PubMed] [Google Scholar]
- 62. Sigler L. 1996. Ajellomyces crescens sp. nov., taxonomy of Emmonsia spp., and relatedness with Blastomyces dermatitidis (teleomorph Ajellomyces dermatitidis). J. Med. Vet. Mycol. 34: 303– 314 [PubMed] [Google Scholar]
- 63. Schrick K, Garvik B, Hartwell LH. 1997. Mating in Saccharomyces cerevisiae: the role of the pheromone signal transduction pathway in the chemotropic response to pheromone. Genetics 147: 19– 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Poggeler S. 2002. Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Curr. Genet. 42: 153– 160 [DOI] [PubMed] [Google Scholar]
- 65. Fisher MC, Koenig GL, White TJ, Taylor JW. 2002. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 94: 73– 84 [PubMed] [Google Scholar]
- 66. Barker BM, Jewell KA, Kroken S, Orbach MJ. 2007. The population biology of coccidioides: epidemiologic implications for disease outbreaks. Ann. N. Y. Acad. Sci. 1111: 147– 163 [DOI] [PubMed] [Google Scholar]
- 67. Anzawa K, Kawasaki M, Mochizuki T, Ishizaki H. 2010. Successful mating of Trichophyton rubrum with Arthroderma simii. Med. Mycol. 48: 629– 634 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.