Abstract
Deletion of ribosomal protein L32 genes resulted in a nonsexual flocculation of fission yeast. Nonsexual flocculation also occurred when two other ribosomal protein genes, rpl21-2 and rpl9-2, were deleted. However, deletion of two nonribosomal protein genes, mpg and fbp, did not cause flocculation. Overall transcript levels of rpl32 in rpl32-1Δ and rpl32-2Δ cells were reduced by 35.9% and 46.9%, respectively, and overall ribosome levels in rpl32-1Δ and rpl32-2Δ cells dropped 31.1% and 27.8%, respectively, compared to wild-type cells. Interestingly, ribosome protein expression levels and ribosome levels were also reduced greatly in sexually flocculating diploid YHL6381/WT (h+/h−) cells compared to a mixture of YHL6381 (h+) and WT (h−) nonflocculating haploid cells. Transcriptome analysis indicated that the reduction of ribosomal levels in sexual flocculating cells was caused by more-extensive suppression of ribosomal biosynthesis gene expression, while the reduction of ribosomal levels caused by deleting ribosomal protein genes in nonsexual flocculating cells was due to an imbalance between ribosomal proteins. We propose that once the reduction of ribosomal levels is below a certain threshold value, flocculation is triggered.
INTRODUCTION
In sexual reproduction, heterothallic haploid fission yeast cells, h+ or h−, require a partner of the opposite mating type to mate (1–4). During the sexual mating, flocculation takes place first among the cells, which allows two different mating types of haploid cells to come together and form zygotes (2). Sexual flocculation is the result of specific recognition of cell surface proteins called “adhesion molecules” from one mating type by sugar molecules or amino acid residues on the surface of the opposite mating cell type (5). Several mating type-specific adhesins from fission yeast have been described (6, 7). Adhesin genes are not constitutively expressed and are subjected to tight transcriptional regulation (8). Carbon and nitrogen limitation activates their expression (9). The differential expression of these adhesin genes allows the cells to better adapt to different environments. Even though different adhesin genes are expressed under different conditions, their functions are the same and are interchangeable (10).
Heterothallic haploid strains of the fission yeast Schizosaccharomyces pombe, h+ or h−, are usually nonflocculent when cultured separately (11). However, Kim et al. (2001) reported that a Lkh1 null mutant of S. pombe exhibited nonsexual flocculation. Lkh1 encodes a LAMMER kinase homolog, which represses expression of adhesions (12). Kang et al. (2010) also showed that deletion of the tup12+ gene, functioning as a Lkh1 binding partner, resulted in a nonsexual flocculation in the fission yeast (13). Recently, Matsuzawa et al. (2011) reported that overexpression of FLO8, encoding a transcriptional activator of the dominant flocculation genes in budding yeast Saccharomyces cerevisiae, induced a nonsexual flocculation in fission yeast (14). Therefore, the relationship between nonsexual flocculation caused by gene manipulation and sexual flocculation needs to be further studied.
We previously reported that ribosomal protein L32-2 in fission yeast could specifically bind to DNA fragments containing GTTGGT sequences and activate reporter gene expression in a GAL4 yeast two-hybrid system (15). To understand its function further, we created deletion mutants for the rpl32 genes in fission yeast. Surprisingly, we found that deleting either of the two rpl32 paralogous genes resulted in a clearly visible nonsexual flocculation in fission yeast, accompanying a reduction of the ribosomal level. Further studies explored whether sexual flocculation in fission yeast cells also displayed a reduction of ribosomal protein expression and ribosomal biogenesis during mating. Thus, we compared the transcriptomics of sexually and nonsexually flocculating cells to that of nonflocculating cells to find out their molecular mechanism.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The fission yeast strains used in this study are listed in Table 1. The YEPD medium consists of yeast extract (10 g/liter), tryptone (20 g/liter), and glucose (20 g/liter). The YEPME mating medium was derived from YEPD medium (16) by replacing glucose with malt extract (containing ∼50% maltose) (1, 17). The synthetic Edinburgh minimal medium (EMM2) and LB medium were made as described by Kim et al. (2001) (12). Different antibiotics were added when needed and were specified in each experiment. Fission yeast cells were usually cultured in 100 ml of medium in an Erlenmeyer flask at 30°C and 220 rpm.
Table 1.
Schizosaccharomyces pombe strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| Wild-type SPQ01 (WT) | h−, leu1-32 | Stratagene |
| WT SPQ01/pESP3 | h−, leu1-32/pESP3 | This report |
| SPQ01 rpl32-1Δ | h−, rpl32-1::kanmx6+, leu1-32 | This report |
| SPQ01 rpl32-2Δ | h−, rpl32-2::kanmx6+, leu1-32 | This report |
| SPQ01 rpl32-1Δ/pESP3 | h−, rpl32-1::kanmx6+, leu1-32/pESP3 | This report |
| SPQ01 rpl32-2Δ/pESP3 | h−, rpl32-2::kanmx6+, leu1-32/pESP3 | This report |
| SPQ01 rpl32-1 rpl32-1Δ | h−, rpl32-1::kanmx6+, leu1-32/pESP3-rpl32-1 | This report |
| SPQ01 rpl32-2 rpl32-2Δ | h−, rpl32-2::kanmx6+, leu1-32/pESP3-rpl32-2 | This report |
| SPQ01 rpl32-1 | h−, leu1-32/pESP3-rpl32-1(leu+) | This report |
| SPQ01 rpl32-2 | h−, leu1-32/pESP3-rpl32-2(leu+) | This report |
| SPQ01 rpl21-2Δ | h−, rpl21-2::kanmx6+, leu1-32 | This report |
| SPQ01 rpl9-2Δ | h−, rpl9-2::kanmx6+, leu1-32 | This report |
| SPQ01 mpgΔ | h−, mpg::kanmx6+, leu1-32 | This report |
| SPQ01 fbpΔ | h−, fbp::kanmx6+, leu1-32 | This report |
| SPQ01 kanmx6 | h−, leu1-32/pESP3-kanmx6 | This report |
| YHL6381 | h+, his3-D1, leu1-32, ura4-D18, ade6-M210 | 19 |
| YHL6381/WT | h+/h−, his3-D1, leu1-32, ura4-D18, ade6-M210/h−, leu1-32 | This report |
To determine growth curves, cells were cultured in YEPD medium, sampled every 2 h, harvested by centrifugation, washed twice in 100 mM EDTA to disperse floc (18), and suspended in the same volume of solution for determination of the cell density at an optical density of 600 nm (OD600), which was converted into the number of cells based on the blood cell count standard curve.
For nonsexual flocculation analysis, haploid yeast cells were cultured to the late exponential phase (about 48 h; OD600 of 5.0) in YEPD medium. For complementation experiments, the haploid yeast cells of deletion mutants transformed with a wild-type (WT) ribosome protein gene were precultured to the late exponential phase (about 48 h; OD600 of 5.0) in EMM2 medium and then harvested by centrifugation, washed twice with double-distilled water (ddH2O), resuspended in the same volume of YEPD medium, and incubated for 3 h. The cells overexpressing the kanmx6+ gene were cultured using the same method.
For sexual flocculation analysis, haploid yeast YHL6381 (h+) and WT (h−) cells were separately precultured in YEPD medium for 48 h until the OD600 reached 5.0. The cells were collected by centrifugation, washed with 30 mM EDTA–ddH2O, resuspended in the same volume of YEPME mating medium or YEPD medium, and then mixed at a 1:1 cell number ratio and incubated for 1.5 h. For the control, precultured YH6381 and WT cells were separately inoculated into identical volumes of YEPME mating medium or YEPD medium and incubated for 1.5 h. For culturing YHL6381 cells, the YEPD medium and YEPME medium mentioned here were supplemented with Ade, His, Leu, and Ura at 225 mg/liter.
Generation of deletion mutants and overexpression transformants.
The rpl32-1, rpl32-2, rpl21-2, rpl9-2, mpg, or fbp-targeting DNA fragments containing kanmx6 were amplified by PCR from genomic DNA of WT cells and plasmid pFA6a-kanmx6 using deletion primer sets (see Table S2 in the supplemental material) and transformed into WT cells to generate deletion mutants by gene replacement (19).
To overexpress the indicated genes or rescue the deleted genes, the gene fragments were amplified from SPQ01 genomic DNA with gene-specific primers (see Table S2 in the supplemental material) and cloned into the NdeI-BamH I sites of plasmid pESP3 (Stratagene, La Jolla, CA). The WT strain or deletion mutants were transformed with the constructed plasmids using the lithium acetate (LiAC) transformation method (20). Transformants were grown on EMM2 plates and confirmed by PCR using appropriate primers (20).
QPCR analysis.
Total RNA was isolated from cells using TRIzol reagent (Invitrogen) and treated with RNase-free DNase I (TaKaRa) to eliminate any genomic DNA. A 10-ng volume of DNA-free RNA was used for the first strand of cDNA synthesis using a SuperScript III first-strand synthesis system (Invitrogen). The amplifications of indicated genes were performed in triplicate, within the same quantitative PCR (QPCR) run, and only one amplicon was produced per reaction. The beta-actin gene was used as an internal control to standardize the transcript level of the genes. The standardized transcript level of the genes in the indicated cells, such as flocculating cells, was compared to the normalized transcript level of the genes in the corresponding control cells, such as nonflocculating cells, for fold change determinations. The entire procedure (RNA extraction, first-strand synthesis, and QPCR) was repeated for a minimum of three times (biological replications). Data were analyzed using Rotor-Gene Q5/6 plex system software (Qiagen) and subsequently exported to Microsoft Excel for further analysis. Primers used in QPCR are listed in Table S2 in the supplemental material.
Preparation of anti-RPL32 antibody.
A RPL32-2 gene fragment was amplified by PCR from WT SPQ01 genomic DNA using the rpl32-2 primer set (see Table S2 in the supplemental material) and cloned into the EcoRI-XhoI sites of plasmid pET28a (Novagen, Madison, WI) to construct plasmid pET28a-rpl32-2+, which expresses RPL32-2 as an N-terminal 6His fusion protein. The plasmid was transformed into Escherichia coli strain BL21-CodonPlus (DE3) Rosetta (Novagen). The transformants were grown to an OD600 of 0.6 in LB broth containing 50 μg/ml kanamycin and 34 μg/ml chloramphenicol at 37°C. Synthesis of RPL32-2 was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to reach a final concentration of 1 mM. Cells were harvested 6 h after the induction. The pellet was suspended in 10 mM Tris-HCl (pH 8.0) buffer containing 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and 100 μg/ml of lysozyme and sonicated. The cell debris was removed by centrifugation, and the extract (10 ml, ∼20 mg) was diluted with an equal volume of binding buffer (20 mM Tris-HCl, 0.5 M NaCl, 5 mM imidazole, pH 7.9) and loaded onto 1-ml nickel-nitrilotriacetic acid (Ni-NTA) agarose columns (Qiagen). After being rinsed with wash buffer (20 mM Tris-HCl, 0.5 M NaCl, 40 mM imidazole, pH 7.9), RPL32-2 protein was eluted from the column with elution buffer (20 mM Tris-HCl, 0.5 M NaCl, 1 M imidazole, pH 7.9) (21). Eluted RPL32-2 proteins were dialyzed in 10 mM Tris-HCl (pH 7.9) buffer and then sent to YingJi Corporation (Shanghai, China) for anti-RPL32-2 antibody preparation in rabbits.
Before use, anti-RPL32-2 antibody was purified by an affinity purification method (22). Briefly, recombinant RPL32-2 proteins were separated using SDS-PAGE and transferred onto a polyvinylidene fluoride (PVDF) membrane. The PVDF membrane was then incubated with RPL32-2 polyclonal antibody (1:3,000 dilution) in TBST buffer (25 mM Tris, 150 mM NaCl, 0.5% Tween 20, pH 7.5) at room temperature for 4 h. The PVDF membrane strips containing RPL32-2 band were cut out, put into 10 ml of 0.2 M glycine–HCl solution (pH 2.7), and subjected to a vortex procedure for 10 min at 4°C. The anti-RPL32-2 antibody was eluted from PVDF strips, and the elution was immediately neutralized with NaOH to pH 8.0. The anti-RPL32-2 antibody can recognize both RPL32-2 and RPL32-1 proteins, since there are only four amino acid differences (96.85% homology in amino acid sequences) between these two proteins.
Western blot analysis.
Denatured protein extracts were prepared by bead-beating cells directly in a solution containing 10 mM Tris-HCl (pH 7.5, 0.5 mM PMSF, and 0.5 mM protease inhibitors, followed by adding the same volume of SDS sample buffer (10 mM Tris-HCl [pH 7.5], 2% SDS, 10% glycerol, 100 mM dithiothreitol [DTT], 0.25% bromophenol blue) and heating for 5 min at 100°C. Proteins were subjected to SDS-PAGE on 12% gradient gels and transferred to a PVDF membrane. Membranes were probed with anti-beta-actin or anti-RPL32-2 antibody at 37°C for 4 h, followed by alkaline phosphatase (AP)-conjugated secondary antibody, in TBS–2% milk–0.5% Tween 20 at 37°C for 2 h. Signal was detected using an alkaline phosphatase detection kit (Qiagen) according to the manufacturer's instructions (23).
Analysis of polyribosome levels.
Yeast cell cultures (OD600 of 1.0) were mixed with cycloheximide to reach a final concentration of 0.1 mg/ml for 20 min at 30°C and then incubated on ice for 10 min. The cells were collected by centrifugation at 4°C and washed with ice-cold polysome lysis buffer (20 mM Tris-HCl [pH 7.5], 50 mM KCl, 10 mM MgCl2, 1 mM DTT, 0.1 mg/ml cycloheximide, 0.2 mg/ml heparin). The cell pellets were resuspended in 1 ml of polysome lysis buffer supplemented with protease inhibitors (Roche, Indianapolis, IN) and RNase inhibitors (Promega). Cell suspensions were transferred to prechilled 15-ml conical tubes containing 0.5 g of glass beads and subjected to 6 to 10 pulses of full-speed vortexing for 100 s followed by 100-s pauses. After the glass beads and cell debris were removed by centrifugation at 3,500 rpm for 5 min at 4°C, extracts were transferred to prechilled microcentrifuge tubes and centrifuged for an additional 15 min at 14,000 rpm and 4°C. A total of 15 A260 units of extracts was layered onto 5% to 50% sucrose gradients in polysome lysis buffer and centrifuged for 4 h at 44,000 rpm and 4°C in a Beckman SW41 rotor. The gradients were then fractionated by upward displacement with microtubes using a peristaltic pump, and the absorbance at 254 nm was continuously monitored using a UV detector (24). Polyribosomes levels were calculated based on the total absorbance tracing of the peak area (25, 26).
Observation and analysis of flocculation.
For observation of flocculation, 2 ml of cell cultures was placed in a petri dish for 10 min. Images were then taken with a Nikon D3100 camera. For micrograph imaging, 5 μl of culture was placed between the slide and coverslip and was observed under a Zeiss Axio Imager A1 microscope (Zeiss, Jena, Germany) with a 63× oil immersion objective differential interference contrast (DIC) lens (numerical aperture [NA], 1.4). Images were taken using Sensicam, a quantum efficiency (QE) cooled digital camera system (Cooke Corp., Romulus, MI).
For determination of flocculation activity, 2 ml of cell culture was transferred to a 2-ml centrifuge tube. Two identical samples and tubes were prepared in this way for each culture. After thorough mixing, the optical density (OD600) of 400 μl of the upper layer of the cells in one of the tubes for each culture was carefully measured as the initial cell density, or At = 0. After 10 min of settling, the optical density (OD600) of 400 μl of the upper layer of the cells from the other tube of each culture was measured as the end cell density, or At = 10. Cell sedimentation rate was calculated as V = 1 − At = 10/At = 0 (4).
Analysis of sexual mating diploid cells.
Mixed YHL6381 (h+) and WT (h−) cultures in YEPME mating medium were streaked on YEPME solid mating medium to form individual colonies. The cells from individual colonies were inoculated into 5 ml of YEPD liquid medium and cultured at 30°C and 220 rpm until the OD600 reached 2.0. YHL6381 (h+) or WT (h−) cells were also separately cultured in 5 ml YEPD liquid medium as controls. Genomic DNA extracted from different types of cells was used for PCR amplification using primers (see Table S2 in the supplemental material) specific for M and P mating factor genes at the Mat1 locus (27).
Microarray analysis using yeast gene chip.
For transcriptome analysis of nonsexual flocculation, haploid rpl32-1Δ rpl32-2Δ cells cultured in YEPD medium were used as experiment groups whereas haploid WT cells cultured in YEPD medium were used as a control. For transcriptome analysis of sexual flocculation, diploid YHL6381/WT cells cultured and induced in YEPME mating medium were used as experiment groups, and mixed haploid WT and YHL6381 cells cocultivated in YEPD medium were used as a control. The above-described cells were harvested by centrifugation and directly frozen and stored at −80°C. For RNA isolation, 5 × 108 cells were disrupted in the presence of liquid nitrogen using a mortar and pestle. RNA isolation, cDNA synthesis, and microarray analysis were done as a fee-based service by Gene-BioTech, Shanghai, China. Microarray analysis was replicated three times using cells from independent biological samples. GeneChip Yeast Genome 2.0 arrays (Affymetrix, Santa Clara, CA) were used, and the biotin-labeled cDNA hybridization products were stained by the use of streptavidin, R-phycoerythrin (SAPE) conjugate. Arrays were scanned after hybridization, and data were collected by the use of a Scanner 3000 7G system (Affymetrix). All 15 raw expression files were normalized using a GC-RMA processor. All normalized expression data were analyzed using the PARTEK genomic suite 6.5 software tool. A t test was applied to detect differences in gene expression between each experimental group and the control group. Two criteria were used to determine whether a gene was differentially expressed: fold change of ±1.2 and P value < 0.05 (using a two-tailed distribution). The value of 1.2 is an accepted cutoff which, with statistical significance, is likely to be validated by real-time PCR (28). Gene ontology and cluster analysis was performed by the use of the dChip software tool.
Microarray data accession numbers.
All the microarray data from these independent fission yeast samples were divided into two sets, a nonsexual flocculation series and a sexual flocculation series, and have been uploaded into the NCBI Gene Expression Omnibus (GEO) database. The GEO accession number for the nonsexual flocculation series is GSE43248, and the GEO accession number for the sexual flocculation series is GSE43250.
RESULTS
Deletion of rpl32 genes reduces the ribosome level and induces nonsexual flocculation in haploid mutant cells.
RPL32 proteins in S. pombe are encoded by two duplicated homologue genes, rpl32-1 (SPB16C6.11) and rpl32-2 (SPAC3H5.10). Deletion mutants were created for individual genes from the wild-type (WT) SPQ01 strain (h−) using the gene replacement method (Fig. 1A) and were confirmed using PCR (Fig. 1B). QPCR analysis showed that the expression of rpl32-1 in rpl32-1Δ cells and the expression of rpl32-2 in rpl32-2Δ cells were completely abolished (Fig. 1C). The total reductions of rpl32 (rpl32-1 rpl32-2) transcript levels were 35.9% and 46.9% in rpl32-1Δ and rpl32-2Δ cells, respectively, compared to WT cells. Western blot analysis also showed a significant decrease in the total amount of RPL32 protein in each deletion mutant (Fig. 1D), consistent with the QPCR results. Reduction of RPL32 protein levels affected ribosome assembly and structure (Fig. 1E). Compared to the WT strain, the rpl32-1Δ and rpl32-2Δ deletion mutants both showed a lowered accumulation of free 60S subunits. This ribosomal subunit imbalance led to 31.1% and 27.8% reductions, respectively, in overall ribosome levels (80s monosome and polysomes) in rpl32-1Δ and rpl32-2Δ cells compared to WT cells (Fig. 1E).
Fig 1.
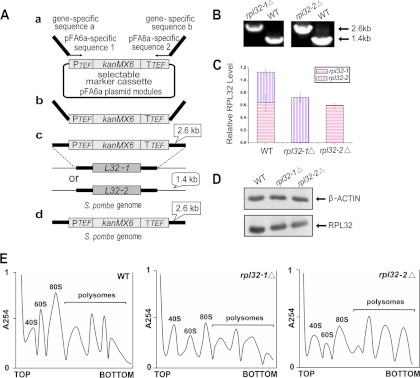
Deletion of rpl32 genes reduced ribosome levels in haploid cells. (A) Deletion of rpl32 paralogous genes in S. pombe using a PCR-based approach with long DNA tracts of flanking homology to the target locus. (B) Deletion mutants were confirmed by PCR with genomic DNA as a template and locus-specific primers flanking the target sites. (C) QPCR quantification of rpl32-1 and rpl32-2 transcript abundance in deletion mutants and WT cells cultured in YEPD medium. beta-actin was used to standardize transcript levels of paralog rpl32-1 and rpl32-2. (D) Western blot analysis of RPL32 in deletion mutants and WT cells cultured in YEPD medium. (E) Polysome profiles of deletion mutants and WT cells cultured to the late exponential phase (OD600 of 5.0) in YEPD medium. A total of 15 A260 units of extracts from each culture was analyzed on 5% to 50% sucrose gradients centrifuged for 4 h at 44,000 rpm (25).
rpl32-1Δ and rpl32-2Δ cells grew much slower, reaching only half of the growth rate of WT cells (Fig. 2A). Surprisingly, when the deletion mutant cells were cultured in a nutrient-rich YEPD medium, they aggregated to form flocculations (Fig. 2B). Flocculation was not observed in WT cells grown in the same medium. Quantitative analysis showed that WT cells in liquid culture came out of the medium slowly with 10.4% sedimentation after 10 min of settling, while rpl32-1Δ and rpl32-2Δ cells showed 48.5% and 49.9% sedimentation, respectively (Fig. 2C). Thus, we cloned rpl32-1 and rpl32-2 genes into the pESP3 expression plasmid and transformed pESP3-rpl32-1 and pESP3-rpl32-2 into rpl32-1Δ and rpl32-2Δ deletion mutants, respectively. Deletion mutant strains, deletion-rescued strains, and WT cells harboring empty vector (as a control) were cultured on EMM2 minimal medium until the stationary phase was reached. Cells were collected from the EMM2 culture, resuspended in the same volume of YEPD medium, and cultured for 3 h. As shown in Fig. 2D, the rpl32-1Δ and rpl32-2Δ cells still showed flocculation whereas no flocculation was seen in rpl32-1 rpl32-1Δ, rpl32-2 rpl32-2Δ, and WT cell cultures, confirming further that the nonsexual flocculation in rpl32-1Δ or rpl32-2Δ cells is caused by rpl32-1 or rpl32-2 deletion.
Fig 2.
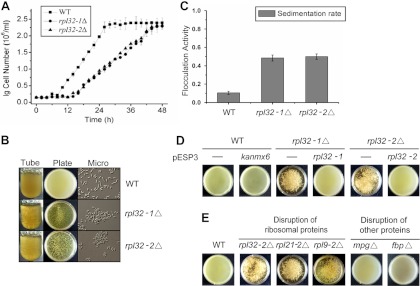
Deletion of rpl32 or some other ribosome protein genes induced nonsexual flocculation. (A) Growth curves of WT, rpl32-1Δ, and rpl32-2Δ cells in YEPD medium. (B) Flocculation of cell cultures of WT, rpl32-1Δ, and rpl32-2Δ strains which were cultured in YEPD medium. (C) Flocculation activity of WT, rpl32-1Δ, and rpl32-2Δ cells cultured in YEPD medium. The flocculation activity was determined by measuring the sedimentation rate as the optical density (OD600) of the top layer of the cell suspension after 10 min of settling. (D) Flocculation of rpl32-1Δ/pESP3 and rpl32-2Δ/pESP3 deletion mutants, rpl32-1Δ/pESP3-rpl32-1 and rpl32-2Δ/pESP3-rpl32-2 complementation strains, and WT/pESP3-kanmx6 and WT/pESP3 strains. Cells were first cultured in EMM2 minimal medium until the stationary phase was reached and then transferred into the same volume of YEPD medium and cultured for 3 h. (E) Flocculation of WT, rpl32-2Δ, rpl21-2Δ, rpl9-2Δ, mpgΔ, and fbpΔ deletion mutants in petri dishes. Images were taken after these cell cultures were placed in petri dishes for 10 min; data represent the means ± standard errors of the results of three independent experiments.
To eliminate the possibility that the flocculation may be caused by the kanmx6 antibiotic resistance gene introduced during the mutant generation as a selection marker (G418 resistance), we expressed the kanmx6 gene in the SPQ01 strain. No flocculation was observed in this recombinant strain expressing the kanmx6 gene (Fig. 2D). Further, we randomly selected two other duplicated ribosomal protein genes, rpl21-2 (SPAC959.08) and rpl9-2 (SPCC613.06), and two nonribosomal protein genes, the mannose-1-phosphate guanyltransferase gene, or mpg (SPBC13G1.02), and the fructose-2,6-bisphosphate 2-phosphatase gene, or fbp (SPAC732.02c), and generated deletion mutants for these genes. rpl21-2Δ and rpl9-2Δ deletion mutants also showed flocculation in YEPD medium whereas mpgΔ and fbpΔ deletion mutants did not (Fig. 2E), suggesting that the nonsexual flocculation was not caused specifically by reduction of the RPL32 protein level but was rather a general response to the reduction of the overall ribosome level.
Sexual flocculation fission yeast cells display a reduction of ribosomal protein expression and ribosomal biogenesis during mating.
Since flocculation in the fission yeast S. pombe is an essential step required for mating and ascospore formation in sexual reproduction (2, 29), we examined whether sexually flocculating yeast cells also exhibited a reduction of ribosomal protein expression and ribosomal biogenesis during mating. The WT S. pombe SPQ01 strain used in this study is a haploid strain of the h− mating type, so an h+ strain, YHL6381, was used as the mating partner for the WT strain. We used YEPD medium, which is YEG medium (29) plus peptone, as a non-flocculation-inducing medium, and YEPME mating medium, in which malt extract containing ∼50% maltose (17) replaced glucose in YEPD, as a flocculation-inducing medium (16).
We first established a sexually flocculating cell system for S. pombe. When haploid WT and YHL6381 cells precultured separately in YEPD medium were mixed at a 1:1 cell number ratio and transferred into YEPME mating medium, flocculation occurred after 90 min of inoculation (Fig. 3A, lower panel, column 3), whereas when the same mixed cells were transferred into YEPD medium, no flocculation was observed (Fig. 3A, upper panel, column 3), indicating that it was only when they were cocultured in YEPME mating medium that heterothallic strains of S. pombe, h− and h+, were able to be sexually coflocculated. When haploid YHL6381 and WT cells precultured separately in YEPD medium were transferred into YEPME mating medium for further individual cultivation, flocculation did not occur either (Fig. 3A, lower panel, columns 1 and 2), indicating that after being cultured separately in YEPME mating medium, the heterothallic strains, h− and h+, were nonflocculent. When WT cells that overexpressed rpl32-1 or rpl32-2 as a consequence of harboring the constructs of pESP3-rpl32-1 and pESP3-rpl32-1 were, respectively, mixed with YHL6381 cells and cocultured in YEPME mating medium, flocculation still took place, indicating that high levels of RPL32 alone did not interfere with sexual flocculation (Fig. 3A, lower panel, columns 4 and 5). When haploid YHL6381 and WT cells were precultured separately in the YEPME mating medium first and then the two culture broths were mixed together, no flocculation was observed (Fig. 3A, column 6), indicating that heterothallic haploid cells, in the absence of opposite mating type cells, did not trigger flocculation even in YEPME mating medium. We were able to amplify only a 729-bp M factor DNA band from the haploid SPQ01 cells and a 987-bp P factor DNA band from the YHL6381 cells, while both 729-bp and 987-bp DNA bands were amplified from each single colony isolated on the plate from YHL6381 (h+)/WT (h−) cells cocultivated in the YEPME mating medium (Fig. 3B), further proving that mating had occurred and that the cells in those colonies were h+/h− diploid.
Fig 3.
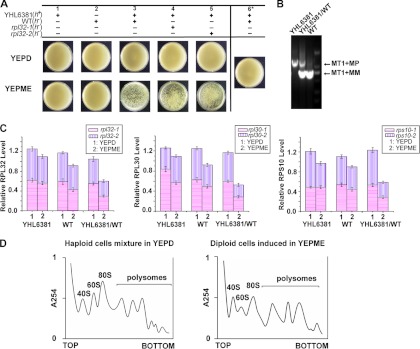
Expression of ribosome protein genes and ribosome levels were reduced in sexually flocculating yeast cells. (A) Sexual flocculation analysis of different combinations of cultures for comparisons between heterothallic haploid WT (h−) and YHL6381 (h+) cells. Images were taken after these cell cultures were placed in petri dishes for 10 min. (B) Diploid cells formed from mating were confirmed by PCR. MT1+MP, MP gene located in mat1; MT1+MM, MM gene located in mat1. (C) QPCR quantification of total rpl32, rpl30, and rps10 transcripts in haploid WT (h−) and YHL6381 (h+) cells cultured separately in YEPME mating medium compared with those in YEPD medium, and total rpl32, rpl30, and rps10 transcripts in the flocculating diploid cells formed in YEPME mating medium compared to those in mixed haploid WT (h−) and YHL6381 (h+) cells cocultivated in YEPD medium. beta-actin was used to standardize transcript levels. (D) Comparison of polysome profiles between the flocculating diploid cells cultured in YEPME mating medium and the nonflocculating haploid cells cocultivated in YEPD medium to the late exponential phase (OD600 of 5.0). A total of 15 A260 units of extracts from each culture was analyzed on 5% to 50% sucrose gradients centrifuged for 4 h at 44,000 rpm (25).
The QPCR analysis showed that the transcript levels of rpl32 genes in nonflocculating haploid YHL6381 (h+) and WT (h−) cells grown in YEPME mating medium were reduced by 12.1% and 23.5%, respectively, compared to the same type of haploid cells grown in YEPD medium. Interestedly, the overall transcript levels of rpl32 genes in the nonflocculating mixed cells of haploid YHL6381 (h+) and WT (h−) cells cocultured in YEPD medium were also reduced by 16.3% and 10.8%, respectively, compared to those of the haploid YHL6381 (h+) and WT (h−) cells cultured separately in the same medium. However, the flocculating diploid YHL6381/WT (h+/h−) cells formed by being cocultured in YEPME mating medium showed a 42.1% decrease in the total rpl32 transcript levels compared to mixed haploid cells of the YHL6381 (h+) and WT (h−) strains cocultivated in YEPD medium (Fig. 3C). We also studied the expression of other ribosomal protein genes. As shown in Fig. 3C, the total transcript levels of the rpl30 and rps10 paralogous genes were reduced by only approximately 13% to 23% in haploid YHL6381 (h+) and WT (h−) cells separately grown in YEPME mating medium compared to the same haploid cells grown in YEPD medium. Similarly, the overall transcript levels of rpl30 and rps10 paralogous genes in the nonflocculating mixed cells of the haploid YHL6381 (h+) and WT (h−) strains cocultured in YEPD medium were also decreased only by 6% and 11%, respectively, compared to those of the haploid YHL6381 (h+) and WT (h−) cells cultured separately in the same medium, while in diploid YHL6381/WT (h+/h−) cells grown in YEPME mating medium they were greatly (approximately 40% to 55%) reduced compared to the haploid cells from a mixed YHL6381 (h+) and WT (h−) culture cogrown in YEPD medium. Polyribosomal analysis demonstrated that flocculating diploid YHL6381/WT (h+/h−) cells grown in YEPME mating medium showed a 37.2% decrease in the total ribosome level compared to the haploid cells from a mixed YHL6381 (h+) and WT (h−) culture cogrown in YEPD medium (Fig. 3D). These results indicated a general trend of reduction in the ribosomal protein gene expression level as well as in the ribosome level during sexual flocculation, as shown in nonsexually flocculating rpl32-1Δ and rpl32-2Δ cells.
Transcriptome analysis indicates that similar molecular mechanisms are involved in nonsexual and sexual flocculation in fission yeast.
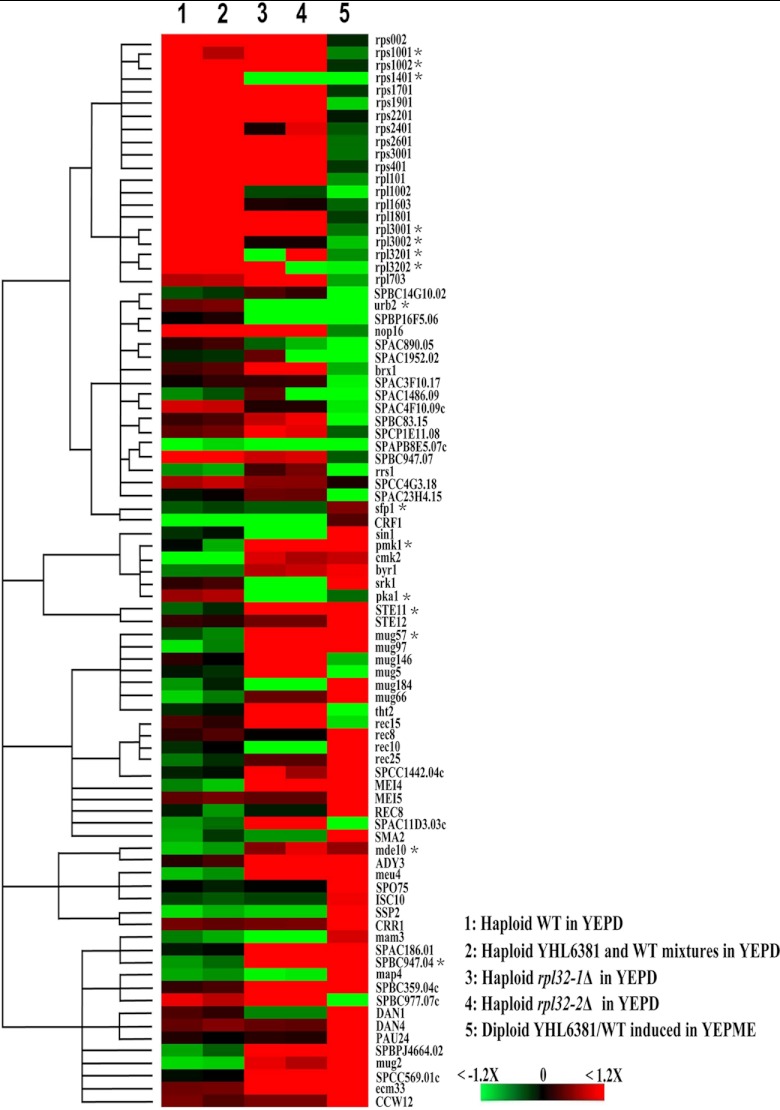
Since both nonsexual and sexual flocculations are associated with the reduction of ribosomal protein gene expression and ribosome level, we wondered whether nonsexual and sexual flocculation shared similar molecular mechanisms. Thus, we compared the transcriptome of nonsexually flocculating haploid rpl32-1Δ and rpl32-2Δ cells to that of nonflocculating haploid WT SPQ01 cells and compared the transcriptome of sexually flocculating diploid YHL6381/WT (h+/h−) cells cocultured and induced in YEPME mating medium to that of nonflocculating mixed haploid cells of YHL6381 (h+) and WT (h−) cocultivated in YEPD medium. Figure 4 presents the comparative transcriptomic patterns of 85 genes which showed a change in expression of over 1.2-fold (P < 0.05) between flocculating and nonflocculating cells. As shown in Fig. 4, the transcription pattern of nonflocculating mixed haploid cells of YHL6381 (h+) and WT (h−) strains was almost as the same as that of nonflocculating WT cells, while the transcription pattern of nonsexually flocculating haploid rpl32-1Δ and rpl32-2Δ cells was similar to that of sexually flocculating diploid YHL6381/WT (h+/h−) cells. Most of genes which were minimally expressed in nonflocculating cells were upregulated in expression level in both nonsexually and sexually flocculating cells, while some parts of genes which were highly expressed in nonflocculating cells were downregulated in both nonsexually and sexually flocculating cells and the other parts of those genes were downregulated only in sexually flocculating cells.
Fig 4.
Transcriptomic patterns of haploid WT, rpl32-1Δ, and rpl32-2Δ cells cultured in YEPD medium, diploid YHL6381/WT cells induced in YEPME mating medium, and mixed haploid WT and YHL6381 cells cocultivated in YEPD medium. The color panel indicates relatively increased (red), decreased (green), and unchanged (black) transcription levels for 85 genes with changes in expression of at least 1.2-fold in flocculating cells. Cutoffs of 1.2-fold change and statistical significance of P < 0.05 were used. *, changes in the transcript levels of these randomly selected genes were confirmed by QPCR.
From the supporting information presented in Table S1 in the supplemental material, we found that 20 different ribosomal protein genes, including rpl32-1 and rpl32-2, showed reduced expression in sexually flocculating diploid YHL6381/WT (h+/h−) cells relative to nonflocculating mixed haploid cells of YHL6381 (h+) and WT (h−) strains cocultivated in YEPD medium and that only 4 of those genes showed reduced expression in both nonsexually flocculating haploid rpl32-1Δ and rpl32-2Δ mutant cells. And 17 ribosomal biogenesis genes showed reduced expression in sexually flocculating diploid cells; among those, only 2 genes showed decreased expression in both rpl32-1Δ and rpl32-2Δ cells. In contrast, expression of sfp1 and crf1, both which are involved in repression of ribosomal biogenesis gene expression (GeneDB), was increased in sexually flocculating diploid YHL6381/WT (h+/h−) cells. However, expression of these two genes was not affected in the nonsexually flocculating haploid cells. It was noticed that rpl21-2 and rpl9-2, deletion of which resulted in nonsexual flocculation of haploid cells (Fig. 2E), were not included in the above-mentioned 20 downregulated ribosomal protein genes.
We found that expression of seven regulatory factor genes (STE11, pmk1, cmk2, byr1, srk1, STE12, and sin1) in the mitogen-activated protein kinase (MAPK) pheromone response pathway (30) was upregulated in sexually flocculating diploid YHL6381/WT (h+/h−) cells and that four of those genes (STE11, pmk1, cmk2, and byr1) were also upregulated in nonsexually flocculating rpl32-1Δ and rpl32-2Δ haploid cells. Moreover, expression of the pka1 gene which encodes a key factor in the cyclic AMP (cAMP)/protein kinase A (PKA) pathway was downregulated in both nonsexually and sexually flocculating cells. Reduced expression of pka1 is often accompanied by increased expression of STE11, which is a key regulatory factor in sexual flocculation (31). Among the downstream genes that are regulated by STE11, 12 genes were upregulated in sexually flocculating diploid cells (YHL6381/WT) and 9 genes were upregulated in nonsexually flocculating rpl32-1Δ and rpl32-2Δ haploid cells. Four of these upregulated genes, mug57, mug97, mei4, and SPCC1442.04c, were common to both sexually and nonsexually flocculating cells. In addition, seven genes involved in sexual spore formation and differentiation were upregulated in the sexual flocculation diploid YHL6381/WT cells, and three genes, meu14, mde10, and ADY3, were also upregulated in nonsexually flocculating rpl32-1Δ and rpl32-2Δ haploid cells.
As shown in Table S1 in the supplemental material, 12 putative cell wall adhesin genes, including known adhesin protein Mam3 and Map4 genes, were upregulated in sexually flocculating diploid YHL6381/WT cells, and 8 putative cell wall adhesin genes were upregulated in nonsexually flocculating haploid rpl32-1Δ and rpl32-2Δ cells; of those, 6 were also upregulated in sexually flocculating cells.
We randomly chose 20 genes with an average transcription fold change > 1.2 in microarray analysis between flocculating and nonflocculating cells from different catalogs for QPCR analysis and proved that these differentially expressed genes really were upregulated or downregulated in flocculating cells compared to nonflocculating cells, consistent with the results from microarray analysis (see Fig. 4 legend; see also Table S1 in the supplemental material).
DISCUSSION
This report shows that deletion of ribosomal proteins as well as a corresponding reduction of overall ribosome levels leads to a nonsexual flocculation of fission yeast. A downregulation of ribosomal protein synthesis and reduction of the ribosomal level were also detected in sexually flocculating diploid YHL6381/WT (h+/h−) cells. A logical interpretation is that nonflocculation caused by deletion of ribosomal proteins may exploit the regulation of the ribosomal level in the onset of sexual flocculation. The transcriptome analysis provided additional data to support this hypothesis. The transcript levels of 20 ribosomal protein genes and 17 ribosomal biogenesis genes in the yeast genome were reduced during sexual flocculation, while expression levels of two known ribosome synthesis repressor genes were increased in sexually flocculating diploid cells. Thus, in sexual flocculation cells, reduction of the ribosome level was caused by extensive downregulation of expression of ribosomal protein genes and ribosomal biogenesis genes and by upregulation of the ribosomal biogenesis gene repressors. This point is supported by the fact that sexual flocculation could not be blocked by overexpression of one specific ribosomal protein alone such as RPL32-1 or RPL32-2 in mating cells (Fig. 3A, lower panel, columns 4 and 5). It is known that ribosome synthesis depends on a specific ratio of each of the ribosome proteins and rRNAs in the ribosome (24, 25). Deletion of individual ribosome protein genes broke this balance between ribosome proteins and therefore led to a reduction in the overall ribosome level, as indicated by the fact that polyribosome analysis (see Results) triggered a nonsexual flocculation of haploid cells. Furthermore, rpl21-2 and rpl9-2, deletion of which resulted in nonsexual flocculation of haploid cells (Fig. 2E), were not included among the 20 downregulated ribosomal protein genes in sexually flocculating cells (see Table S1 in the supplemental material), further implying that as long as ribosomal protein levels as well as the corresponding ribosomal levels were reduced, flocculation might be triggered in haploid cells, as occurred in sexual flocculation cells. Therefore, deleting ribosomal protein genes may lead to nonsexual flocculation as a consequence of mimicking the reduction of the ribosomal level in the sexual flocculation process.
However, though the haploid YHL6381 (h+) and WT (h−) cells grown in YEPME mating medium showed overall rpl32 gene transcription that was reduced by 12.1% and 23.5%, respectively, compared to the levels seen with the same haploid cells grown in YEPD medium, and overall rpl32 gene transcription of the mixed haploid cells of YHL6381 (h+) and WT (h−) cocultured in YEPD medium also decreased by 16.3% and 10.8%, respectively, compared to the transcription levels seen with the haploid YHL6381 (h+) and WT (h−) cells cultured separately in the same medium, none of these cells formed flocculates. This may be related to the extent of reduction of the ribosomal protein transcript level in fission yeast, since the flocculating diploid YHL6381/WT (h+/h−) cells formed in YEPME mating medium showed a 42.1% decrease in the overall rpl32 transcript level compared to nonflocculating mixed haploid YHL6381 (h+) and WT (h−) cells cocultivated in YEPD medium. This idea is further supported by the nonsexual flocculation of haploid rpl32-1Δ and rpl32-2Δ deletion mutant cells grown in YEPD medium, which showed 35.9% and 46.9% reductions in the overall rpl32 transcript level, respectively, compared to nonflocculating WT cells grown in the same medium. Therefore, flocculation may occur only when the reduction of ribosomal protein levels is below a certain threshold value (probably below 35%, based on the present study); that is, triggering of flocculation may be dependent on the extent of the reduction of the transcript level of the ribosome proteins.
Current data suggest that, once past the ribosome checkpoint level, nonsexually and sexually flocculating cells share some common mechanism for initiation of flocculation. First, deleting of either of the ribosomal protein L32 paralogs upregulated the expression level of putative adhesin genes in nonsexual flocculating cells, while expression levels of putative adhesin genes were enhanced in sexual flocculating cells also, accompanied by a decrease in the ribosomal protein expression level. And six upregulated adhesin genes were shared by the nonsexually and sexually flocculating cells. These results suggest that a decrease in expression of ribosomal proteins stimulates expression of adhesin proteins, leading to flocculation. Second, both nonsexually flocculating haploid cells of rpl32 deletion mutants and sexually flocculating diploid cells showed that some major components in the MAPK pheromone response pathway, such as STE11, Mei4, Byr1, and Pka1, were significantly upregulated (STE11, Mei4, and Byr1) or downregulated (Pka1) at the transcript level. In S. pombe, STE11 is a key transcription factor in sexual reproduction (31); Mei4 is a meiosis-specific transcription factor that plays an important role in the progression of meiosis and sporulation (32); byr1 resembles byr2 in many respects, overexpression of byr2 increases cell agglutinability, and interfering forms of byr2 block the increased agglutinability (33); and a defect of the pka1 gene readily initiates sexual development in rich medium (34, 35). It has been reported that the MAPK pheromone response pathway can trigger adhensin FLO11-mediated cell surface flocculation in S. cerevisiae cells, such as pseudohyphal development in diploids, or filament and invasive growth or mating in haploids, in response to stress and nutrient limitation (36–38). Taking these data together, reductions of ribosomal protein levels may trigger adhensin synthesis for flocculation and mating via the STE11-mediated MAPK pheromone response pathway. Third, genes involved in meiosis and spore formation, such as meu14 (39), mde10 (40), and ADY3 (41), were also upregulated in both nonsexually and sexually flocculating cells. Generally, when carbon or nitrogen sources are low, S. pombe initiates sexual development, and different haploid mating types (h+ and h−) conjugate (flocculate), producing diploid zygotes. The zygotes enter meiosis, form haploid tetrads, and eventually sporulate (42, 43). However, in our experiments, haploid deletion mutants, rpl32-1Δ and rpl32-2Δ cells, were grown separately in YEPD rich medium and they were not induced by pheromones or cells of the opposite mating type, while they exhibited high expression of meiosis and spore formation genes, and formed nonsexual flocculate such as occurs in sexual mating. Since the STE11-mediated MAPK pheromone response pathway is also activated in the rpl32 deletion mutants, we conclude that ribosome level reduction caused by deletion of ribosomal protein can initiate a series of physiological processes related to the sexual reproduction which mimic a trigger function of the ribosomal level in sexual flocculation and mating.
Why do fission yeast utilize the ribosome level as a trigger to initiate flocculation for control of the sexual mating process? The answer is that yeast sexual reproduction is a cellular response to unhealthy environments (44). We found that heterothallic strains of S. pombe, WT (h−), and YHL6381 (h+) were able to be sexually coflocculated, shortly after they were cocultivated in YEPME mating medium, which contains only low concentrations of maltose and no glucose, while they were not in YEPD medium, which contains a high concentration of glucose. However, if S. pombe WT (h−) and YHL6381 (h+) strains were to be cocultivated in YEPD medium for 24 h when the carbon resource was depleted, they would sexually flocculate (data not shown). Apparently, the onset of sexual flocculation is dependent on a low sugar concentration in culture media, just as seen in S. cerevisiae (9, 16). It has been reported that ribosomal protein synthesis and ribosome assembly in yeast cells are affected by nutrient availability in the environment (45–48). Nutrient deficiency may reduce the synthesis of amino acids, which results in the accumulation of uncharged tRNA, eventually leading to the inhibition of transcription of ribosomal protein. In consequence, the ribosome level is decreased in cells. This inhibition is called the “stringent response” (49–51). Bastidas et al. (2009) even reported that, in Candida albicans, inhibition of TOR signaling by rapamycin (which mimics nutrient limitation) led to adhesin expression and flocculation of cells (52). Taking these results together, we propose that, by linking the ribosome level to the onset of flocculation, yeast cells can sense environmental changes and initiate sexual reproduction on time.
Supplementary Material
ACKNOWLEDGMENTS
This research is supported by the National Science Foundation of China (no. 30670025 and no. 31070060) and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Footnotes
Published ahead of print 25 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00321-12.
REFERENCES
- 1. Egel R. 1971. Physiological aspects of conjugation in fission yeast. Planta 98: 89–96 [DOI] [PubMed] [Google Scholar]
- 2. Johnson BF, Walker T, Miyata M, Miyata H, Calleja GB. 1987. Sexual co-flocculation and asexual self-flocculation of heterothallic fission yeast cells (Schizosaccharomyces pombe). Can. J. Microbiol. 33: 684–688 [Google Scholar]
- 3. Miyata M, Doi H, Miyata H, Johnson BF. 1997. Sexual co-flocculation by heterothallic cells of the fission yeast Schizosaccharomyces pombe modulated by medium constituents. Antonie van Leeuwenhoek 71: 207–215 [DOI] [PubMed] [Google Scholar]
- 4. Miyata M, Matsuoka M, Inada T. 1997. Induction of sexual co-flocculation of heterothallic fission yeast (Schizosaccharomyces pombe) cells by mating pheromones. J. Gen. Appl. Microbiol. 43: 169–174 [DOI] [PubMed] [Google Scholar]
- 5. Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. 2007. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 71: 282–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sharifmoghadam MR, Bustos-Sanmamed P, Valdivieso MH. 2006. The fission yeast Map4 protein is a novel adhesin required for mating. FEBS Lett. 580: 4457–4462 [DOI] [PubMed] [Google Scholar]
- 7. Linder T, Gustafsson CM. 2008. Molecular phylogenetics of ascomycotal adhesins—a novel family of putative cell-surface adhesive proteins in fission yeasts. Fungal. Genet. Biol. 45: 485–497 [DOI] [PubMed] [Google Scholar]
- 8. Verstrepen KJ, Reynolds TB, Fink GR. 2004. Origins of variation in the fungal cell surface. Nat. Rev. Microbiol. 2: 533–540 [DOI] [PubMed] [Google Scholar]
- 9. Verstrepen KJ, Derdelinckx G, Verachtert H, Delvaux FR. 2003. Yeast flocculation: what brewers should know. Appl. Microbiol. Biotechnol. 61: 197–205 [DOI] [PubMed] [Google Scholar]
- 10. Guo B, Styles CA, Feng QH, Fink GR. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. U. S. A. 97: 12158–12163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tanaka N, Awai A, Bhuiyan MSA, Fujita K, Fukui H, Takegawa K. 1999. Cell surface galactosylation is essential for nonsexual flocculation in Schizosaccharomyces pombe. J. Bacteriol. 181: 1356–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim KH, Cho YM, Kang WH, Kim JH, Byun KH, Park YD, Bae KS, Park HM. 2001. Negative regulation of filamentous growth and flocculation by Lkh1, a fission yeast LAMMER kinase homolog. Biochem. Biophys. Res. Commun. 289: 1237–1242 [DOI] [PubMed] [Google Scholar]
- 13. Kang WH, Park YH, Park HM. 2010. The LAMMER kinase homolog, Lkh1, regulates Tup transcriptional repressors through phosphorylation in Schizosaccharomyces pombe. J. Biol. Chem. 285: 13797–13806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsuzawa T, Morita T, Tanaka N, Tohda H, Takegawa K. 2011. Identification of a galactose-specific flocculin essential for non-sexual flocculation and filamentous growth in Schizosaccharomyces pombe. Mol. Microbiol. 82: 1531–1544 [DOI] [PubMed] [Google Scholar]
- 15. Wang J, Yuan S, Jiang SB. 2006. The ribosomal protein L32-2 (RPL32-2) of S. pombe exhibits a novel extraribosomal function by acting as a potential transcriptional regulator. FEBS Lett. 580: 1827–1832 [DOI] [PubMed] [Google Scholar]
- 16. Sampermans S, Mortier J, Soares EV. 2005. Flocculation onset in Saccharomyces cerevisiae: the role of nutrients. J. Appl. Microbiol. 98: 525–531 [DOI] [PubMed] [Google Scholar]
- 17. Crandall M, Lawrence LJ. 1980. Sporulation in Hansenula wingei is induced by nitrogen starvation in maltose-containing media. J. Bacteriol. 142: 276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stratford M. 1989. Yeast flocculation: calcium specificity. Yeast 5: 487–496 [Google Scholar]
- 19. Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Philippsen P, Pringle JR. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- 20. Keeney JB, Boeke JD. 1994. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136: 849–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang Y, McGillicuddy E, Weindel M, Dong S, Maraia RJ. 2003. The fission yeast TFIIB-related factor limits RNA polymerase III to a TATA-dependent pathway of TBP recruitment. Nucleic Acids Res. 31: 2108–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Olmsted JB. 1981. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J. Biol. Chem. 256: 11955–11957 [PubMed] [Google Scholar]
- 23. Powers T, Walter P. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bachand F, Silver PA. 2004. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 23: 2641–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rotenberg MO, Moritz M, Woolford JL. 1988. Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Gene Dev. 2: 160–172 [DOI] [PubMed] [Google Scholar]
- 26. Kraig E, Haber JE. 1980. Messenger ribonucleic acid and protein metabolism during sporulation of Saccharomyces cerevisiae. J. Bacteriol. 144: 1098–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Willer M, Hoffmann L, Styrkarsdottir U, Egel R, Davey J, Nielsen O. 1995. Two-step activation of meiosis by the mat1 locus in Schizosaccharomyces pombe. Mol. Cell. Biol. 15: 4964–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shehadeh LA, Yu K, Wang L, Guevara A, Singer C, Vance J, Papapetropoulos S. 2010. SRRM2, a potential blood biomarker revealing high alternative splicing in Parkinson's disease. PLoS One 5: e9104 doi:10.1371/journal.pone.0009104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Calleja GB, Johnson BF. 1971. Flocculation in a fission yeast: an initial step in the conjugation process. Can. J. Microbiol. 17: 1175–1177 [DOI] [PubMed] [Google Scholar]
- 30. Muthuvijayan V, Marten MR. 2004. In silico reconstruction of nutrient-sensing signal transduction pathways in Aspergillus nidulans. In Silico Biol. 4: 605–631 [PubMed] [Google Scholar]
- 31. Sugimoto A, Irno Y, Maeda T, Watanabe Y, Yamamoto M. 1991. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Gene Dev. 5: 1990–1999 [DOI] [PubMed] [Google Scholar]
- 32. Horie S, Watanabe Y, Tanaka K, Nishiwaki S, Fujioka H, Abe H, Yamamoto M, Shimoda C. 1998. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol. Cell. Biol. 18: 2118–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y, Xu HP, Riggs M, Rodgers L, Wigler M. 1991. byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol. Cell. Biol. 11: 3554–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamawaki-Kataoka Y, Tamaoki T, Choe HR, Tanaka H, Kataoka T. 1989. Adenylate cyclases in yeast: a comparison of the genes from Schizosaccharomyces pombe and Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 86: 5693–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kawamukai M, Ferguson K, Wigler M, Young D. 1991. Genetic and biochemical analysis of the adenylyl cyclase of Schizosaccharomyces pombe. Cell. Regul. 2: 155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roberts RL, Fink GR. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8: 2974–2985 [DOI] [PubMed] [Google Scholar]
- 37. van Dyk D, Pretorius IS, Bauer FF. 2005. Mss11p is a central element of the regulatory network that controls FLO11 expression and invasive growth in Saccharomyces cerevisiae. Genetics 169: 91–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Verstrepen KJ, Klis FM. 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60: 5–15 [DOI] [PubMed] [Google Scholar]
- 39. Okuzaki D, Satake W, Hirata A, Nojima H. 2003. Fission yeast meu14+ is required for proper nuclear division and accurate forespore membrane formation during meiosis II. J. Cell Sci. 116: 2721–2735 [DOI] [PubMed] [Google Scholar]
- 40. Nakamura T, Abe H, Hirata A, Shimoda C. 2004. ADAM family protein Mde10 is essential for development of spore envelopes in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 3: 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moreno-Borchart AC, Strasser K, Finkbeiner MG, Shevchenko A, Knop M. 2001. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 20: 6946–6957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nielsen O. 2004. Mating-type control and differentiation, p 281–296 In Egel R. (ed), The molecular biology of Schizosaccharomyces pombe. Springer, Heidelberg, Germany [Google Scholar]
- 43. Yamamoto M, Imai I, Watanabe Y. 1997. Mating and sporulation in Schizosaccharomyces pombe, p 1035–1106 In Pringle JR, Broach JR, Jones EW. (ed), The molecular and cellular biology of the yeast Saccharomyces: life cycle and cell biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 44. Paul SK, Goldar MM, Yakura M, Oowatari Y, Kawamukai M. 2009. Glutamyl tRNA synthetases and glutamic acid induce sexual differentiation of Schizosacchromyces pombe. Biosci. Biotechnol. Biochem. 73: 1339–1347 [DOI] [PubMed] [Google Scholar]
- 45. Warner JR. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437–440 [DOI] [PubMed] [Google Scholar]
- 46. Ju Q, Warner JR. 1994. Ribosome synthesis during the growth cycle of Saccharomyces cerevisiae. Yeast 10: 151–157 [DOI] [PubMed] [Google Scholar]
- 47. Werner-Washburne M, Braun E, Johnston GC, Singer RA. 1993. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 57: 383–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Levy S, Ihmels J, Carmi M, Weinberger A, Friedlander G, Barkai N. 2007. Strategy of transcription regulation in the budding yeast. PLoS One 2: e250 doi:10.1371/journal.pone.0000250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Warner JR, Gorenstein C. 1978. Yeast has a true stringent response. Nature 275: 338–339 [DOI] [PubMed] [Google Scholar]
- 50. Moehle CM, Hinnebusch AG. 1991. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 2723–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Woolford JL, Warner JR. 1991. Genome dynamics, protein synthesis, and energetics, p 587–626 In Pringle JR, Broach JR, Jones EW. (ed), The molecular and cellular biology of the yeast Saccharomyces: life cycle and cell biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 52. Bastidas RJ, Heitman J, Cardenas ME. 2009. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog. 5: e1000294 doi:10.1371/journal.ppat.1000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.