Abstract
The ascomycete Trichoderma reesei is a paradigm for the regulation and production of plant cell wall-degrading enzymes, including xylanases. Four xylanases, including XYN1 and XYN2 of glycosyl hydrolase family 11 (GH11), the GH10 XYN3, and the GH30 XYN4, were already described. By genome mining, we identified a fifth xylanase, XYN5, belonging to GH11. Transcriptional analysis reveals that the expression of all xylanases but xyn3 is induced by d-xylose, dependent on the cellulase and xylanase regulator XYR1 and negatively regulated by the carbon catabolite repressor CRE1. Impairment of d-xylose catabolism at the d-xylose reductase and xylitol dehydrogenase step strongly enhanced induction by d-xylose. Knockout of the l-xylulose reductase-encoding gene lxr3, which connects the d-xylose and l-arabinose catabolic pathways, had no effect on xylanase induction. Besides the induction by d-xylose, the T. reesei xylanases were also induced by l-arabinose, and this induction was also enhanced in knockout mutants in l-arabinose reductase (xyl1), l-arabitol dehydrogenase (lad1), and l-xylulose reductase (lxr3). Induction by l-arabinose was also XYR1 dependent. Analysis of intracellular polyols revealed accumulation of xylitol in all strains only during incubation with d-xylose and accumulation of l-arabitol only during incubation with l-arabinose. Induction by l-arabinose could be further stimulated by addition of d-xylose. We conclude that the expression of the T. reesei xylanases can be induced by both d-xylose and l-arabinose, but independently of each other and by using different inducing metabolites.
INTRODUCTION
Current attempts to use plant biomass for production of advanced biofuels and high-value chemicals in biorefineries have fortified the interest in the enzymatic hydrolysis of its polysaccharide components. Besides cellulose, hemicelluloses can make up to 30% of the plant dry matter, of which xylan is the major hemicellulose polymer in cereals and hardwood. Xylan consists of a β-1,4-linked d-xylose backbone, to which other residues such as l-arabinose, 4-O-methyl-glucuronate, and acetyl side chains can be attached, thus resulting in a wide variety of xylan structures.
The sordariomycete Trichoderma reesei (teleomorph Hypocrea jecorina) is a paradigm for research on cellulases and hemicellulases and used as a producer of these enzymes by various companies (1, 2). With regard to its xylanases, four have been purified and characterized: two members of glycosyl hydrolase family 11 (GH11) (XYN1 and XYN2) (3, 4), the GH10 member XYN3 (5, 6), and the GH30 member XYN4 (7). While the first three are all endo-β-1,4-xylanases, XYN4 is classified as a xylan 1,4-β-xylosidase, because it produces d-xylose as the main end product from xylan. It also displays greater activity toward unsubstituted xylans or acetylated methylglucuronic acid xylans than the GH10 and GH11 xylanases (7).
The formation of the enzymes needed for degradation of cellulose and hemicelluloses in T. reesei is adaptive and occurs only in the presence of an inducer. While the cellulases of T. reesei are known to be coordinately regulated by cellulose, lactose, and the β-1,2-diglucoside sophorose (8, 9), differences in the induction of the xylanases were reported: expression of xyn1 is induced by d-xylose, whereas expression of xyn2 is induced by xylobiose and the cellulase-inducing carbohydrates cellulose and sophorose (10). xyn3 expression was found only in a mutant strain (T. reesei PC-3-7) and was induced only by cellulase inducers and not by d-xylose (11). No data on the expression of xyn4 are available. Xylanase expression by d-xylose is regulated via the transcriptional activator XYR1 or its orthologue XlnR and by general carbon catabolite (de)repression in T. reesei and other fungi, including, e.g., Aspergillus spp., Neurospora crassa, and Fusarium spp., although species-specific adaptations are found (12–18). In T. reesei, xyn1 and xyn2 respond to carbon catabolite repression in different ways (19).
Triggering of expression of polysaccharide-hydrolyzing enzymes is often achieved by different mono- or disaccharides arising from the hydrolysis of the polysaccharide (20). However, whether these compounds or metabolites derived from them are the actual inducers is mostly not known. d-Xylose is converted via d-xylose reductase, xylitol dehydrogenase, and xylulokinase to d-xylulose 5-phosphate to enter the pentose phosphate pathway. The pathway is interconnected to l-arabinose catabolism, which involves an l-arabinose reductase, l-arabitol dehydrogenase, and l-xylulose reductase to form xylitol, the first common intermediate (cf. Fig. 4; reviewed in reference 15). Mach-Aigner et al. (21, 22) studied induction of the xylanase genes xyn1 and xyn2 in T. reesei strains blocked in specific steps in the d-xylose and l-arabinose catabolic pathways. They concluded that the first step in d-xylose catabolism catalyzed by the d-xylose reductase XYL1 is necessary for (full) induction of xyn1 and xyn2 and hypothesized that l-arabitol, formed from d-xylose, would be the true inducer of xylanase expression.
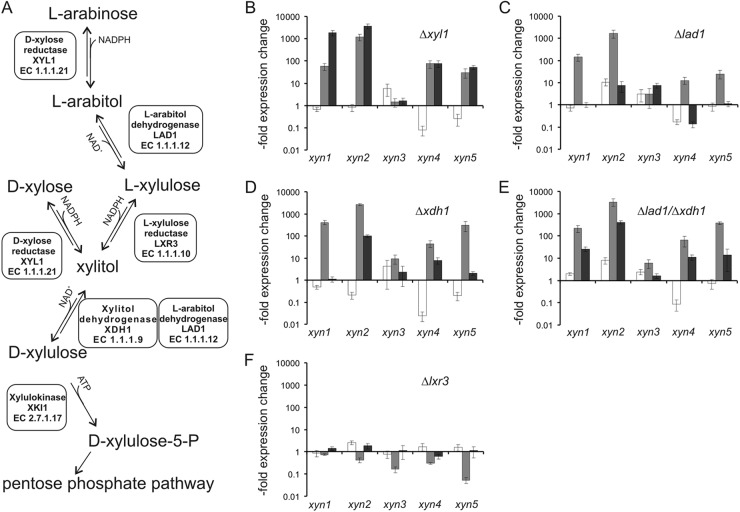
Fig 4.
Xylanase induction by d-xylose in strains with deletions for specific steps of the d-xylose and l-arabinose pathways. (A) Scheme of the d-xylose and l-arabinose catabolic pathways in T. reesei. (B to F) Relative expression levels of the five xylanases (xyn1 to xyn5) in T. reesei strains with deletions of the d-xylose/aldose reductase (Δxyl1) (B), the l-arabitol dehydrogenase (Δlad1) (C), the xylitol dehydrogenase (Δxdh1) (D), both dehydrogenases (Δlad1/Δxdh1) (E), and the l-xylulose reductase (Δlxr3) (F). Strains were placed on medium containing 1 mM d-xylose, and samples were taken 2 h (white bars), 4 h (gray bars), and 6 h (black bars) after addition of the carbon source. Expression values, plotted on a logarithmic scale, are relative to the value for strain QM9414 at the respective time point on inducing medium.
Here we have studied the regulation of all xylanase-encoding genes of T. reesei, using mutants in the d-xylose and l-arabinose catabolic pathways, to obtain a hint toward the inducer of xylanase gene expression. We show that, in contrast to the findings presented above, the absence of XYL1 enhances induction of the inducible xylanolytic enzyme system (XYN1, XYN2, XYN4, and XYN5) by d-xylose. In addition, we show that l-arabinose also induces xylanase transcription but does so independently of d-xylose.
MATERIALS AND METHODS
Strains and cultivation conditions.
T. reesei strain QM9414 (ATCC 26921) (23), which served as the reference strain, and strains with the knockouts Δxyl1 (24), Δlad1, Δxdh1, Δlad1/Δxdh1 (25), Δlxr3 (Metz et al., submitted), Δxyr1 (26), and Δcre1 (27) were precultured for 24 h in 250 ml Mandels-Andreotti (MA) medium (28) containing 1% (wt/vol) glycerol as the sole carbon source on a rotary shaker (250 rpm) at 28°C. Subsequently, mycelia were collected, washed, and transferred to 250 ml MA medium without a carbon source. After 30 min of incubation, the inducing carbon source (d-xylose, xylitol, or l-arabinose) was added to the cultures. Samples of mycelia were taken directly before adding d-xylose and after 2 h, 4 h, and 6 h of induction. As a control, all strains were cultured in MA medium without d-xylose.
Quantification of xylanase gene expression.
The mRNA was extracted following a phenol-chloroform-based approach (29). cDNA was synthesized using a RevertAid H minus first-strand cDNA synthesis kit (Fermentas), following the manufacturer's protocol. Quantitative real-time PCRs (qPCRs) were performed on a Bio-Rad iQ thermal cycler. The reaction mix contained 12.5 μl SYBR green Supermix (Bio-Rad), 8.5 μl pure water (Roth), 1 μl forward primer (160 mM), 1 μl reverse primer (160 mM), and 2 μl of 1:100-diluted template cDNA. Oligonucleotides are listed in Table S1 in the supplemental material, except for the oligonucleotide for xyn1 (22). The tef1 gene (encoding transcription elongation factor 1α) was used as an internal standard. Expression data were evaluated using REST software (30). Reactions were performed in triplicate. Data correspond to at least two biological replicates.
Analysis of phylogeny and evolution.
DNA and protein sequences were visually aligned by using Genedoc (version 2.6) software (31). Phylogenetic trees were constructed by the neighbor-joining method (27), using the computer program MEGA, version 5.0 (32). Unalignable N- and C-terminal regions in the amino acid sequences were omitted from the analyses, and gaps and missing data were pairwise deleted. The pairwise Ka/Ks ratio was determined with the DNASp (version 5.0) program (33). Codon-based Fisher's test and the codon-based Z test implemented in MEGA (version 4.0) (34) were used to directly test the hypotheses of evolutionary models.
Sugar and polyol quantification.
Carbohydrates in the medium were analyzed by high-pressure liquid chromatography essentially as described previously (35) using 5 mM sulfuric acid as the eluent at 40°C. For analysis of the intracellular sugars and polyols, mycelia were collected by centrifugation (3 min, 8,000 rpm), washed with double-distilled H2O (ddH2O), and resuspended in 1 ml of ddH2O. This suspension was heated at 100°C for 10 min and centrifuged (10 min, 14,000 rpm), and the supernatant was used for analysis by gas chromatography (GC), using d-sorbitol as a standard. Samples were vacuum dried and then converted to methylsilyl derivatives by addition of 50% pyridine, 35% hexamethyldisilazane, and 15% (vol/vol) trimethylchlorosilane following incubation at room temperature overnight. Samples were measured in an Agilent 7890A GC system using an HP-5 column (length, 30 m; diameter, 0.32 mm; film thickness, 0.25 μm) with a flame ionization detector (Agilent Technologies, Santa Clara, CA). The temperature program was 100°C for 1 min, followed by a temperature increase of 5°C/min to 220°C and an increase of 35°C/min to 320°C, and the temperature was then kept constant at 320°C for 5 min. The helium flow rate was set to 1.4 ml/min, the injector temperature was 260°C, and the detector temperature was 300°C. Data are related to an intracellular volume of 2.4 ml per g of dry biomass (36).
RESULTS
Identification of a fifth T. reesei xylanase, XYN5.
In order to study the regulation of the complete xylanolytic system of T. reesei, we mined its genome database for eventual yet undescribed xylanases by using a BLASTP search with different fungal xylanases. This led to the identification of, indeed, one further xylanase gene, xyn5, whose deduced protein sequence encoded a third GH11 member. XYN5 is annotated incorrectly as a GH18 family member on the T. reesei genome home page (http://genome.jgi.doe.gov/Trire2/Trire2.info.html) and in GenBank (GenBank accession number EGR44310). To learn its relationship to the two other GH11 members, XYN1 and XYN2, we used it as a query in BLAST search and picked out the 38 best hits (E value, <−100) for phylogenetic analysis by the maximum likelihood method. As can be seen in Fig. 1, the xylanases were grouped into two major clades, one containing T. reesei XYN1 and the other containing T. reesei XYN2. A small clade between them containing xylanases from Myceliophthora thermophila and related proteins could not be safely aligned with either of these two clades, but as this was not relevant to the present investigation, it was not further elucidated. The new XYN5 and its orthologues from T. virens and Trichoderma atroviride formed a sister clade to XYN1 in the respective clade. Interestingly, the XYN1 clade lacked any relationship to the respective species phylogeny and contained enzymes from several Penicillium/Talaromyces spp. It also contained a further clade consisting of Trichoderma virens and T. atroviride GH11 xylanases that were lacking from T. reesei. Since the accumulation of paralogs for this gene family could indicate a selective advantage of these genes for Trichoderma spp., we examined the evolutionary forces driving these gene duplications: to this end we calculated the Ka/Ks ratio for all pairwise combinations of xylanase exons in the XYN1 clade for all three Trichoderma spp. In all cases, we obtained only ratios that were significantly less than 1 with a mean Tajima's D test value of 0.254, implying that nucleotide sequence differences between genes have primarily occurred at synonymous sites. Plotting Ks versus Ka showed that the Ks values for some gene-to-gene comparisons are very high (up to 1.8) and have apparently reached the saturation level (37). These findings suggest that the XYN1 clade evolves by purifying selection, which could be proven by the codon-based Z test and the codon-based Fisher's exact test (data not shown).
Fig 1.
Phylogenetic relationship of T. reesei xylanase XYN5 (GenBank accession number EGR44310) to other fungal xylanases of the GH11 family. A further clade consisting of additional GH11 xylanases lacking in T. reesei but present in T. atroviride and T. virens is marked by a dotted box. The numbers below nodes indicate the bootstrap value. The bar marker indicates the genetic distance, which is proportional to the number of amino acid substitutions. The GenBank accession numbers of the respective proteins are indicated.
XYN1, XYN2, XYN4, and XYN5 are coregulated by d-xylose, XYR1, and CRE1.
In order to identify optimal inducing conditions for the five T. reesei xylanases, we tested three different concentrations (0.5, 1, and 5 mM) of d-xylose in precultivated mycelia of T. reesei over a period of 6 h (data not shown; Fig. 2B). Consistent with earlier reports (8), 1 mM d-xylose provided the highest xylanase transcript levels, whereas 5 mM d-xylose already delayed induction. No induction was observed with xylitol, which was not readily taken up under the present conditions (data not shown).
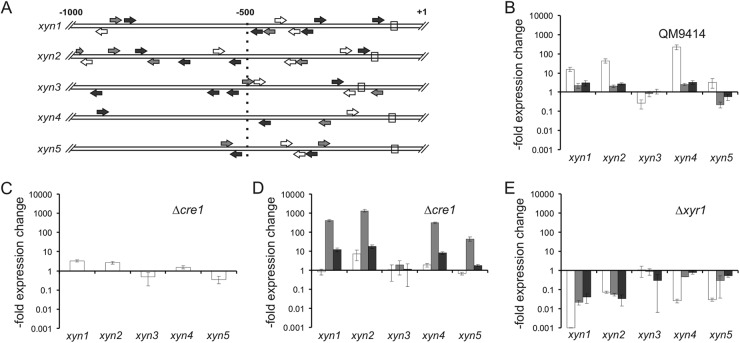
Fig 2.
Regulation of T. reesei xylanase gene expression. (A) Comparative analysis of regulatory motifs present in the different xylanase promoter regions. Consensus sequences for XYR1 (white arrows; 5′-GGCTAA-3′ or 5′-GGGTAA-3′, as in the case of xyn2), HAP2/3/5 (gray arrows; 5′-CCAAT-3′), CRE1 (black arrows; 5′-SYGGRG-3′), and the TATA box (white rectangle) are plotted on a region of 1,000 bp upstream of the start codon. Arrows indicate the orientation of the respective motifs present on either the sense strand or the antisense strand. (B to E) Relative expression levels of the five xylanases in T. reesei QM9414 and strains with deletion of the genes for the carbon catabolite repressor CRE1 (Δcre1) (C and D) or the cellulase and xylanase regulator XYR1 (Δxyr1) (E). Precultivated mycelia were placed on medium with 1 mM d-xylose (B, D, and E; inducing conditions) or without a carbon source (C). Samples were taken 2 h (white bars), 4 h (gray bars), and 6 h (black bars) after addition of the carbon sources. Relative expression values are plotted on a logarithmic scale where 1 indicates the transcription level of the reference strain QM9414 on medium without a carbon source (B) or at the respective time point on inducing medium (C to E).
xyn1 and xyn2 were previously reported to be subject to regulation by the C2H2 carbon catabolite repressor CRE1 via a double-lock mechanism (19, 38) that controls xylanase transcription at two different levels, by direct repression through CRE1, and indirectly through the CRE1-mediated repression of the transcriptional activator XYR1 (39). Binding sites for the carbon catabolite repressor CRE1 are present in all xylanase genes (Fig. 2A), but only in xyn1 they occur as an inverted repeat which has been proposed to be essential for carbon catabolite repression (27). We therefore tested the expression of all five xylanases in a Δcre1 strain: in the absence of an inducer, xyn1, xyn2, and xyn4 were derepressed in the Δcre1 strain (Fig. 2C). Addition of the inducer d-xylose led to a further increase in transcript abundance for xyn1, xyn2, xyn4, and xyn5 (Fig. 2D) compared to that for the parent strain QM9414, indicating that xyn1, xyn2, and xyn4 are subject to the double-lock mechanism mentioned above but that xyn5 is not.
The induction by d-xylose of all inducible xylanases was also coordinately dependent on the Zn2Cys6-type transcriptional activator XYR1, as the expression under d-xylose-inducing conditions of all of them was undetectable in a Δxyr1 strain (Fig. 2E). This coordinated regulation of transcription was reflected by the presence of consensus binding motifs for XYR1 in all five xylanase upstream sequences (Fig. 2A). Also, all contained binding sites for the HAP2/3/5 complex in the vicinity of the XYR1 binding motif. A further binding site, 5′-AGAA-3′, was abundantly present in all five promoter regions. This site is bound by the putative XPP1 repressor (40), but its relevance for xylanase transcription, if any, is not clear at the moment and the data are therefore not included in Fig. 2A.
Impairment in d-xylose metabolism enhances induction by d-xylose.
Having identified that d-xylose is able to coordinately induce four out of five xylanases in T. reesei, we wondered how a manipulation of the flux through the d-xylose catabolic pathway would influence uptake, intracellular polyol formation, and their induction. This pathway involves the d-xylose/aldose reductase XYL1 (24) and the xylitol dehydrogenase XDH1 (see Fig. 4A). The function of the latter can be partially replaced by the l-arabitol dehydrogenase LAD1 (25). LXR3 (B. Metz et al., submitted for publication) connects d-xylose catabolism with l-arabinose catabolism via the reversible conversion of l-xylulose to xylitol, the first common intermediate.
We used isogenic knockout strains in the respective genes (i.e., Δxyl1, Δlad1, Δxdh1, Δlad1/Δxdh1, Δlxr3) and followed d-xylose uptake and intracellular sugar/polyol accumulation. As can be seen for some selected strains in Fig. 3, d-xylose was taken up at the highest rate in strain QM9414, while d-xylose could still be detected in the medium of the Δlad1/Δxdh1 strain after 4 h and in that of the Δxyl1 strain even after 6 h. Uptake remained unaltered in the l-arabinose catabolic pathway mutant Δlxr3. An analysis of the intracellular sugar and polyol pool of samples taken after 1 and 3 h of incubation showed that xylitol—but no other polyol (such as l-arabitol)—accumulated intracellularly (Fig. 3). Its concentration was elevated in the Δlad1/Δxdh1 double-deletion strain. The fact that xylitol accumulation is not completely abolished in the Δxyl1 strain can be explained by the finding that there is still a minor residual d-xylose reductase activity detectable in this strain (25).
Fig 3.
d-Xylose and l-arabinose uptake and intracellular accumulation of their corresponding polyols in T. reesei. d-Xylose (A) and l-arabinose (B) concentrations in the medium (◆, QM9414; •, Δxyl1; ■, Δlad1/Δxdh1; ▲, Δlxr3). (C to F) Intracellular accumulation of d-xylose (white bars), xylitol (gray bars), l-arabinose (not detectable, therefore no bars), and l-arabitol (dark gray bars) in T. reesei strains QM9414 (C), Δlad1/Δxdh1 (D), Δxyl1 (E), and Δlxr3 (F). Strains were placed on medium containing 1 mM d-xylose or l-arabinose.
As shown in Fig. 4B to E, all the T. reesei strains bearing knockouts in xyl1, xdh1, lad1, and xdh1/lad1 displayed significantly increased expression of xyn1, xyn2, xyn4, and xyn5 and also—albeit less strongly—increased xyn3 expression. In contrast, T. reesei Δlxr3, which has a deletion of the l-xylulose reductase LXR3 and is therefore restricted in the conversion of xylitol to l-xylulose and further to l-arabitol, displayed xylanase expression levels comparable to those found in QM9414 (Fig. 4F).
These results indicate that the d-xylose reductase step is not essential for xylanase induction, as reported earlier (21). They also show that the actual inducing component that mediates d-xylose induction of the five xylanases is a metabolite accumulating prior to the xylitol oxidation step, as the Δlad1/Δxdh1 double-deletion strain completely blocks further catabolism of xylitol to d-xylulose. This strain is also not able to produce l-arabitol, as the step from l-xylulose to l-arabitol is blocked. This conclusion is consistent with the fact that l-arabitol was not detected in the mycelia during these experiments, indicating that l-arabitol cannot be the inducer formed from d-xylose under these conditions.
Xylanases are induced by l-arabinose, and their transcription is enhanced in l-arabinose pathway mutants.
Our data (see above) disproved the hypothesis that l-arabitol is the true inducer for d-xylose-mediated induction of xylanases in T. reesei. However, this does not exclude the possibility that xylanases could also be induced by the hemicellulosic pentose l-arabinose or a metabolite of its catabolism. Again, we used our set of isogenic knockout strains to investigate how the carbon flux through these individual steps would influence induction. During these experiments, l-arabinose was taken up much more slowly than d-xylose, and only the polyol l-arabitol—and not xylitol—was found to accumulate in the mycelia (Fig. 3).
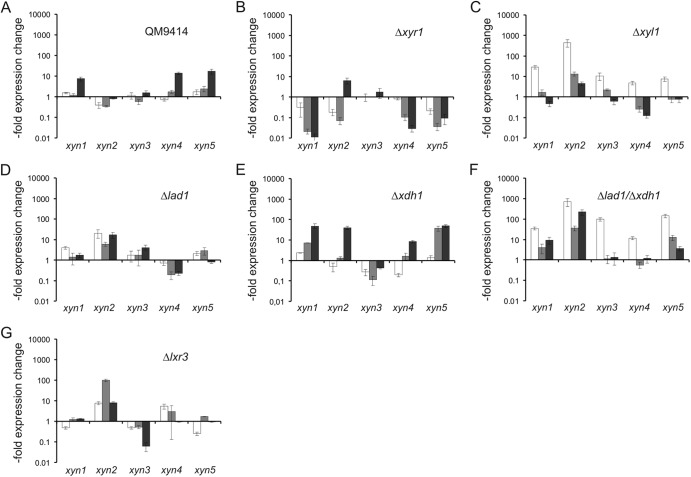
Figures 5A and B show that the xylanase genes xyn1, xyn4, and xyn5 are indeed induced by l-arabinose and that this induction is also fully dependent on the general cellulase and hemicellulase regulator XYR1. In contrast to d-xylose induction, xylanase transcript levels were in the beginning lower and were the highest after 6 h. This is obviously the result of the much slower uptake of l-arabinose. Using our set of d-xylose/l-arabinose pathway mutants, we found that the induction by l-arabinose was again stimulated in the Δxyl1 and Δlad1 strains and was stimulated the most strongly in the Δlad1/Δxdh1 strains, including also xyn2. In the Δlxr3 strain, the expression of only xyn2 and xyn4 was enhanced. Data obtained with the Δlad1, Δxdh1, and Δlad1/Δxdh1 strains, although different in the levels of accumulated transcript (probably due to the different substrate affinities of XDH1 and LAD1, leading to different steady-state intermediate concentrations), indicate that induction by l-arabinose must be due to a metabolite that accumulates before the l-arabitol dehydrogenase step and that l-arabinose thus induces xylanase in a manner independent of that of d-xylose. In support of this, induction by l-arabinose could be further stimulated by addition of d-xylose, and in the case of xyn1, xyn2, and xyn4, the final induction was higher than the sum of individual inductions, demonstrating that their action is synergistic (Fig. 6).
Fig 5.
Xylanase induction by l-arabinose. (A to G) Relative expression levels of the five xylanase genes xyn1 to xyn5 in T. reesei QM9414 (A) and the deletion strains Δxyr1 (B), Δxyl1 (C), Δlad1 (D), Δxdh1 (E), Δlad1/Δxdh1 (F), and Δlxr3 (G) placed on medium containing 1 mM l-arabinose plotted on a logarithmic y scale. Samples were taken 2 h (white bars), 4 h (gray bars), and 6 h (black bars) after addition of the carbon source. The values shown are related to the expression levels detected in QM9414 on medium without a carbon source (A) or the transcriptional level of QM9414 at the respective time point on inducing medium (B to G).
Fig 6.
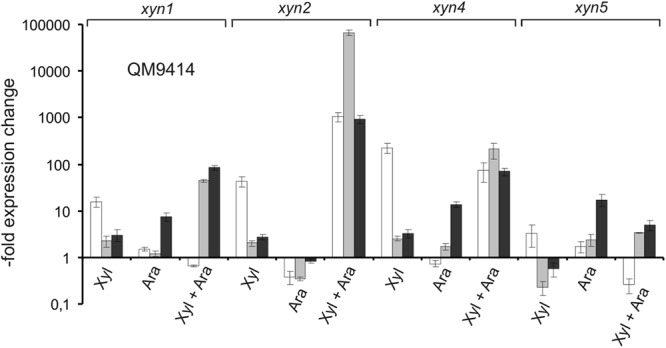
Induction of xylanases by simultaneous presence of d-xylose and l-arabinose. Relative expression levels of the xylanase genes xyn1, xyn2, xyn4, and xyn5 in the T. reesei reference strain QM9414 placed on medium containing 1 mM l-arabinose, 1 mM d-xylose, or both. Samples were taken 2 h (white bars), 4 h (gray bars), and 6 h (black bars) after addition of the carbon source. The values shown are related to the expression levels detected in QM9414 on medium without a carbon source.
DISCUSSION
T. reesei contains three GH11 xylanases (XYN1, XYN2, and XYN5), one GH10 xylanase (XYN3), and at least one GH30 xylanase (XYN4). On the basis of the present results, most of the xylanases can be induced by d-xylose, with the exception being XYN3. The l-arabinose-induced xylanases are XYN1, XYN4, and XYN5. XYN3 has previously been described to be expressed only in the QM9414 mutant strain PC-3-7 on cellulose, l-sorbose, and sophorose (6). While our data confirmed the absence of d-xylose-induced expression in the parent strain QM9414, we nevertheless found notable expression upon induction by l-arabinose in strains blocked in either the reduction of l-arabinose (Δxyl1) or oxidation of l-arabitol (Δlad1/Δxdh1). We also observed induction by lactose in the parent strain QM9414 (C. Ivanova et al., unpublished data). Hence, this is the only one out of the five xylanases that escaped regulation by d-xylose or l-arabinose. The reason for this is unknown, as the enzymatic properties of XYN3—which may shed a light on its natural substrate—have not been investigated as yet. We should also like to note that our present data were obtained with the low-level cellulase producer QM9414, which is a two-step mutant from QM6a. Although the differences in the genome sequence between QM9414 and QM6a are not present in genes that could be hypothesized to play a role in xylanase regulation (41), we cannot rule out the possibility that the regulation in wild-type strains of T. reesei could differ in minor aspects. XYN5 showed only a low level of induction during the early phase of induction by d-xylose (2 h) and was subsequently downregulated. On the other hand, higher xyn5 levels were found upon l-arabinose induction or in a carbon catabolite-derepressing background (Δcre1) upon induction by d-xylose. It is therefore possible that the d-xylose concentration is more critical for the expression of xyn5 than for the expression of other xylanases. Generally, this study revealed that the inducing concentration of d-xylose appears to be by far more critical for induction than it is in other fungi, such as Aspergillus niger (39) and Neurospora crassa (17), as d-xylose leads to a repression of xylanase formation already at comparatively low concentrations in T. reesei.
Our data provide a new model for xylanase regulation with respect to the potential inducer(s) and the contribution of the enzymes of the catabolic pathways for d-xylose and l-arabinose in inducer formation. We clearly show that d-xylose and l-arabinose induce xylanases via different compounds: while some enzymes of the catabolic pathways for l-arabinose and d-xylose can act in both pathways (e.g., XYL1, XDH1), LXR3 is the specific step that connects both pathways. Thus, the phenotype of the Δlxr3 strain allows one to distinguish between the contributions of the two pathways, as it blocks the interconversion of the two sugars. Consequently, the observed induction by d-xylose and l-arabinose in the Δlxr3 strain must be due to different inducing metabolites. Further support for this conclusion comes from the Δlad1/Δxdh1 double-deletion strain, which completely blocks xylitol to d-xylulose and l-xylulose to l-arabitol formation when d-xylose is used as the carbon source. When induced by l-arabinose, it blocks the step from l-arabitol to l-xylulose. This strain shows a strong stimulation of xylanase transcription for both pentoses. The strong inducing effect observed by d-xylose and l-arabinose in a Δxyl1 strain where the major enzyme for d-xylose and l-arabinose reductase activity is missing would support the conclusion that both d-xylose and l-arabinose are the true inducers, since both pentoses stimulate induction without being efficiently metabolized. Such an interpretation would be in accordance with data from N. crassa, where a deletion of the xyl1 orthologue impairs growth on d-xylose but stimulates expression of an endoxylanase (17). The equally strong stimulation of induction by d-xylose and l-arabinose in the Δlad1/Δxdh1 strains, however, makes the identification of the true inducer more difficult and would argue in favor of xylitol or l-arabitol. It is therefore possible that the protein interacting with the inducer can bind both components. Akel et al. (42) have recently demonstrated that the expression of α-l-arabinofuranosidase genes in T. reesei requires a cross talk between l-arabinose or l-arabitol and the aldose reductase XYL1, and the present data are also compatible with the operation of such a mechanism.
The data presented above (i.e., that the absence of d-xylose reductase or xylitol/l-arabitol dehydrogenases leads to increased xylanase gene expression) contradict recent data by Mach-Aigner et al. (21, 22). One argument to explain these differences is that for transcript analysis they used only a single time point at which expression in the Δxyl1 and Δxdh1/Δlad1 strain had not yet taken place and was therefore overlooked. However, their claim that l-arabitol is the inducer of xylanases in the presence of d-xylose can clearly not be maintained: l-arabitol did not accumulate in any of the mutant strains when incubated on d-xylose, and this appears to be logical, because strains with a nonfunctional LXR3 or LAD1/XDH1 cannot metabolize d-xylose to l-arabitol.
In this paper, we have also shown that l-arabinose can act as an inducer of xylanase gene expression and that this induction is also dependent on XYR1. While this finding is new, it was previously shown that a blockage of the l-arabinose pathway results in the upregulation of l-arabinose-inducible α-l-arabinofuranosidases or β-xylosidases in Aspergillus nidulans or T. reesei (42, 43). In T. reesei, l-arabinose-induced expression of abf2 and bxl1 is dependent on XYR1, while the expression of two genes, abf1 and abf3, was only slightly affected. In aspergilli, a second transcriptional regulator, AraR, responds to the presence of l-arabinose and activates l-arabinose-releasing enzyme transcription (44). AraR is, however, specific for the Eurotiales, and no orthologues are present in T. reesei. In T. reesei, the inducing effects by l-arabinose on xylanase gene expression are part of the XYR1 circuit, and future research in aspergilli will show if xylanases are induced by l-arabinose and if this regulation is via XlnR or AraR. Our data also show that this induction is not simply due to the formation of the same inducing metabolite from d-xylose and l-arabinose, as suggested earlier (21), as further supported by the additive effect of induction by d-xylose and l-arabinose. The advantage of having two inducers may come from the fact that there are two major types of xylans: one occurs in the cell wall of cereals, which often contain large quantities of l-arabinose and are consequently termed arabinoxylans. In contrast, the second major type occurs in hardwood xylans, which contain large amounts of d-glucuronic acid linked to the backbone and are named glucuronoxylans (45). T. reesei appears to be more adapted to the latter because of its amplification of GH67 α-methyl-glucuronidases and GH30 glucuronyl-xylanases in its genome (20). The availability of an arabinoxylan may therefore be signaled by l-arabinose removed from the arabinoxylan side chains, whose removal will make the xylan backbone available for hydrolysis.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by grants from the Austrian Science Fund (FWF) to C.P.K. (P 21266) and B.S. (P 19421, P24219). S.H. is member of the Ph.D. school program Applied Bioscience Technologies AB-Tec, financed by the Vienna University of Technology.
Footnotes
Published ahead of print 4 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00182-12.
REFERENCES
- 1. Beg QK, Kapoor M, Mahajan L, Hoondal GS. 2001. Microbial xylanases and their industrial applications: a review. Appl. Microbiol. Biotechnol. 56: 326– 338 [DOI] [PubMed] [Google Scholar]
- 2. Dashtban M, Schraft H, Qin W. 2009. Fungal bioconversion of lignocellulosic residues; opportunities & perspectives. Int. J. Biol. Sci. 5: 578– 595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tenkanen M, Puls J, Poutanen K. 1992. Two major xylanases of Trichoderma reesei. Enzyme Microb. Technol. 14: 566– 574 [Google Scholar]
- 4. Törrönen A, Mach RL, Messner R, Gonzalez R, Kalkkinen N, Harkki A, Kubicek CP. 1992. The two major xylanases from Trichoderma reesei: characterization of both enzymes and genes. Nat. Biotechnol. 10: 1461– 1465 [DOI] [PubMed] [Google Scholar]
- 5. Ogasawara W, Shida Y, Furukawa T, Shimada R, Nakagawa S, Kawamura M, Yagyu T, Kosuge A, Xu J, Nogawa M, Okada H, Morikawa Y. 2006. Cloning, functional expression and promoter analysis of xylanase III gene from Trichoderma reesei. Appl. Microbiol. Biotechnol. 72: 995– 1003 [DOI] [PubMed] [Google Scholar]
- 6. Xu J, Takakuwa N, Nogawa M, Okada H, Morikawa Y. 1998. A third xylanase from Trichoderma reesei PC-3-7. Appl. Microbiol. Biotechnol. 49: 718– 724 [Google Scholar]
- 7. Saloheimo M, Siika-Aho M, Tenkanen M, Penttilä M. April 2003. Xylanase from Trichoderma reesei, method for production thereof, and methods employing this enzyme. U.S. patent 6,555,335 B1 [Google Scholar]
- 8. Foreman PK, Brown D, Dankmeyer L, Dean R, Diener S, Dunn-Coleman NS, Goedegebuur F, Houfek TD, England GJ, Kelley AS, Meerman HJ, Mitchell T, Mitchinson C, Olivares HA, Teunissen PJM, Yao J, Ward M. 2003. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J. Biol. Chem. 278: 31988– 31997 [DOI] [PubMed] [Google Scholar]
- 9. Kubicek CP, Mikus M, Schuster A, Schmoll M, Seiboth B. 2009. Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnol. Biofuels 2: 19 doi:10.1186/1754-6834-2-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zeilinger S, Mach RL, Schindler M, Herzog P, Kubicek CP. 1996. Different inducibility of expression of the two xylanase genes xyn1 and xyn2 in Trichoderma reesei. J. Biol. Chem. 271:25624– 25629 [DOI] [PubMed] [Google Scholar]
- 11. Xu J, Nogawa M, Okada H, Morikawa Y. 2000. Regulation of xyn3 gene expression in Trichoderma reesei PC-3-7. Appl. Microbiol. Biotechnol. 54: 370– 375 [DOI] [PubMed] [Google Scholar]
- 12. Calero-Nieto F, Di Pietro A, Roncero MI, Hera C. 2007. Role of the transcriptional activator xlnR of Fusarium oxysporum in regulation of xylanase genes and virulence. Mol. Plant Microbe Interact. 20: 977– 985 [DOI] [PubMed] [Google Scholar]
- 13. de Vries RP, Visser J, de Graaff LH. 1999. CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res. Microbiol. 150: 281– 285 [DOI] [PubMed] [Google Scholar]
- 14. Marui J, Tanaka A, Mimura S, de Graaff LH, Visser J, Kitamoto N, Kato M, Kobayashi T, Tsukagoshi N. 2002. A transcriptional activator, AoXlnR, controls the expression of genes encoding xylanolytic enzymes in Aspergillus oryzae. Fungal Genet. Biol. 35: 157– 169 [DOI] [PubMed] [Google Scholar]
- 15. Seiboth B, Herold S, Kubicek CP. 2012. Metabolic engineering of inducer formation for cellulase and hemicellulase gene expression in Trichoderma reesei. Subcell. Biochem. 64: 367– 390 [DOI] [PubMed] [Google Scholar]
- 16. Stricker AR, Mach RL, de Graaff LH. 2008. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei). Appl. Microbiol. Biotechnol. 78: 211– 220 [DOI] [PubMed] [Google Scholar]
- 17. Sun J, Tian C, Diamond S, Glass NL. 2012. Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryot. Cell 11: 482– 493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Peij NN, Gielkens MM, de Vries RP, Visser J, de Graaff LH. 1998. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 64: 3615– 3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mach RL, Strauss J, Zeilinger S, Schindler M, Kubicek CP. 1996. Carbon catabolite repression of xylanase I (xyn1) gene expression in Trichoderma reesei. Mol. Microbiol. 21: 1273– 1281 [DOI] [PubMed] [Google Scholar]
- 20. Kubicek CP. 2012. Fungi and lignocellulosic biomass. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 21. Mach-Aigner AR, Gudynaite-Savitch L, Mach RL. 2011. l-Arabitol is the actual inducer of xylanase expression in Hypocrea jecorina (Trichoderma reesei). Appl. Environ. Microbiol. 77: 5988– 5994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mach-Aigner AR, Pucher ME, Mach RL. 2010. d-Xylose as a repressor or inducer of xylanase expression in Hypocrea jecorina (Trichoderma reesei). Appl. Environ. Microbiol. 76: 1770– 1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mandels M, Weber J, Parizek R. 1971. Enhanced cellulase production by a mutant of Trichoderma viride. Appl. Microbiol. 21: 152– 154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seiboth B, Gamauf C, Pail M, Hartl L, Kubicek CP. 2007. The d-xylose reductase of Hypocrea jecorina is the major aldose reductase in pentose and d-galactose catabolism and necessary for β-galactosidase and cellulase induction by lactose. Mol. Microbiol. 66: 890– 900 [DOI] [PubMed] [Google Scholar]
- 25. Seiboth B, Hartl L, Pail M, Kubicek CP. 2003. d-Xylose metabolism in Hypocrea jecorina: loss of the xylitol dehydrogenase step can be partially compensated for by lad1-encoded l-arabinitol-4-dehydrogenase. Eukaryot. Cell 2: 867– 875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stricker AR, Grosstessner-Hain K, Würleitner E, Mach RL. 2006. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 5: 2128– 2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Portnoy T, Margeot A, Linke R, Atanasova L, Fekete E, Sandor E, Hartl L, Karaffa L, Druzhinina IS, Seiboth B, Le Crom S, Kubicek CP. 2011. The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: a master regulator of carbon assimilation. BMC Genomics 12: 269 doi:10.1186/1471-2164-12-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mandels M, Andreotti RE. 1978. Problems and challenges in the cellulose to cellulase fermentation. Process Biochem. 13:6 [Google Scholar]
- 29. Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156– 159 [DOI] [PubMed] [Google Scholar]
- 30. Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30: e36 doi:10.1093/nar/30.9.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nicholas KB, Nicholas HB., Jr 1997. Genedoc: a tool for editing and annotating multiple sequence alignments. NSRBC, Pittsburgh, PA [Google Scholar]
- 32. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731– 2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451– 1452 [DOI] [PubMed] [Google Scholar]
- 34. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596– 1599 [DOI] [PubMed] [Google Scholar]
- 35. Metz B, de Vries RP, Polak S, Seidl V, Seiboth B. 2009. The Hypocrea jecorina (syn. Trichoderma reesei) lxr1 gene encodes a d-mannitol dehydrogenase and is not involved in l-arabinose catabolism. FEBS Lett. 583: 1309– 1313 [DOI] [PubMed] [Google Scholar]
- 36. Slayman CW, Tatum EL. 1964. Potassium transport in Neurospora. I. Intracellular sodium and potassium concentrations, and cation requirements for growth. Biochim. Biophys. Acta 88: 578– 592 [PubMed] [Google Scholar]
- 37. Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, Inc., New York, NY [Google Scholar]
- 38. Mach-Aigner AR, Pucher ME, Steiger MG, Bauer GE, Preis SJ, Mach RL. 2008. Transcriptional regulation of xyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina. Appl. Environ. Microbiol. 74: 6554– 6562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mach-Aigner AR, Omony J, Jovanovic B, van Boxtel AJ, de Graaff LH. 2012. d-Xylose concentration-dependent hydrolase expression profiles and the function of CreA and XlnR in Aspergillus niger. Appl. Environ. Microbiol. 78: 3145– 3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Würleitner E, Pera L, Wacenovsky C, Cziferszky A, Zeilinger S, Kubicek CP, Mach RL. 2003. Transcriptional regulation of xyn2 in Hypocrea jecorina. Eukaryot. Cell 2: 150– 158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vitikainen M, Arvas M, Pakula T, Oja M, Penttilä M, Saloheimo M. 2010. Array comparative genomic hybridization analysis of Trichoderma reesei strains with enhanced cellulase production properties. BMC Genomics 11: 441 doi:10.1186/1471-2164-11-441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Akel E, Metz B, Seiboth B, Kubicek CP. 2009. Molecular regulation of arabinan and l-arabinose metabolism in Hypocrea jecorina (Trichoderma reesei). Eukaryot. Cell 8:1837– 1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de Vries RP, Flipphi MJ, Witteveen CF, Visser J. 1994. Characterization of an Aspergillus nidulans l-arabitol dehydrogenase mutant. FEMS Microbiol. Lett. 123: 83– 90 [DOI] [PubMed] [Google Scholar]
- 44. Battaglia E, Hansen SF, Leendertse A, Madrid S, Mulder H, Nikolaev I, de Vries RP. 2011. Regulation of pentose utilisation by AraR, but not XlnR, differs in Aspergillus nidulans and Aspergillus niger. Appl. Microbiol. Biotechnol. 91: 387– 397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scheller HV, Ulvskov P. 2010. Hemicelluloses. Annu. Rev. Plant Biol. 61: 263– 289 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.