Abstract
Zebrafish (Danio rerio) is a popular vertebrate model organism largely deployed using outbred laboratory animals. The nonisogenic nature of the zebrafish as a model system offers the opportunity to understand natural variations and their effect in modulating phenotype. In an effort to better characterize the range of natural variation in this model system and to complement the zebrafish reference genome project, the whole genome sequence of a wild zebrafish at 39-fold genome coverage was determined. Comparative analysis with the zebrafish reference genome revealed approximately 5.2 million single nucleotide variations and over 1.6 million insertion–deletion variations. This dataset thus represents a new catalog of genetic variations in the zebrafish genome. Further analysis revealed selective enrichment for variations in genes involved in immune function and response to the environment, suggesting genome-level adaptations to environmental niches. We also show that human disease gene orthologs in the sequenced wild zebrafish genome show a lower ratio of nonsynonymous to synonymous single nucleotide variations.
Introduction
Vertebrate model organisms used for investigating human biology are predominantly inbred and are traditionally studied in the context of near-isogenic genetic backgrounds1,2 despite the fact that modern humans represent genetic admixture from diverse populations, with their genomes shaped by social, ethnic, and environmental factors.3–5 The zebrafish (Danio rerio) is a prominent and increasingly genetically tractable vertebrate model organism6 that is most commonly studied using nonisogenic backgrounds, strains that are commonly maintained as outbred populations. Traditional laboratory strains of zebrafish are derived from a number of wild collected zebrafish (for a complete list, see http://zfin.org/action/feature/wildtype-list) and are usually propagated using a selected number of founding animals. This founding process of bringing hobby fish into the laboratory has occurred several times over the past half-century, resulting in significant genetic diversity among common lab lines because of distinct geographic and temporal origins. Compared to most isogenic model systems, this genetic diversity of zebrafish is thus well represented at both individual and population level in these laboratory strains and in the reference genome project (Ensembl Zv9 build); however, how this compares to that of the wild population has been previously unknown.7–9 Importantly, the impact of captivity could be significant on the kinds and level of genetic diversity found in lab strains. For example, inbreeding and small population sizes of captive zebrafish leads to reduced variation within, and divergence among, zebrafish strains,9 and this effect may influence penetrance of the phenotypes and traits such as learning, behavior, and response to pharmacological agents.10–14 Geographical origins of wild zebrafish have also been suggested to have a potential influence on genetic makeup.15 This study presents the whole genome sequence of an adult male wild zebrafish at a comprehensive 39-fold genome coverage. Comparative analysis with the zebrafish reference genome revealed approximately 5.2 million single nucleotide variations and over 1.6 million insertion–deletion variations. This dataset thus represents a catalog of genetic variations in the wild zebrafish genome. The genome of a wild zebrafish provides insight into the diversity of naturally selected genetic variations and provides a starting point for genome-wide studies on genes influencing natural phenotypic variation and the effects of domestication.15
Materials and Methods
Ethics statement
Fish experiments were performed in strict accordance with the recommendations and guidelines laid down by the Council of Scientific and Industrial Research (CSIR)–Institute of Genomics and Integrative Biology India. The protocol was approved by the Institutional Animal Ethics Committee (IAEC) of the CSIR-Institute of Genomics and Integrative Biology, Delhi, India. All efforts were made to minimize animal suffering.
Zebrafish collection and maintenance
An adult wildtype zebrafish (hereafter referred to as the Assam, abbreviated as ASWT) was collected from its natural habitat in Northeastern India (Supplementary Fig. S1; Supplementary Data available online at www.liebertpub.com/zeb). The fish was transported and maintained in a recirculation system at the CSIR–Institute of Genomics and Integrative Biology following institutional animal ethics committee approved protocols and procedures.
DNA library preparation and sequence generation
A visibly healthy zebrafish with minimal acclimatization to laboratory conditions was selected for genomic DNA isolation. Genomic DNA was isolated using the protocol described previously.16 The DNA Library was prepared from one adult male fish, and sequenced using Genome Analyzer IIx (Illumina, Inc.) according to standard manufacturer protocols. Single and pair-end flow cells were used for generating the 36- or 76-base-pair sequences according to standard manufacturer protocols. The output images from each cycle were processed for converting signal intensities, base calling, and for calculating quality scores using Illumina Pipeline Software version 1.3.
Alignment of short sequence reads and data processing
We used “Mapping and Assembly with Quality” v0.7 (MAQ)17 to align the read-pairs onto the zebrafish reference genome (Zv9) and generated a consensus sequence from the alignment. For calling the consensus sequence, MAQ uses a statistical model and also considers the Phred quality score at each position along the consensus.
Single nucleotide variation detection and validation
Single nucleotide variation (SNVs) was called using the MAQ cns2snp option followed by a single nucleotide polymorphism (SNP) filter script on the consensus sequence. To call an SNV, a minimum of 32 reads and a maximum of 54 reads with nonoverlapping start site supporting the nonreference allele had to be present. The lower limit of reads was based on average coverage at all polymorphic loci, and the upper limit was set at the geometric mean plus one standard deviation to avoid potential miscalling from potential copy number variations.
In addition, the minimum and maximum Phred quality scores required at the polymorphic loci and its adjacent loci was set at 30 and 40, respectively. Only those polymorphic loci that did not have any variations in the adjacent five base pairs were considered for all possible single-base substitutions. Only SNVs with unique placements were retained for our analysis. After determination of unique SNVs, we compared these with those reported in dbSNP18 and Ensembl database for zebrafish.19 This led to the identification of putative novel SNVs. Annotation of the SNVs was done using the variant effect predictor tool.20
Mass spectroscopy–based Sequenome-iPLEX Assay (MassARRAY Analyzer 4) was used for validating 395 SNVs that were called in the ASWT genome.21 In addition, a custom SNP array was designed based on 201,917 SNPs predicted to be polymorphic in the SATmap cross based on comparisons of the Illumina sequence of the homozygous AB and Tübingen founders (http://zfin.org/action/genotype/genotype-detail?zdbID=ZDB-GENO-100413-1). This custom array is from Affymetrix (item number 520747, array name ZFSNP200m520747F, array format 49-7875). The chip was used according to the manufacturer's instructions for an NspI whole genome sampling array (WGSA) to genotype-known homozygous and heterozygous samples from the SATmap panel, as well as duplicate samples of the ASWT fish. The Bayesian robust linear model with Malaanobis distance and perfect match probes (BRLMM-P) clustering algorithm in the Affymetrix Power Tools software (APT) was used to determine genotypes from the resulting cell intensity files (CEL), whereas the ASWT samples were defined as unknowns, and a hints file was used to supply assumed genotypes (e.g., G0 [homozygous] and F1 [heterozygous] SATmap individuals processed alongside the ASWT CEL files), thus providing a genotype training set according to the manufacturer's recommendations. For further details of the cross, sequencing, SNP selection, genotyping array and processing, see M. Clark et al. (unpublished data). The genotype calls that concurred in the genotyping chips run in duplicates were further filtered, and the results were used to form the “intersection” set. The results of the intersection set were compared to the polymorphic base calls generated from the genomic alignments of the reads to assess the quality of our sequencing.
Insertion–deletion detection and validation
Insertion–deletions (indels) were identified using MAQ's indelpe option and by detecting abnormal alignment patterns around indels. A minimum of five paired-end reads with different start sites was required for supporting an indel. In addition, indels occurring within five bases of each other were not considered. Only indels with unique genomic placements were retained for analysis. Indels were isolated using indel-specific primers and amplified using polymerase chain reaction (PCR) using DNA as template. Amplified regions were sequenced using BigDye Terminator chemistry (Applied Biosystems). The sequences of these amplicons were further analyzed for confirming the existence of indels.
Structural variation prediction and validation
Structural variations including insertions, deletions, inversions, and chromosomal translocation were called using the BreakDancer software.22 The boundaries of the structural variations were identified based on abnormally aligned read pairs that have improper orientation or span sizes between the Zv9 and ASWT sequences. Structural variants with minimum sequence length of 100 bp and those with 20 paired-end reads with different start sites were only considered in our study. Structural variations with unique genomic placements were specifically retained for analysis. Primers were designed flanking the putative structural variants and amplified using polymerase chain reaction using DNA as template. Amplified regions were sequenced using BigDye Terminator chemistry (Applied Biosystems). The sequence of these amplicons were further aligned and compared to the reference sequence for confirming the existence of the structural variants.
Distribution and effects of variations in the genome
RefSeq gene and protein coding gene datasets corresponding to zebrafish genome build Zv9 were retrieved from the University of California Santa Cruz (UCSC)23 and Ensembl database,24 respectively. The distribution of variations in exons, introns, 5′ and 3′ untranslated regions (UTRs), and splice sites in known protein coding genes were analyzed using custom-built scripts. Analyses were focused on variations leading to premature truncation or loss of termination signal, thereby altering the length of predicted protein coding genes. The distribution of variations in noncoding genes was also analyzed.
Substitution rates, selection, and human homologs
Variations were mapped with respect to the gene locations, and the effect of variations was predicted based on annotations derived from Ensembl. Ensembl version 66 and the Variant Effect Predictor (VEP)20 pipeline were used for the analysis. Synonymous and nonsynonymous substitutions were tabulated for each of the 30,587 genes in zebrafish. Briefly, genes with no synonymous SNVs were removed from further analyses. The resulting dataset comprised 19,885 genes. These genes and their respective ratios of nonsynonymous to synonymous SNVs were plotted across the zebrafish chromosome using Haploview software25 and custom scripts. Human–zebrafish homolog genes were retrieved from Homologene26 by parsing for clusters with both human and zebrafish genes. Human disease genes were downloaded from the Online Mendelian Inheritance in Man (OMIM) database,27 and their homologs in zebrafish were identified. Functional annotation and enrichment of gene ontology terms were analyzed using the DAVID functional annotation tool.28
Results
This study reports the whole genome sequencing of an adult male zebrafish collected from its natural habitat in Northeast India. The genome was sequenced to over 39× coverage (Table 1) using 36–75 base-pair, single, and paired-end reads with an insert size of approximately 300 bases (Supplementary Table S1). Sequence reads were aligned on the zebrafish reference genome (Ensembl Zv9 build; hereafter called Zv9) using the base-quality aware reference mapping software, MAQ.17 Approximately 87.4% of the reads (∼55.81 Gb) were successfully aligned to the reference genome derived from lab strains. Uniquely placed reads covered 97.21% of the ∼1.4 Gb in the zebrafish reference genome (Supplementary Fig. S2). The remaining ∼2.7% of the reference genome not covered by this sequencing and annotation process potentially represent repetitive sequences, ambiguous bases, or gaps. The resulting consensus ASWT genome sequence was used to identify single nucleotide variations (SNVs), insertions–deletions (indels), and structural variations (Supplementary Fig. S3).
Table 1.
Data Production and Alignment Results for ASWT Zebrafish Genome
| Data type | Number of raw reads | Number of mapped reads | Total bases (Gb) | Number of mapped bases (Gb) | Effective depth (fold) |
|---|---|---|---|---|---|
| Single end reads | 763,983,931 | 320,717,132 | 30.77 | 22.87 | 16.23 |
| Paired end reads | 991,318,622 | 567,946,436 | 37.63 | 33.43 | 23.72 |
| Total | 1,755,302,553 | 888,663,568 | 68.40 | 56.30 | 39.96 |
Summary of sequencing reads from five independent sequencing experiments. The sequencing reads were aligned back to the zebrafish reference genome (Zv9). The effective depth was calculated by dividing the mapped bases by the length of Zv9 (excluding “N” bp in the length). Details of the individual sequencing runs are provided in Supplementary Table S1.
Over 5.2 million uniquely placed SNVs were identified in the ASWT genome using a stringent selection criteria (Table 2). The average sequence coverage and chromosomal distribution of these SNVs are presented in Supplementary Table S2. Comparison of the ASWT-derived SNVs, with those available at dbSNP (v130) and Ensembl (SNP called by Stemple Lab, SATMap Project, and available at Ensembl), revealed that the majority of the SNVs (>97%) have not been previously reported in the zebrafish reference genome (Table 2). A subset of over 25,000 SNVs was assessed using independent genotyping approaches (Supplementary Table S3), and showed high concordance (heterozygous SNVs, 96.2 %, and homozygous SNVs, 98.6 %, respectively).
Table 2.
List of Single Nucleotide Variations and Insertion–Deletion in the Adult Wildtype Zebrafish (ASWT) Genome
| List of variations | Number of variations |
|---|---|
| Total number of single nucleotide variants (SNVs) | 5,289,829 |
| Homozygous SNVsa | 1,179,274 |
| Heterozygous SNVsb | 4,110,555 |
| SNVs mapping to dbSNP (v130)c | 96,600 |
| SNVs mapping to Sanger single nucleotide polymorphism (SNP) datasetd | 43,775 |
| Novel SNVse | 5,149,454 |
| SNVs mapping within genesf | 3,514,884 |
| SNVs mapping to intergenic regionsg | 1,774,945 |
| Total number of insertion–deletions (indels) | 1,658,655 |
| Total number of insertions | 765,131 |
| Total number of deletions | 893,524 |
| Indels within genesh | 438,748 |
| Indels in the intergenic regioni | 1,219,907 |
Where both the alleles differ from the reference.
Where only one allele differs from the reference.
SNVs identical to dbSNP dataset v130 for zebrafish.
SNVs overlapping with Sanger SNV dataset for Zebrafish (variants called by Stemple Lab, SATMap project, and available at Ensembl database).
Novel SNVs identified from the ASWT zebrafish genome.
SNVs present in protein-coding genes available at Ensemble and RefSeq databases as predicted by variant effect predictor tool.
Variations present between protein-coding genes as predicted by variant effect predictor tool.
Indels present within the RefSeq genes.
Indels present between RefSeq genes.
The genomic location and potential functional consequences of the SNVs were further examined using a computational approach. Of the ∼5.2 million SNVs identified in the ASWT genome sequence, 3,514,884 were located to genes of which 145,679 SNVs fall in gene exons distributed as follows: 102,866 synonymous, 43,059 nonsynonymous, 226 non-sense, and 43 variations abolished a stop codon (Table 2 and Supplementary Table S4). An amino acid substitution matrix revealed that the exonic SNVs were not biased toward the encoding of any amino acids (Supplementary Table S5).
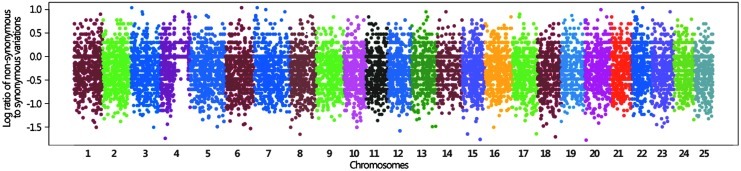
The ratio of nonsynonymous to synonymous substitution rate of single nucleotide variations has been used to compare intraspecies variability and selection.29 Synonymous and nonsynonymous substitutions were tabulated for each of the 30,587 genes in zebrafish. Genes with no synonymous SNVs were removed from further analyses. The resulting dataset comprised 19,885 genes. The nonsynonymous to synonymous substitution rate of SNVs was determined for each of these 19,885 zebrafish genes to identify genes potentially under positive selection (ratios of nonsynonymous to synonymous SNVs ≥1.0) (Fig. 1 and Supplementary Table S6). Over 3,800 genes in the ASWT genome exhibit a ratio of nonsynonymous to synonymous SNVs greater than or equal to 1.0 (Supplementary Table S7). The functional categories of these genes were assessed using gene ontology annotations. This analysis revealed enrichment of genes related to the immune response, response to stress, and the cellular response to stimulus (Supplementary Table S8). Further analysis of gene function and conservation revealed that these genes possessed significantly fewer human homologs (Chi-square 343.495 and p-value<0.001). This effect included human disease gene homologs (Chi-square 322.104 and p-value<0.001) (Supplementary Tables S9 and S10).
FIG. 1.
Manhattan plot of the ratios of nonsynonymous to synonymous substitution rates for 19,885 genes in the zebrafish genome is shown. The x-axis represents chromosomes in the zebrafish genome. Each dot represents the log ratio of nonsynonymous to synonymous SNVs of one gene.
Indels were identified by mapping paired-end reads to the reference genome (Supplementary Table S11). Of the 1,658,655 small indels identified in the ASWT genome, the majority (over 99%) had not been previously described (Supplementary Table S12). A subset of indels (n=28) was independently validated using targeted sequencing, and all confirmed the presence of the specific indel (Supplementary Table S13). Of the total number of small indels identified in the ASWT genome sequence, 330 insertions and 426 deletions fall within gene exons, of which 203 are predicted to cause a frame-shift (Supplementary Table S12); 1,329 structural variations were also identified in the ASWT genome (Supplementary Tables S14 and S16 to S20). A subset (n=25) of the structural variations was experimentally tested using targeted sequencing approaches. Of the 25 structural variations tested, 19 (76%) showed concordance with bioinformatics prediction (Supplementary Table S15). The remaining six genomic loci also displayed structural variation; however, the extent of the structural variation differed from this bioinformatic prediction.
Discussion
Genomic variations have been extensively studied for their association with phenotypic outcomes in humans30 and several organisms,31–34 including zebrafish.7,8 Previous estimations of variations in zebrafish populations were derived through candidate SNP approaches7,9,35 or by investigating specific variation subsets.8 These studies suggest substantial genetic variation, primarily single nucleotide variations, among zebrafish in the wild, consistent with their wide geographic distribution. The present study describes genome-wide, sequence-based genetic variations including over 5.2 million SNVs and 1.6 million indels. This catalog provides a starting point toward a more comprehensive description of genetic variations in this model organism, which could be used in the future for development of marker panels.
Detailed analysis reveals potential positive selection of genes associated with immune function and response to the environment. Positive selection of immune genes have been previously reported in fish species such as fugu36 and Atlantic cod37 and in other organisms including wild flies.32 A positive selection for immune genes has also been extensively studied in human populations.38 Zebrafish homologs of human disease genes in general displayed a lower ratio of nonsynonymous to synonymous SNVs. This observation was statistically significant, suggesting conserved sequence and function as the driving force in evolution of these genes. An alternative explanation for the lower number of homolog disease genes between human and zebrafish could also be the longer branch length in positively selected genes. Nevertheless, well-studied genes such as Selenocysteine lyase (SCLY) and Cannabinoid receptor-2 (CNR2) showed high ratios of nonsynonymous to synonymous SNVs, suggesting the possibility of positive selection in these specific loci (SCLY=5 and CNR2=4) (Supplementary Table S10).
Selenium is a micronutrient essential for normal brain function in humans and other vertebrates.39 Several neurological disorders and behavioral changes such as altered motor, learning, anxiety, confusion, and hostility have been linked to availability of selenium in humans and animals.14,39 The altered metabolism of selenium among human and zebrafish populations are likely due to genetic variants in biochemical pathways that are involved in the processing of selenium.14,40 The positive selection of SCLY, a key gene involved in selenoprotein biosynthesis, suggests that genetic heterogeneity and selection operating in wild populations of zebrafish may influence the physiological response to selenium metabolism, and this metabolic process may be important in modulating behavior in the wild.
The understanding of endocannabinoid signaling is rapidly expanding. The cannabinoid receptor family of G-protein-coupled receptor primarily works as signal modifiers, in particular as neuromodulators and immunomodulators.41,42 Cannabinoid receptor-2 (CNR2) plays important roles in balancing immune responses. As such, CNR2 is a key regulator of immune response in the gut, shifting the balance from an immuno-vigilant state to a more permissive one that can help beneficial microbes flourish.41,43 Variants in CNR2 are likely going to affect the local interaction of the innate immune system with microbiota.
In conclusion, the present study documents a genome-scale map of variations at single nucleotide resolution in a wild zebrafish. Genes involved in the immune response and environmental stimuli were determined to be potentially under positive selection, which corroborates earlier observations in other organisms.
This study is not without caveats. This work is limited to analysis of only one wild zebrafish genome, and as multiple zebrafish genomes from diverse geographic niche become available, the catalogue of genetic variation is likely to be enriched further. The availability of only one reference zebrafish genome to compare with has been a major limitation, which precludes several potentially informative genomic analyses on selection. The gene annotations in the zebrafish reference genomes have been largely derived from expressed sequence tag (EST) information and computational methods. Recent reannotation efforts using deep sequencing of the tissue and cell-type transcriptomes in humans and other model organisms have not been largely applied to zebrafish, which might have implications in the estimation of gene and exon boundaries and transcript isoforms and variations in noncoding RNAs. Furthermore, the limited availability of epigenomic data sets for zebrafish precludes us from understanding potentially functional regulatory variations. As more genome-scale datasets on zebrafish, including whole-genome resequencing, becomes available online, these limitations are likely to be overcome.
Notes
The raw sequence datasets described in this study are available under accession number ERP001723. ASWT progeny fish are available from the CSIR-IGIB upon request.
Supplementary Material
Acknowledgments
The authors acknowledge funding from the Council of Scientific and Industrial Research (CSIR), India, through the FAC002 Grant (SS and VS). Computational analyses were performed at the CSIR Center for in silico Biology at IGIB. We thank Sunny Malhotra for assistance with zebrafish maintenance. We thank Drs. Dwaipayan Bharadwaj and Chetana Sachidanandan for comments on the manuscript. AP acknowledges senior research fellowship from CSIR, India. We acknowledge Sourav Ghosh for help in the figure preparation. We acknowledge support from the Mayo Foundation and NIH grants GM63904 (SCE) and DA032194 (KJC).
Author Contributions
SS, VS, and SCE conceived the study and directed the research. Genome sequencing and experimental validation of the SNVs, indels, and SVs was conducted by AP with assistance from M. Singh, RC, ARS, and M. Swarnkar. Bioinformatic analyses were conducted by RP and AP. Scientific support for smooth performance of the sequencing facility and datacenter was provided by NS and VP respectively. MDC, CT, and DLS designed the Affymetrix genotyping chip and conducted the genotyping analysis. EWK and JPK contributed to the bioinformatics analysis. KJC interpreted the results for the CNR2 gene. SS, VS, and SCE wrote the article together with contributions from EWK, JPK, KJC, DLS, MDC, and AP, who contributed sections and edited the drafts to yield the final version of the manuscript.
Disclosure Statement
No competing financial interests exist.
References
- 1.Beck JA. Lloyd S. Hafezparast M. Lennon-Pierce M. Eppig JT. Festing MF, et al. Genealogies of mouse inbred strains. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 2.Festing MF. Fisher EM. Mighty mice. Nature. 2000;404:815. doi: 10.1038/35009167. [DOI] [PubMed] [Google Scholar]
- 3.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 4.Indian Genome Variation Consortium. Genetic landscape of the people of India: a canvas for disease gene exploration. J Genet. 2008;87:3–20. doi: 10.1007/s12041-008-0002-x. [DOI] [PubMed] [Google Scholar]
- 5.Hayden EC. International genome project launched. Nature. 2008;451:378–379. doi: 10.1038/451378b. [DOI] [PubMed] [Google Scholar]
- 6.Lieschke GJ. Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 7.Guryev V. Koudijs MJ. Berezikov E. Johnson SL. Plasterk RH. van Eeden FJ, et al. Genetic variation in the zebrafish. Genome Res. 2006;16:491–497. doi: 10.1101/gr.4791006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown KH. Dobrinski KP. Lee AS. Gokcumen O. Mills RE. Shi X, et al. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc Natl Acad Sci USA. 2012;109:529–534. doi: 10.1073/pnas.1112163109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whiteley AR. Bhat A. Martins EP. Mayden RL. Arunachalam M. Uusi-Heikkilä S, et al. Population genomics of wild and laboratory zebrafish (Danio rerio) Mol Ecol. 2011;20:4259–4276. doi: 10.1111/j.1365-294X.2011.05272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coe TS. Hamilton PB. Griffiths AM. Hodgson DJ. Wahab MA. Tyler CR, et al. Genetic variation in strains of zebrafish (Danio rerio) and the implications for ecotoxicology studies. Ecotoxicology. 2009;18:144–150. doi: 10.1007/s10646-008-0267-0. [DOI] [PubMed] [Google Scholar]
- 11.Engeszer RE. Ryan MJ. Parichy DM. Learned social preference in zebrafish. Curr Biol. 2004;14:881–884. doi: 10.1016/j.cub.2004.04.042. [DOI] [PubMed] [Google Scholar]
- 12.Lockwood B. Bjerke S. Kobayashi K. Guo S. Acute effects of alcohol on larval zebrafish: a genetic system for large-scale screening. Pharmacol Biochem Behav. 2004;77:647–654. doi: 10.1016/j.pbb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Sanders LH. Whitlock KE. Phenotype of the zebrafish masterblind (mbl) mutant is dependent on genetic background. Dev Dyn. 2003;227:291–300. doi: 10.1002/dvdy.10308. [DOI] [PubMed] [Google Scholar]
- 14.Benner MJ. Drew RE. Hardy RW. Robison BD. Zebrafish (Danio rerio) vary by strain and sex in their behavioral and transcriptional responses to selenium supplementation. Comp Biochem Physiol A Mol Integr Physiol. 2012;157:310–318. doi: 10.1016/j.cbpa.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engeszer RE. Patterson LB. Rao AA. Parichy DM. Zebrafish in the wild: a review of natural history and new notes from the field. Zebrafish. 2007;4:21–40. doi: 10.1089/zeb.2006.9997. [DOI] [PubMed] [Google Scholar]
- 16.Davidson AE. Balciunas D. Mohn D. Shaffer J. Hermanson S. Sivasubbu S, et al. Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev Biol. 2003;263:191–202. doi: 10.1016/j.ydbio.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Li H. Ruan J. Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherry ST. Ward MH. Kholodov M. Baker J. Phan L. Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flicek P. Amode MR. Barrell D. Beal K. Brent S. Carvalho-Silva D, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLaren W. Pritchard B. Rios D. Chen Y. Flicek P. Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabriel S. Ziaugra L. Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;60:2.12.1–2.12.16. doi: 10.1002/0471142905.hg0212s60. [DOI] [PubMed] [Google Scholar]
- 22.Chen K. Wallis JW. McLellan MD. Larson DE. Kalicki JM. Pohl CS, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dreszer TR. Karolchik D. Zweig AS. Hinrichs AS. Raney BJ. Kuhn RM, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinsella RJ. Kähäri A. Haider S. Zamora J. Proctor G. Spudich G, et al. Database (Oxford) 2011. Ensembl BioMarts: a hub for data retrieval across taxonomic space; p. bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrett JC. Fry B. Maller J. Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Wheeler DL. Church DM. Lash AE. Leipe DD. Madden TL. Pontius JU, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;29:11–16. doi: 10.1093/nar/29.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamosh A. Scott AF. Amberger J. Valle D. McKusick VA. Online Mendelian Inheritance in Man (OMIM) Hum Mutat. 2000;15:57–61. doi: 10.1002/(SICI)1098-1004(200001)15:1<57::AID-HUMU12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 28.Huang dW. Sherman BT. Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Higashino A. Sakate R. Kameoka V. Takahashi I. Hirata M. Tanuma R, et al. Whole-genome sequencing and analysis of the Malaysian cynomolgus macaque (Macaca fascicularis) genome. Genome Biol. 2012;13:R58. doi: 10.1186/gb-2012-13-7-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frazer KA. Murray SS. Schork NJ. Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 31.Keane TM. Goodstadt L. Danecek P. White MA. Wong K. Yalcin B, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lazzaro BP. Sceurman BK. Clark AG. Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science. 2004;303:1873–1876. doi: 10.1126/science.1092447. [DOI] [PubMed] [Google Scholar]
- 33.Ossowski S. Schneeberger K. Clark RM. Lanz C. Warthmann N. Weigel D, et al. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hillier LW. Marth GT. Quinlan AR. Dooling D. Fewell G. Barnett D, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 35.Bradley KM. Elmore JB. Breyer JP. Yaspan BL. Jessen JR. Knapik EW, et al. A major zebrafish polymorphism resource for genetic mapping. Genome Biol. 2007;8:R55. doi: 10.1186/gb-2007-8-4-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aparicio S. Chapman J. Stupka E. Putnam N. Chia JM. Dehal P, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 37.Star B. Nederbragt AJ. Jentoft S. Grimholt U. Malmstrøm M. Gregers TF, et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011;477:207–210. doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bairagya BB. Bhattacharya P. Bhattacharya SK. Dey B. Dey U. Ghosh T, et al. Genetic variation and haplotype structures of innate immunity genes in eastern India. Infect Genet Evol. 2008;8:360–366. doi: 10.1016/j.meegid.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rayman MP. The importance of selenium to human health. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 40.Méplan C. Crosley LK. Nicol F. Beckett GJ. Howie AF. Hill KE, et al. Genetic polymorphisms in the human selenoprotein P gene determine the response of selenoprotein markers to selenium supplementation in a gender-specific manner (the SELGEN study) FASEB J. 2007;21:3063–3074. doi: 10.1096/fj.07-8166com. [DOI] [PubMed] [Google Scholar]
- 41.Cluny NL. Reimer RA. Sharkey KA. Cannabinoid signalling regulates inflammation and energy balance: The importance of the brain-gut axis. Brain Behav Immun. 2012;26:691–698. doi: 10.1016/j.bbi.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Kano M. Ohno-Shosaku T. Hashimotodani Y. Uchigashima M. Watanabe M. Endocannabinoid-mediated control of synaptic transmission. Physiol Rev. 2009;89:309–380. doi: 10.1152/physrev.00019.2008. [DOI] [PubMed] [Google Scholar]
- 43.Muccioli GG. Naslain D. Bäckhed F. Reigstad CS. Lambert DM. Delzenne NM, et al. The endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol. 2010;6:392. doi: 10.1038/msb.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.