Abstract
Inhaled glucocorticoids, such as beclomethasone dipropionate (BDP), are the mainstay treatment of asthma. However, ∼30% of patients exhibit little to no benefit from treatment. It has been postulated that glucocorticoid resistance, or insensitivity, is attributable to individual differences in glucocorticoid receptor-mediated processes. It is possible that variations in cytochrome P450 3A enzyme-mediated metabolism of BDP may contribute to this phenomenon. This hypothesis was explored by evaluating the contributions of CYP3A4, 3A5, 3A7, and esterase enzymes in the metabolism of BDP in vitro and relating metabolism to changes in CYP3A enzyme mRNA expression via the glucocorticoid receptor in lung and liver cells. CYP3A4 and CYP3A5 metabolized BDP via hydroxylation ([M4] and [M6]) and dehydrogenation ([M5]) at similar rates; CYP3A7 did not metabolize BDP. A new metabolite [M6], formed by the combined action of esterases and CYP3A4 hydroxylation, was also characterized. To validate the results observed using microsomes and recombinant enzymes, studies were also conducted using A549 lung and DPX2 liver cells. Both liver and lung cells produced esterase-dependent metabolites [M1–M3], with [M1] correlating with CYP3A5 mRNA induction in A549 cells. Liver cells produced both hydroxylated and dehydrogenated metabolites [M4, M5, and M6], but lung cells produced only the dehydrogenated metabolite [M5]. These studies show that CYP3A4 and CYP3A5 metabolize BDP to inactive metabolites and suggest that differences in the expression or function of these enzymes in the lung and/or liver could influence BDP disposition in humans.
Introduction
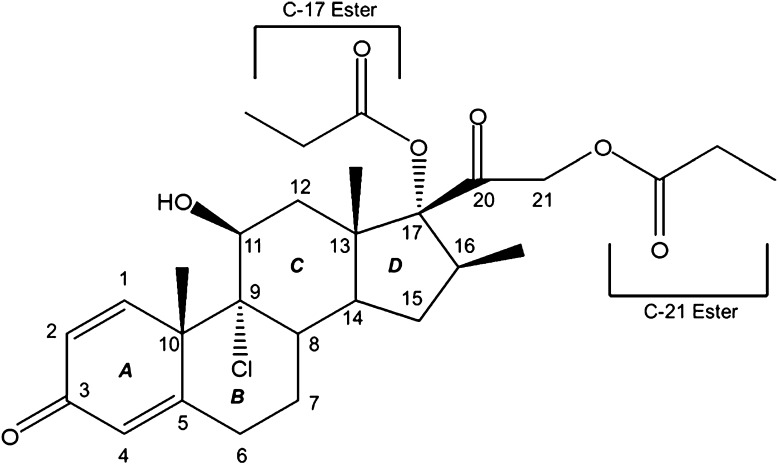
Beclomethasone dipropionate (BDP) (Fig. 1) is a glucocorticoid administered by inhalation to treat asthma. BDP is administered as a prodrug that requires cleavage of the C-21 ester by esterase enzymes to be pharmacologically active (Wilcox and Avery, 1973; Brogden et al., 1984). Prior studies have described the production and pharmacokinetic properties of beclomethasone-17-monopropionate ([M1] in Fig. 2) and the pharmacologically less active metabolites beclomethasone 21-monopropionate [M2] and beclomethasone [M3] in human plasma and human lung homogenates (Foe et al., 1998a,b, 2000a,b; Daley-Yates et al., 2001). All three BDP metabolites are glucocorticoid receptor (GR) agonists, but beclomethasone 17-monopropionate [M1] exhibits 30-fold greater affinity for GR than BDP, whereas beclomethasone 21-monopropionate [M2] has ∼50-fold lower affinity than BDP (Wurthwein and Rohdewald, 1990; Chanoine et al., 1991).
Fig. 1.
Structure of and atom numbering for BDP.
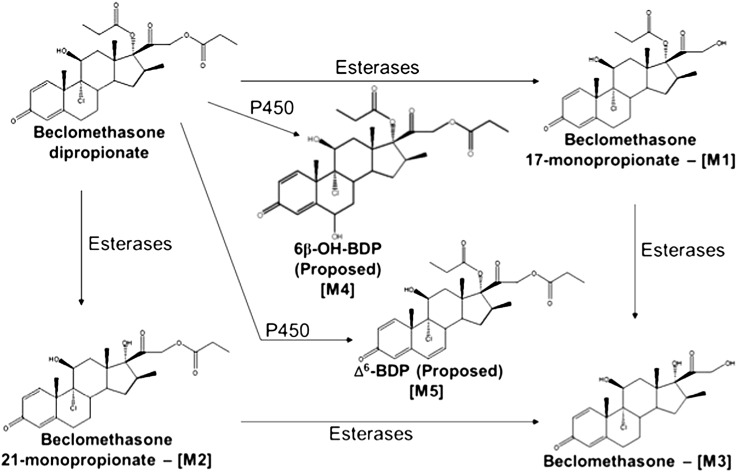
Fig. 2.
Proposed metabolic scheme for beclomethasone dipropionate. [M1] is the active form of the drug. [M1], [M2], and [M3] metabolites are produced by esterases, and [M4] and [M5] are produced by cytochrome P450 enzymes.
Prior studies demonstrated that de-esterification and activation of BDP was principally mediated by esterases (Mutch et al., 2007). Additionally, three other metabolites, designated as D-1, D-2, and D-3, have been documented (Foe et al., 1998a,b). D-2 and D-3 contained an epoxide on the C-ring at positions 9 and 11 (Fig. 1), presumably arising from cytochrome P450-mediated oxygenation and hydrolysis of both the 17- and 21-esters (D-2) or the 21-ester only (D-3); these metabolites are presumably inactive/clearance metabolites. Metabolism arising from the A- and B-rings of BDP (Fig. 1) has not been reported in the literature, nor has the production of specific metabolites by individual cytochrome P450 enzymes.
Xenobiotic metabolism in humans and animals frequently involves the action of cytochrome P450 enzymes. The most abundant subfamily of cytochrome P450 enzymes contributing to the clearance of the largest number of different xenobiotics is the CYP3A enzymes (Thummel and Wilkinson, 1998). CYP3A4, 3A5, and 3A7 are the most pertinent cytochrome P450 enzymes for this study because they are the most prominent cytochrome P450 enzymes involved in glucocorticoid metabolism (Jonsson et al., 1995; Pearce et al., 2006). CYP3A4 is the most abundant CYP3A enzyme in the liver and intestines (Jonsson et al., 1995; Westlind-Johnsson et al., 2003; Leclerc et al., 2010), and CYP3A5 is preferentially expressed in the lung (Hukkanen et al., 1997, 2003; Leclerc et al., 2010). CYP3A7 is expressed in fetal liver, but diminishes after birth as CYP3A4 becomes the dominant CYP3A enzyme (Schuetz et al., 1994; Lacroix et al., 1997). To date, the metabolism of BDP by these three human CYP3A enzymes and the contribution of cytochrome P450-dependent metabolism to the overall metabolism of BDP in lung and liver cells have not been reported.
The purpose of this study was to evaluate CYP3A-mediated metabolism of BDP to provide insights into how cytochrome P450 enzymes might affect the disposition of BDP in lung cells, which are the target for inhaled glucocorticoids. These studies are part of a larger study having an overarching hypothesis that variations in pulmonary metabolic clearance of glucocorticoids by CYP3A enzymes may impact the therapeutic efficacy of inhaled glucocorticoids in humans.
Materials and Methods
Chemicals.
Beclomethasone dipropionate (BDP), prednisolone, NADPH, ammonium acetate, eserine, benzoic acid, ketoconazole, and methanol were purchased from Sigma-Aldrich Chemical Company (St. Louis, MO). Paraoxon was purchased from Chem Service (West Chester, PA). Recombinant cytochrome P450 3A4, 3A5, and 3A7 enzymes were purchased from Becton Dickinson and Company (Franklin Lakes, NJ). Human liver microsomes were purchased from Celsis In Vitro Technologies (Baltimore, MD). Beclomethasone 17-monopropionate, beclomethasone 21-monopropionate, and beclomethasone were purchased from Steraloids, Inc. (Newport, RI).
Liquid Chromatography-Tandem Mass Spectrometry.
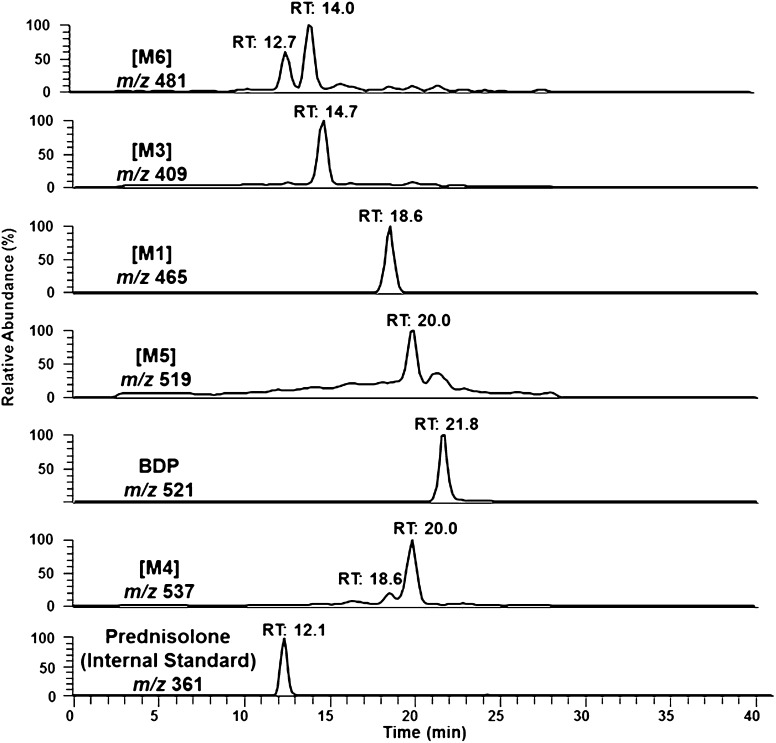
Liquid chromatography-tandem mass spectrometry (LC/MS/MS) was conducted using a Thermo LCQ Advantage Max ion trap instrument equipped with a Finnigan Surveyor LC pump, Surveyor Autosampler, and universal Ion Max source operated with Thermo Xcalibur software version 2.0 (Thermo Fisher Scientific, Waltham, MA). Positive electrospray ionization (ESI) was used. The mass spectrometer was optimized for the detection of BDP. The source temperature was 290°C, ionization voltage was 4.5 kV, capillary voltage was 6 V, and the sheath gas (N2) was 60 units. Parameters for MS/MS analysis were as follows: a collision energy of 27.5% for BDP, [M1], [M2], [M3], and prednisolone (internal standard), and 30% for [M4], [M5], and [M6]; activation Q = 0.25; activation time = 30 ms; and an isolation width of 2 amu. BDP, BDP metabolites, and prednisolone (internal standard) were resolved on a 150 × 2-mm Gemini 5-μm C18 reverse-phase HPLC column (Phenomenex Inc., Torrance, CA) and eluted with a linear gradient of 45–90% methanol over 17 minutes and holding at 90% methanol for 8 minutes. The aqueous solvent was 2 mM ammonium acetate (pH 6.4), the flow rate was 0.2 ml/min, and the column temperature was 30°C. BDP, BDP metabolites, and prednisolone (internal standard) were identified by the [M+H]+ ions m/z 521 (BDP), m/z 465 ([M1] and [M2]), m/z 409 ([M3]), m/z 537 ([M4]), m/z 519 ([M5]), m/z 481 ([M6]), and m/z 361 (prednisolone), in addition to the presence or absence of diagnostic product ions in the MS/MS spectra, LC retention time, and comparison with authentic standards ([M1] and [M2] only). A representative chromatogram obtained from the analysis of BDP, BDP metabolites, and prednisolone recovered from an in vitro incubation of BDP with CYP3A4 is shown for reference as Fig. 3. Comparison of the rates and extent of BDP metabolism and metabolite formation by CYP3A enzymes was based on the analyte-to-internal standard (prednisolone) peak area ratios obtained from data analysis using Thermo Xcalibur 2.0 software.
Fig. 3.
Representative LC/MS selected-ion monitoring (SIM) chromatograms showing the analyte peaks corresponding to prednisolone (the internal standard), BDP, and BDP metabolites generated by CYP3A4 in vitro. Individual SIM traces are labeled with the corresponding metabolite identities, m/z filter, and analyte elution time.
Characterization of BDP Metabolites.
In vitro incubations using human liver microsomes contained 100 pmol of cytochrome P450, 20 μM BDP, and 2 mM NADPH in 50 mM potassium phosphate buffer, pH 7.4 (0.5 ml total volume). Esterase inhibitors (eserine, paraoxon, and benzoic acid in a 1:1:1 mixture, each at 35 μM) were added to inhibit esterase-mediated metabolism and to ascertain the relative contributions of cytochrome P450 enzymes and esterases in BDP metabolism. Reactions were initiated by addition of NADPH and continued at 37°C for 20 minutes. Reactions were terminated by adding 0.5 ml methanol containing prednisolone (5 nM). The samples were cooled on ice for 5 minutes, and the insoluble material was pelleted by centrifugation for 10 minutes at 21,000g. The supernatant was collected, loaded onto C18 Sep-Pak cartridges (Waters, Taunton, MA), washed with water, and eluted with 100% methanol. The eluates containing BDP and metabolites were dried under forced air, reconstituted in 60 μl H2O:methanol (1:1 v/v), centrifuged at 21,000g for 5 minutes, and transferred to auto sampler vials for analysis.
Metabolism of BDP by Recombinant CYP3A Enzymes.
Incubations contained 2.5 pmol recombinant CYP3A enzyme, 1 μM BDP, and 1.3 mM NADPH in 30 mM potassium phosphate buffer, pH 7.4 (0.5 ml total volume). Control incubations did not contain NADPH. The esterase inhibitors described above were also included in selected incubations, except at concentrations of 28 μM to account for the lower esterase content associated with the recombinant CYP3A microsomes relative to human lung and liver microsomes. Incubations were performed at 37°C, with aliquots of 50 μl removed at 0, 5, 10, 15, 20, 30, 40, and 60 minutes. Aliquots were mixed with an equal volume of methanol containing prednisolone (1 nM) and prepared for LC/MS/MS analysis as described above. The ratio of the peak area for BDP relative to the internal standard at each time point was plotted versus time. Data were fit using a one-phase exponential decay model [Y = Span(e–kobsX) + Plateau; plateau = 0, t1/2 = 0.69/kobs] using GraphPad Prism 4.02 software for Windows (San Diego, CA) and approximate t1/2 values are reported.
Characterization of BDP Metabolites Produced by Individual CYP3A Enzymes.
Incubations were performed as described above, except 25 pmol of recombinant CYP3A4, 3A5, or 3A7 and 50 μM BDP (0.5 ml total volume) were used.
Analysis of BDP Metabolites in Cell Culture.
A549 cells (human lung adenocarcinoma) (ATCC, Manassas, VA) were cultured in Dulbecco’s modified Eagle’s medium containing 5% fetal bovine serum (Life Technologies, Grand Island, NY). DPX2 cells (HepG2 background with human PXR stably overexpressed to drive the expression of a CYP3A4 reporter gene construct by PXR agonists) were provided by Dr. Judy Raucy (Puracyp Inc., Carlsbad, CA). DPX2 cells were cultured in Puracyp media (Puracyp Inc.) (Lamb et al., 2010). Both cell lines were plated in six-well plates and treated with BDP (10 μM) in Opti-MEM 1 reduced serum media (Life Technologies) at 70% confluence, with and without esterase inhibitors (paraoxon and eserine 1:1; 175 μM), for 24 hours. Cells treated with esterase inhibitors were pretreated for 2 hours, followed by addition of BDP for 22 hours. The cell culture medium was extracted using 2× volume (6 ml) methyl tert-butyl ether containing prednisolone (1 nM). Samples were centrifuged and the organic fraction was collected, dried under air at room temperature, reconstituted in 60 μl H2O:methanol (1:1 v/v), clarified again by centrifugation, and the supernatant transferred to auto sampler vials for LC/MS/MS analysis. Treatments were also performed in the presence of ketoconazole (1 µM), a selective CYP3A inhibitor, to determine if the metabolites observed in cell culture media were due to CYP3A enzyme activity.
Quantification of CYP3A Enzyme Expression in A549 and DPX2 Cells.
Cells were treated as described above using dimethyl sulfoxide as the negative control. TRIzol reagent (Life Technologies) was used to extract total RNA, as per the manufacturer protocol. Total RNA was used to synthesize cDNA using iScript Reverse Transcription Supermix for RT-qPCR (Bio Rad, Hercules, CA), as per the manufacturer's protocol. The sequences of PCR primers for CYP3A4, 3A5, 3A7, and β2-macroglobulin (B2M) are provided in Table 1. Real-time quantitative PCR (qPCR) was performed using LightCycler 480 Probes Master (CYP3A5) or LightCycler 480 SYBR Green I Master Mix (CYP3A4, CYP3A7, and β2 macroglobulin) (Roche, Indianapolis, IN) on a Light-Cycler 480 Instrument. The PCR program for assays using the probe mix consisted of a 5-minute incubation at 95°C, followed by 45 cycles of 95°C for 10 seconds, 55°C for 30 seconds, then 72°C for 1 second. The PCR program for samples using SYBR Green I mix consisted of a 5-minute incubation at 95°C, followed by 40 cycles of 95°C for 10 seconds, 63°C for 5 seconds, then 72°C for 10 seconds. Experiments were performed on six biologic replicates (n = 6), mRNA copy number was determined from a standard curve, and the data normalized to the copy number for B2M.
TABLE 1.
qPCR primer sequences for the CYP3A genes and B2M
| Gene | Primer Sequence |
|---|---|
| CYP3A5 | Forward, 5′-CCTATCGTCAGGGTCTCTGGAA-3′ |
| Reverse, 5′-TGATGGCCAGCACAGGGA-3′ | |
| Probe [6FAM]ATGTGGGGAACGTATGAA[BHQ1] | |
| CYP3A4 | Forward, 5′-GAAAGTCGCCTCGAAGATAC-3′ |
| Reverse, 5′-ACGAGCTCCAGATCGGACAG-3′ | |
| CYP3A7 | Forward, 5′-TTCCGTAAGGGCTATTGGAC-3′ |
| Reverse, 5′-TCTGTGATAGCCAGCATAGG-3′ | |
| B2M | Forward, 5′ GATGAGTATGCCTGCCGTGTG-3′ |
| Reverse, 5′-CAATCCAAATGCGGCATCT-3′ |
Results
Metabolism of BDP In Vitro.
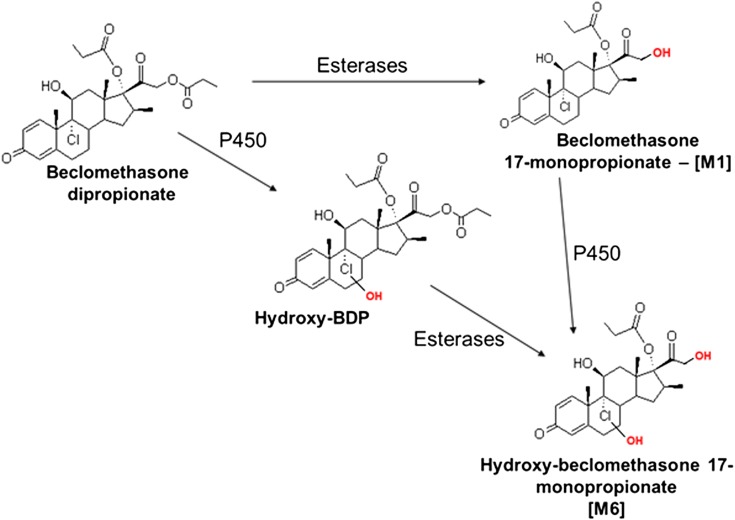
The known esterase-dependent BDP metabolites [M1], [M2], and [M3] (Foe et al., 1998a,b, 2000a,b; Daley-Yates et al., 2001; Mutch et al., 2007) were detected in all incubations lacking esterase inhibitors; formation of [M1–M3] was inhibited when esterase inhibitors were used, with the caveat that trace quantities of [M1] were detected at the level observed in control samples where only BDP was added (i.e., the BDP source material contained trace quantities of [M1] or an [M1] like compound). Additionally, three NADPH- and cytochrome P450-dependent metabolites, [M4] (m/z 537 at 20 minutes), [M5] (m/z 519 at 20 minutes), and [M6] (m/z 481 at 12.7 and 14 minutes), were observed in incubations containing either human liver microsomes or recombinant CYP3A4 or CYP3A5 but not CYP3A7. The proposed metabolite identities are depicted in Fig. 4.
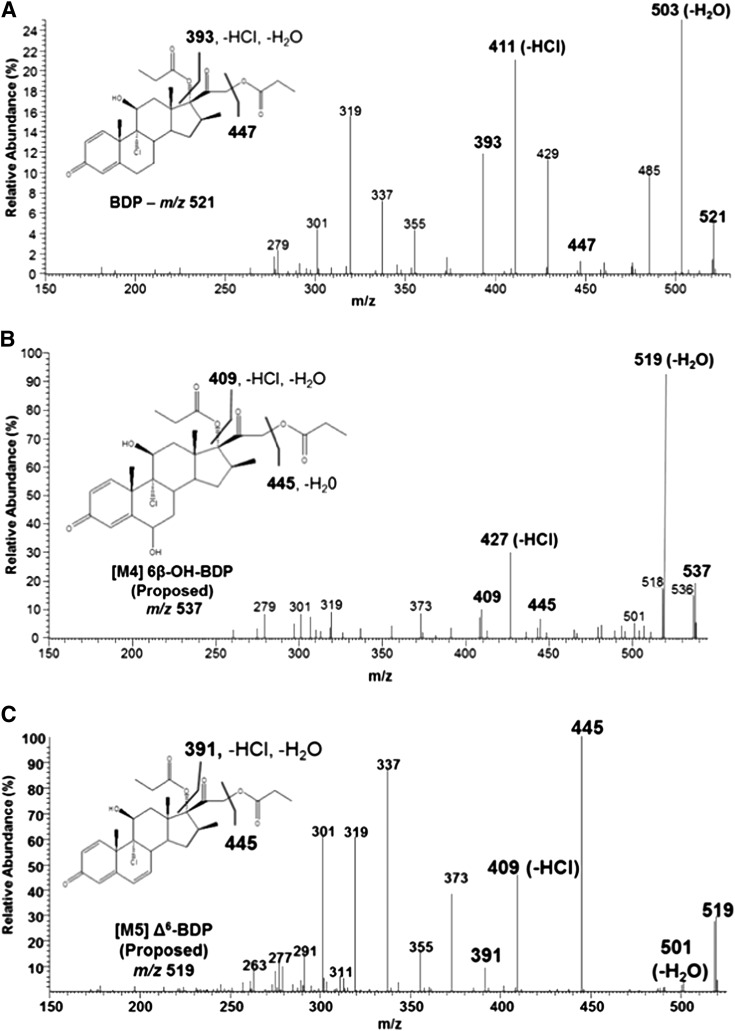
Fig. 4.
(A) MS/MS spectra for BDP, (B) [M4], and (C) [M5]. Diagnostic fragment ions for the metabolites [M4] and [M5] versus the corresponding fragments for BDP are highlighted in bold text. For [M4], these ions were +16 amu and for [M5] they were −2 amu, relative to BDP. Insets show the proposed metabolite structure and neutral loss events leading to the formation of the diagnostic fragment ions highlighted for each metabolite.
Because authentic [M4], [M5], and [M6] were not available as standards, these metabolites were characterized by comparing the metabolite MS/MS spectra to those of BDP, [M1], [M2], and comparable metabolites produced by CYP3A enzymes using other structurally similar glucocorticoids (Moore et al., 2013). The MS/MS spectrum for [M4] (m/z 537 at 20 minutes; Fig. 3) was characterized by an MH+ ion at m/z 537 (i.e., +16 amu relative to BDP; Fig. 4, A and B). This mass shift and corresponding 16-amu shifts in several diagnostic fragment ions of BDP, most notably m/z 409, 427, and 445, which corresponded to the neutral loss of the D-ring substituents on C-17 and C-21, loss of water, and HCl, indicated hydroxylation of the core structure of BDP. The fragmentation pattern for [M4] excluded oxygenation on the propionate groups, suggesting that [M4] was most likely 6β-OH-BDP based on the fact that steroids and structurally similar glucocorticoids preferentially undergo 6β-hydroxylation by CYP3A enzymes and [M4] was a common metabolite for both CY3A4 and 3A7 (Jonsson et al., 1995; Teng et al., 2003; Peet et al., 2005; Pearce et al., 2006; Hughes et al., 2008).
The MS/MS spectrum for [M5] (Fig. 4C; m/z 519 at 20 minutes in Fig. 3) was characterized by a molecular ion at m/z 519 (versus m/z 521 for BDP; Fig. 4A), consistent with desaturation of BDP. Cytochrome P450 3A enzymes dehydrogenate the C-6/7 bond of multiple steroid molecules and other glucocorticoids (Teitelbaum et al., 1981; Edsbacker et al., 1987; Moore et al., 2013). Therefore, [M5] is presumed to be Δ6-BDP. Consistent with this identification, the fragment ions m/z 501 (MH+-H2O), 409 (loss of the ester group on C-21), and 393 (loss of the ester group on C-17, H2O, and HCl) were 2 amu less than the corresponding fragment ions for BDP (Fig. 4, A and C). Of note, [M5] was detected in cell culture incubations when [M4] was not present, indicating that [M5] was not the product of [M4] dehydration during LC/MS analysis (i.e., it is a cytochrome P450-dependent metabolite). Additionally, glutathione did not reduce [M5] formation as shown for the dehydrogenated metabolites of other glucocorticoids (Moore et al., 2013), indicating that [M5] was not electrophilic (unpublished data).
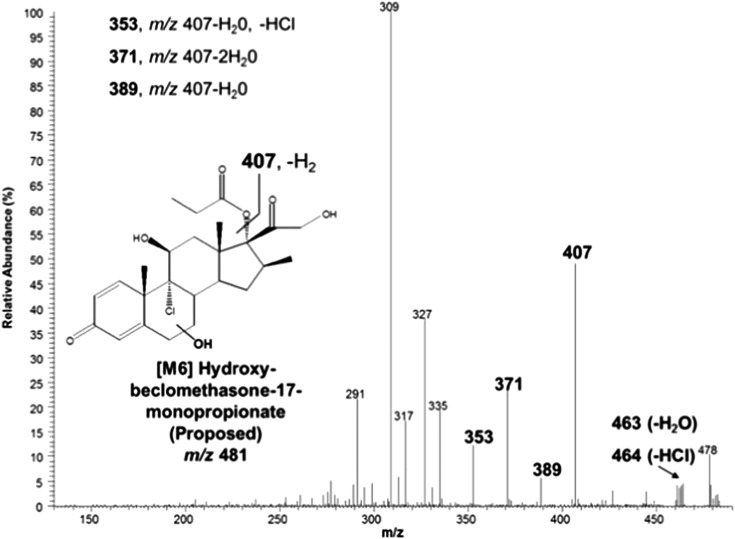
Two additional analyte peaks/metabolites, designated [M6] (m/z 481 at 12.7 and 14 minutes in Fig. 3) were also observed. The MS/MS spectra of the two [M6] chromatographic peaks were indistinguishable. The MS/MS spectrum for the [M6] metabolite peak eluting at 14 minutes is shown in Fig. 5 and is characterized by the addition of oxygen, presumably to the B-ring as described above for [M4], in addition to the removal of an ester group at the C-21 position. On the basis of the MS/MS data, the two [M6] component peaks are believed to arise from oxygen addition at two different sites on the steroid core structure, as discussed for [M4] and CYP3A4 below, and not from differential de-esterification at the C-17 and C-21 positions. The formation of [M6] could theoretically occur through two routes: oxygenation of the B-ring by cytochrome P450 enzymes followed by esterase-mediated cleavage at the C-21 position or the reverse sequence, as depicted in Fig. 6. Later studies showed that only CYP3A4 could produce [M6] in the presence of active esterases.
Fig. 5.
MS/MS spectrum of the [M6] peak eluting at 14 minutes (Fig. 3), with the proposed structure and neutral loss events leading to the diagnostic fragment ions depicted in the inset.
Fig. 6.
Proposed metabolic scheme for the production of [M6] by esterase and CYP3A4 enzymes.
Differential Metabolism of BDP by CYP3A4, 3A5, and 3A7.
Half-lives for the disappearance of BDP in incubations containing recombinant CYP3A4, 3A5, or 3A7 were determined using identical experimental conditions. CYP3A4 (t1/2 = 55 ± 15 minutes) and CYP3A5 (t1/2 = 43 ± 12 minutes) displayed similar kinetics for BDP decomposition, resulting in an approximate 25% decrease in BDP concentration over the course of a 20-minute incubation. CYP3A7 did not metabolize BDP, even at longer time periods.
CYP3A4 and CYP3A5 Produced [M4] and [M5] In Vitro.
Incubations were performed to determine which CYP3A enzymes produced which BDP metabolites. Recombinant CYP3A5 produced both [M4] and [M5], whereas CYP3A4 produced [M4], [M5], and [M6] as well as an additional hydroxylated metabolite eluting at 18.6 minutes in the m/z 537 chromatogram of Fig. 3. The MS/MS spectrum of this minor CYP3A4-specific metabolite was identical to [M4] (Fig. 4B), indicating oxygenation of the core steroid structure similar to [M4]. It is postulated that this metabolite could represent a C-7 hydroxylated metabolite arising from the same metabolic intermediate as [M4] and [M5] or a metabolite oxygenated at either the C-9 or C-11 position, as described previously (Foe et al., 1998a,b). No de-esterified metabolites (i.e., [M1], [M2], or [M3]) were produced by the CYP3A enzymes.
Metabolism of BDP by CYP3A Enzymes in Cells.
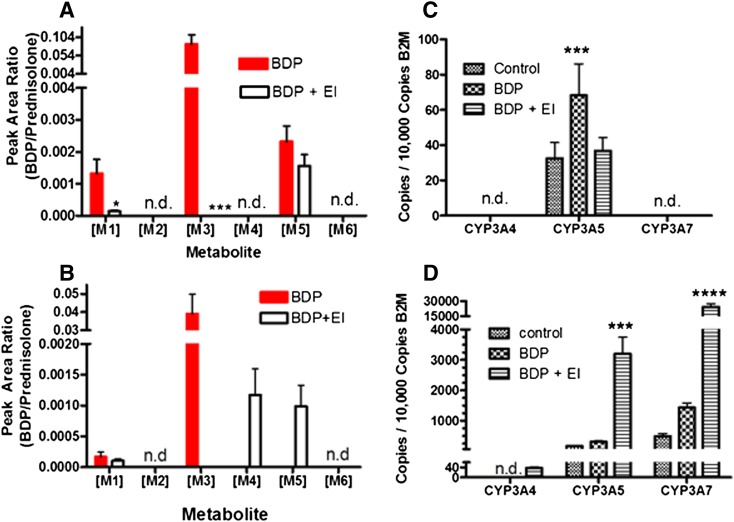
BDP metabolite formation was evaluated in A549 (Fig. 7A) and DPX2 (Fig. 7B) cells. [M1] and [M3] were produced by both cell lines when esterases were active, albeit to a greater extent in A549 cells. Consistent with the role of esterases in [M1] and [M3] production, including esterase inhibitors in the cell treatment solutions inhibited [M1] and [M3] formation, low levels of [M1] were attributed to [M1] (or a similar compound) occurring in the source BDP reagent. The production of the cytochrome P450-dependent metabolite [M5] was also observed in both A549 and DPX2 cells, but was not detected when A549 cells were pretreated and cotreated with the CYP3A inhibitor ketoconazole (unpublished data). These data confirm that [M5] was produced by CYP3A enzymes in A549 cells, presumably CYP3A5 based on Fig. 7C showing that only CYP3A5 mRNA was expressed. DPX2 cells, unlike A549 cells, also produced the cytochrome P450-generated metabolite [M4] (Fig. 7B), but both [M4] and [M5] were only observed when esterase activity was inhibited, presumably the result of preventing further metabolism of [M4] and [M5] to de-esterified products. Neither [M2] nor [M6] was detected in incubations from A549 or DPX2 cells.
Fig. 7.
(A) Quantification of BDP metabolites produced by A549 cells treated with BDP or BDP with esterase inhibitors (+EI). (B) Relative quantification of BDP metabolites produced by DPX2 cells treated with BDP or BDP with esterase inhibitors (+EI). Data are the mean and standard deviation from six replicates. n.d. Signifies that the metabolite was not detected. (C and D) CYP3A enzyme mRNA abundance, measured by qPCR in A549 (C) and DPX2 (D) cells. Data are represented as the number of mRNA copies per 10,000 copies of β2-macroglobulin (a “housekeeping” gene). Statistics used for A549 cell data analysis were one-way analysis of variance with Dunnett’s post-hoc test. For DPX2 cell data analysis two-way ANOVA with Bonferronni post-hoc testing was used. Data are the mean and standard deviation from 6 replicates. n.d. Signifies that mRNA was not detected. *P < 0.05; ***P < 0.001; ****P < 0.0001.
Expression of CYP3A Enzymes in A549 and DPX2 Cells.
qPCR was used to determine the expression of CYP3A4, 3A5, and 3A7 mRNA in A549 cells (Fig. 7C) and DPX2 cells (Fig. 7D). Only CYP3A5 mRNA was detected in A549 cells, which was significantly (∼2-fold) induced by BDP treatment, presumably via [M1] binding to the glucocorticoid receptor (GR), as previously suggested (Dvorak and Pavek, 2010). In DPX2 cells, only mRNA for CYP3A5 and 3A7 were detectable under basal conditions, but 2- to 3-fold induction of mRNA for all three cytochrome P450 enzymes was observed following BDP treatment in the presence of esterase inhibitors (Fig. 7D), presumably the result of PXR activation by BDP, as previously described (Dvorak and Pavek, 2010).
Discussion
Inhaled glucocorticoids are the mainstay treatment of asthma. Although therapy is effective for the majority of patients, ∼30% do not achieve adequate asthma control (Mjaanes et al., 2006). A possible explanation for ineffectiveness of treatment by glucocorticoids, referred to as steroid insensitivity, may be variations in the metabolic clearance capacity of therapeutically active glucocorticoids from lung cells involving CYP3A enzymes. Thus, the goal of this study was to evaluate whether the CYP3A family of enzymes, which are known to metabolize glucocorticoids, contributed to the pharmacological activation of BDP by hydrolyzing the ester at the C-21 position to form [M1] or if cytochrome P450 metabolism only produced pharmacologically less active or inactive metabolites such as [M2],[M3], and presumably [M4–6].
Characterization of BDP metabolism using human liver microsomes, recombinant CYP3A enzymes, lung, and liver cells demonstrated that cytochrome P450 enzymes were not involved in the production of the pharmacologically active metabolite [M1] but produced a number of previously uncharacterized and presumed inactive clearance metabolites [M4–6]. Further evaluation of BDP metabolism by individual CYP3A enzymes indicated that CYP3A4 and CYP3A5 metabolized BDP at similar overall rates but that BDP was not a substrate for CYP3A7 in vitro. Characterization of individual metabolites of BDP generated by CYP3A4 and CYP3A5 revealed the production of previously uncharacterized oxygenated [M4] and dehydrogenated [M5] metabolites, as well as [M6], a product of both CYP3A4-mediated oxygenation and esterase-mediated ester cleavage. Collectively, these results support the hypothetical concept that BDP efficacy may be impacted by the rate and extent to which BDP is metabolized to pharmacologically inactive metabolites in lung cells and/or elsewhere in the body by CYP3A enzymes.
In support of this idea, a recent publication by Stockmann et al. (2013) demonstrated that the expression of the CYP3A4*22 allele, which codes for an inactive enzyme in the liver (Elens et al., 2011a,b, 2012, 2013; Kitzmiller et al., 2013), coupled with treatment of these patients with the CYP3A4 and 3A5 mechanism-based inhibitor fluticasone propionate (Murai et al., 2010), correlated with improved asthma control. As such, a decrease in CYP3A-mediated metabolism of fluticasone propionate (and possibly other glucocorticoids) in the lung and liver may increase the bioavailability of the active drug in the lung, leading to greater efficacy. Similarly, several genetic polymorphisms of CYP3A5 influence the expression levels of functional enzyme. The most common polymorphism is CYP3A5*3, which codes for an inactive form of CYP3A5 (Kuehl et al., 2001; Westlind-Johnsson et al., 2003). The majority of Caucasians are homozygotes for the CYP3A5*3 allele and therefore do not express active CYP3A5 in the lung or elsewhere in the body. However, individuals that express the CYP3A5*1 allele, express an active form of CYP3A5 (Kuehl et al., 2001). Following the logic above, such individuals may exhibit increased clearance capacity for BDP in lung cells and elsewhere in the body, potentially contributing to glucocorticoid insensitivity. However, this intriguing concept is speculative.
Results from his study may also suggest that developmental differences in CYP3A activity could influence BDP (and other glucocorticoid) activity. Specifically, because CYP3A7 is the dominant CYP3A enzyme expressed in newborn patients (Schuetz et al., 1994; Lacroix et al., 1997) and CYP3A7 does not efficiently metabolize BDP, infants could have decreased clearance of BDP in lung and liver cells relative to children and adults, potentially leading to greater exposure to active drug at lower doses. If this idea is confirmed by future studies, the ability to use lower doses to achieve therapeutic benefit could potentially reduce the risk for toxicity, such as adrenal crisis in patients receiving high doses of inhaled glucocorticoids (Newman, 2003).
Finally, the relationship between esterase and CYP3A-dependent metabolism and glucocorticoid receptor activity were explored using lung (A549) and liver (DPX2) cells (Fig. 7). In A549 and DPX2 cells, BDP was metabolized by esterases and CYP3A enzymes to yield [M1], [M3], [M4], and [M5], albeit to varying levels (Fig. 7, A and B). In A549 cells, the formation of [M1] from BDP was dependent upon esterases (Fig. 7A). Ketoconazole, a selective CYP3A enzyme inhibitor, blocked [M5] production in A549 cells but not [M1] production (data not shown), confirming that CYP3A5 did not contribute to the activation of BDP in lung cells. Exploring the induction of CYP3A enzymes in response to BDP showed that only CYP3A5 was induced in A549 cells when esterases were active (Fig. 7B). These data show that the production of [M1], likely acting through the glucocorticoid receptor, was responsible for the induction of CYP3A5. In DPX2 cells, however, CYP3A4, 3A5, and 3A7 mRNA expression was significantly induced by BDP, but only when esterase activity was inhibited (Fig. 7D). These results suggest that rapid esterase-mediated clearance of BDP in the absence of esterase inhibitors limits CYP3A4 mRNA induction in liver cells and that the induction of CYP3A enzyme mRNA was primarily mediated by PXR activation by BDP, not [M1], as documented in the literature (Dvorak and Pavek, 2010). These metabolic profiles and induction results suggest two different pathways regulate CYP3A gene expression in the lung and the liver.
The induction of CYP3A enzymes in response to glucocorticoid treatment has been extensively studied in the liver. At micromolar concentrations, induction of CYP3A enzymes occurs through PXR (which is not expressed in lung cells) (Dvorak and Pavek, 2010). At submicromolar concentrations, glucocorticoid receptor ligands promote glucocorticoid receptor homodimer assembly and translocation into the nucleus where the transcription of the constitutive androstane receptor is induced. The constitutive androstane receptor nuclear receptor then binds to the retinoid X receptor α, forming a constitutive androstane receptor:retinoid X receptor α heterodimer, which ultimately promotes the transcription of CYP3A genes (Dvorak and Pavek, 2010). This pathway, however, has not been fully evaluated in lung cells, and the results presented in Fig. 7 indicating a role for the glucocorticoid receptor in regulating CYP3A5 expression by BDP, and presumably other glucocorticoids, suggest that more studies are needed to determine not only how CYP3A5 is induced in the lung by glucocorticoids, but also to assess the pharmacological significance of increased CYP3A gene/enzyme expression in the lung, relative to the efficiency of glucocorticoids.
In summary, this study expands our knowledge of cytochrome P450-mediated metabolism of the glucocorticoid BDP, including the preliminary identification of several new cytochrome P450-specific metabolites: [M4], [M5], and [M6]. Although these studies do not link variations in CYP3A gene expression and/or function in lung cells, or elsewhere in the body, to variations in the disposition and efficacy of BDP in humans, these data support such a hypothesis. Further research into the ideas generated by this study may ultimately help advance our understanding of glucocorticoid resistance among asthmatics and improve selection of the most effective inhaled glucocorticoid for specific patients.
Acknowledgments
The authors thank Dr. Christopher Orton for initiating this research, Dr. Judy Raucy (Puracyp Inc.) for the kind donation of DPX2 cells, Dr. John G. Lamb for assistance in culturing the DPX2 cells, and Dr. Roger Gaedigk for the CYP3A5 probe and primer sequences.
Abbreviations
- A549
human lung adenocarcinoma
- B2M
β2 macroglobulin
- BDP
beclomethasone dipropionate
- DPX2
HepG2 background with PXR and a CYP3A4 reporter construct stably expressed
- GR
glucocorticoid receptor
- LC/MS/MS
liquid chromatography-tandem mass spectrometry
- NADPH
nicotinamide adenine dinucleotide phosphate
- PXR
pregnane X receptor
- qPCR
real-time quantitative polymerase chain reaction
Authorship Contributions
Participated in research design: Roberts, Moore, Reilly, Ward, Yost.
Conducted experiments: Roberts, Moore.
Performed data analysis: Roberts, Moore.
Wrote or contributed to the writing of the manuscript: Roberts, Moore, Reilly, Ward, Yost.
Footnotes
This work was supported by the National Institutes of Health Eunice Kennedy Shriver National Institute of Child Health and Human Development [Grant R01HD060559]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health and Human Development or the National Institutes of Health. This work was also supported by the National Institutes of Health National Institute of General Medical Sciences [Grant GM074249]; and the Howard Hughes Medical Institute under their HHMI Med to Grad initiative [Grant 56006777].
References
- Brogden RN, Heel RC, Speight TM, Avery GS. (1984) Beclomethasone dipropionate. A reappraisal of its pharmacodynamic properties and therapeutic efficacy after a decade of use in asthma and rhinitis. Drugs 28:99–126 [DOI] [PubMed] [Google Scholar]
- Chanoine F, Grenot C, Heidmann P, Junien JL. (1991) Pharmacokinetics of butixocort 21-propionate, budesonide, and beclomethasone dipropionate in the rat after intratracheal, intravenous, and oral treatments. Drug Metab Dispos 19:546–553 [PubMed] [Google Scholar]
- Daley-Yates PT, Price AC, Sisson JR, Pereira A, Dallow N. (2001) Beclomethasone dipropionate: absolute bioavailability, pharmacokinetics and metabolism following intravenous, oral, intranasal and inhaled administration in man. Br J Clin Pharmacol 51:400–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak Z, Pavek P. (2010) Regulation of drug-metabolizing cytochrome P450 enzymes by glucocorticoids. Drug Metab Rev 42:621–635 [DOI] [PubMed] [Google Scholar]
- Edsbäcker S, Andersson P, Lindberg C, Paulson J, Ryrfeldt A, Thalén A. (1987) Liver metabolism of budesonide in rat, mouse, and man. Comparative aspects. Drug Metab Dispos 15:403–411 [PubMed] [Google Scholar]
- Elens L, Becker ML, Haufroid V, Hofman A, Visser LE, Uitterlinden AG, Stricker BCh, van Schaik RH. (2011a) Novel CYP3A4 intron 6 single nucleotide polymorphism is associated with simvastatin-mediated cholesterol reduction in the Rotterdam Study. Pharmacogenet Genomics 21:861–866 [DOI] [PubMed] [Google Scholar]
- Elens L, Bouamar R, Hesselink DA, Haufroid V, van der Heiden IP, van Gelder T, van Schaik RH. (2011b) A new functional CYP3A4 intron 6 polymorphism significantly affects tacrolimus pharmacokinetics in kidney transplant recipients. Clin Chem 57:1574–1583 [DOI] [PubMed] [Google Scholar]
- Elens L, Hesselink DA, van Schaik RH, van Gelder T. (2012) The CYP3A4*22 allele affects the predictive value of a pharmacogenetic algorithm predicting tacrolimus predose concentrations. Br J Clin Pharmacol [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elens L, Nieuweboer A, Clarke SJ, Charles KA, de Graan AJ, Haufroid V, Mathijssen RH, van Schaik RH. (2013) CYP3A4 intron 6 C>T SNP (CYP3A4*22) encodes lower CYP3A4 activity in cancer patients, as measured with probes midazolam and erythromycin. Pharmacogenomics 14:137–149 [DOI] [PubMed] [Google Scholar]
- Foe K, Brown KF, Seale JP. (1998a) Decomposition of beclomethasone propionate esters in human plasma. Biopharm Drug Dispos 19:1–8 [DOI] [PubMed] [Google Scholar]
- Foe K, Brown KF, Seale JP. (2000a) Comparative kinetics of metabolism of beclomethasone propionate esters in human lung homogenates and plasma. J Pharm Sci 89:1143–1150 [DOI] [PubMed] [Google Scholar]
- Foe K, Cheung HT, Tattam BN, Brown KF, Seale JP. (1998b) Degradation products of beclomethasone dipropionate in human plasma. Drug Metab Dispos 26:132–137 [PubMed] [Google Scholar]
- Foe K, Cutler DJ, Brown KF, Seale JP. (2000b) Metabolism kinetics of beclomethasone propionate esters in human lung homogenates. Pharm Res 17:1007–1012 [DOI] [PubMed] [Google Scholar]
- Hughes SC, Shardlow PC, Hollis FJ, Scott RJ, Motivaras DS, Allen A, Rousell VM. (2008) Metabolism and disposition of fluticasone furoate, an enhanced-affinity glucocorticoid, in humans. Drug Metab Dispos 36:2337–2344 [DOI] [PubMed] [Google Scholar]
- Hukkanen J, Hakkola J, Anttila S, Piipari R, Karjalainen A, Pelkonen O, Raunio H. (1997) Detection of mRNA encoding xenobiotic-metabolizing cytochrome P450s in human bronchoalveolar macrophages and peripheral blood lymphocytes. Mol Carcinog 20:224–230 [DOI] [PubMed] [Google Scholar]
- Hukkanen J, Väisänen T, Lassila A, Piipari R, Anttila S, Pelkonen O, Raunio H, Hakkola J. (2003) Regulation of CYP3A5 by glucocorticoids and cigarette smoke in human lung-derived cells. J Pharmacol Exp Ther 304:745–752 [DOI] [PubMed] [Google Scholar]
- Jönsson G, Aström A, Andersson P. (1995) Budesonide is metabolized by cytochrome P450 3A (CYP3A) enzymes in human liver. Drug Metab Dispos 23:137–142 [PubMed] [Google Scholar]
- Kitzmiller JP, Sullivan DM, Phelps MA, Wang D, Sadee W. (2013) CYP3A4/5 combined genotype analysis for predicting statin dose requirement for optimal lipid control. Drug Metabol Drug Interact 28:59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, et al. (2001) Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 27:383–391 [DOI] [PubMed] [Google Scholar]
- Lacroix D, Sonnier M, Moncion A, Cheron G, Cresteil T. (1997) Expression of CYP3A in the human liver—evidence that the shift between CYP3A7 and CYP3A4 occurs immediately after birth. Eur J Biochem 247:625–634 [DOI] [PubMed] [Google Scholar]
- Lamb JG, Hathaway LB, Munger MA, Raucy JL, Franklin MR. (2010) Nanosilver particle effects on drug metabolism in vitro. Drug Metab Dispos 38:2246–2251 [DOI] [PubMed] [Google Scholar]
- Leclerc J, Tournel G, Courcot-Ngoubo Ngangue E, Pottier N, Lafitte JJ, Jaillard S, Mensier E, Lhermitte M, Broly F, Lo-Guidice JM. (2010) Profiling gene expression of whole cytochrome P450 superfamily in human bronchial and peripheral lung tissues: Differential expression in non-small cell lung cancers. Biochimie 92:292–306 [DOI] [PubMed] [Google Scholar]
- Mjaanes CM, Whelan GJ, Szefler SJ. (2006) Corticosteroid therapy in asthma: predictors of responsiveness. Clin Chest Med 27:119–132, vii [DOI] [PubMed] [Google Scholar]
- Moore CD, Roberts JK, Orton CR, Murai T, Fidler TP, Reilly CA, Yost GS. (2013) Metabolic pathways of inhaled glucocorticoids by the CYP3A enzymes. Drug Metab Dispos 41:379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai T, Reilly CA, Ward RM, Yost GS. (2010) The inhaled glucocorticoid fluticasone propionate efficiently inactivates cytochrome P450 3A5, a predominant lung P450 enzyme. Chem Res Toxicol 23:1356–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutch E, Nave R, McCracken N, Zech K, Williams FM. (2007) The role of esterases in the metabolism of ciclesonide to desisobutyryl-ciclesonide in human tissue. Biochem Pharmacol 73:1657–1664 [DOI] [PubMed] [Google Scholar]
- Newman SP. (2003) Deposition and effects of inhaled corticosteroids. Clin Pharmacokinet 42:529–544 [DOI] [PubMed] [Google Scholar]
- Pearce RE, Leeder JS, Kearns GL. (2006) Biotransformation of fluticasone: in vitro characterization. Drug Metab Dispos 34:1035–1040 [DOI] [PubMed] [Google Scholar]
- Peet CF, Enos T, Nave R, Zech K, Hall M. (2005) Identification of enzymes involved in phase I metabolism of ciclesonide by human liver microsomes. Eur J Drug Metab Pharmacokinet 30:275–286 [DOI] [PubMed] [Google Scholar]
- Schuetz JD, Beach DL, Guzelian PS. (1994) Selective expression of cytochrome P450 CYP3A mRNAs in embryonic and adult human liver. Pharmacogenetics 4:11–20 [DOI] [PubMed] [Google Scholar]
- Stockmann C, Fassl B, Gaedigk R, Nkoy F, Uchida D, Monson S, Reilly CA, Leeder JS, Yost GS, Ward R. (2013) Fluticasone propionate pharmacogenetics: CYP3A4*22 polymorphism and pediatric asthma control. Pediatrics [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum PJ, Chu NI, Cho D, Tökés L, Patterson JW, Wagner PJ, Chaplin MD. (1981) Mechanism for the oxidative defluorination of flunisolide. J Pharmacol Exp Ther 218:16–22 [PubMed] [Google Scholar]
- Teng XW, Cutler DJ, Davies NM. (2003) Mometasone furoate degradation and metabolism in human biological fluids and tissues. Biopharm Drug Dispos 24:321–333 [DOI] [PubMed] [Google Scholar]
- Thummel KE, Wilkinson GR. (1998) In vitro and in vivo drug interactions involving human CYP3A. Annu Rev Pharmacol Toxicol 38:389–430 [DOI] [PubMed] [Google Scholar]
- Westlind-Johnsson A, Malmebo S, Johansson A, Otter C, Andersson TB, Johansson I, Edwards RJ, Boobis AR, Ingelman-Sundberg M. (2003) Comparative analysis of CYP3A expression in human liver suggests only a minor role for CYP3A5 in drug metabolism. Drug Metab Dispos 31:755–761 [DOI] [PubMed] [Google Scholar]
- Wilcox JB, Avery GS. (1973) Beclomethasone dipropionate corticosteroid inhaler: a preliminary report of its pharmacological properties and therapeutic efficacy in asthma. Drugs 6:84–93 [DOI] [PubMed] [Google Scholar]
- Würthwein G, Rohdewald P. (1990) Activation of beclomethasone dipropionate by hydrolysis to beclomethasone-17-monopropionate. Biopharm Drug Dispos 11:381–394 [DOI] [PubMed] [Google Scholar]