Abstract
Estrogen receptor (ER) antagonists are generally thought to inhibit estrogen action through competitive inhibition, resulting in receptor binding to antagonist rather than agonist. However, microarray analyses reveal a group of genes for which ER agonist and antagonist cooperatively regulate expression, suggesting additional models of combined agonist/antagonist action must exist. In conjunction with a chimeric reporter gene and two modified ERs, one [ERα(GSCKV)] with a mutation in the DNA-binding domain and the other (ERα-G521R) with a ligand-binding specificity mutation, we herein demonstrate that ER agonist and antagonist cooperatively activate gene expression through an ER heteroligand dimer complex (ER-HLD) consisting of one subunit of the receptor dimer bound to agonist and another occupied by antagonist. Coimmunoprecipitation experiments confirmed interaction between the agonist-bound and antagonist-bound receptors. This cooperative activation of gene expression was enhanced by steroid receptor coactivator 3 coactivator, and required each ligand-bound subunit of the dimer to bind to DNA, as well as both activation function 1 domains for maximal transcriptional activity. Ligand combinations able to induce ER-HLD transcriptional activity include the agonists 17β-estradiol or conjugated estrogens with the antagonists tamoxifen, raloxifene, bazedoxifene, or fulvestrant. Moreover, ER-HLD can activate transcription in the context of a natural promoter. Taken together, these findings broaden our understanding of the complex relationship between ER agonist and antagonist, and suggest a novel model by which cell and tissue selective effects of antiestrogens may be achieved.
Introduction
Estrogen receptors (ERs) belong to the nuclear receptor (NR) superfamily of transcription factors, and are modular proteins consisting of six domains designated A–F (Heldring et al., 2007). The A/B domain contains activation function 1 (AF-1), which influences transcriptional activity in a ligand-independent manner. The DNA-binding domain (DBD; domain C) defines the response element specificity, whereas the ligand-binding domain (LBD; domain E) mediates ligand binding and dimerization, and contains a ligand-dependent transactivation function referred to as activation function 2 (AF-2). The hinge region (domain D) is located between the DBD and LBD, whereas the F domain is located at the extreme C terminus. There are two ER genes, ERα and ERβ, and the corresponding proteins share approximately 95 and 55% homology in the DBD and LBD, respectively (Thomas and Gustafsson, 2011). In the classic model, both receptors bind to estradiol with high affinity, whereupon they undergo changes in conformation, dimerize as either homodimers (ERα/ERα or ERβ/ERβ) or heterodimers (ERα/ERβ), bind to estrogen response elements (EREs) in the regulatory region of estrogen target genes, and recruit coactivators to modulate gene expression (Heldring et al., 2007; Thomas and Gustafsson, 2011).
The biologic functions of estrogens are important in many tissues, including the breast, prostate, bone, brain, and reproductive tract, and pharmacological regulation of ER function is important in pre- and postmenopausal women. In addition to ER agonists such as 17β-estradiol (E2), there are two classes of ER antagonists. The selective estrogen receptor modulators (SERMs), including tamoxifen, raloxifene, and bazedoxifene, exert estrogen-like and antiestrogen-like activities in a tissue-selective manner, whereas the selective estrogen receptor degraders such as fulvestrant (ICI 182,780) downregulate ERα and inhibit ER function in most contexts. Agonists and antagonists for ER bind to the same site within the LBD (Brzozowski et al., 1997; Shiau et al., 1998), and antagonists are therefore able to competitively block estrogens from binding to the receptor and inducing gene expression. Moreover, antagonist-bound ERs adopt a distinct conformation that enables them to preferentially interact with corepressors rather than coactivators (Huang et al., 2010), thereby reinforcing their negative regulatory properties.
In contrast to the model of agonist and antagonist competing for binding to the LBD, ligand binding to heterodimeric NRs such as retinoid X receptor (RXR) partnered with retinoic acid receptor (RAR), thyroid hormone receptor, vitamin D receptor (VDR), or peroxisome proliferator–activated receptor (PPAR), and regulation of their transcriptional activities is more complex (Germain et al., 2002, 2006; Pérez et al., 2012). Some heterodimers (PPAR/RXR, liver X receptor/RXR, farnesoid X receptor/RXR) are “permissive,” whereby an RXR-selective ligand (“rexinoid”) and an NR partner ligand can independently or synergistically activate the transcriptional activity of the heterodimer (Kliewer et al., 1992; Willy et al., 1995; Leblanc and Stunnenberg, 1995). In contrast, “nonpermissive” heterodimers (including RAR/RXR, VDR/RXR, and thyroid hormone receptor /RXR) are unresponsive to rexinoids alone, and can only be stimulated by ligands that bind to the RXR partner receptor (Kurokawa et al., 1994; Forman et al., 1995; Westin et al., 1998), although rexinoids synergize with partner agonists to activate gene transcription when both ligands are present (Roy et al., 1995; Shulman et al., 2004). In addition, an RXR homodimer antagonist functions as an agonist when RXR is paired to specific partners, including PPAR and RAR (Lala et al., 1996). Thus, the ability of a given receptor ligand to activate or repress gene expression can be influenced by other ligands bound to the dimer partner.
Unlike RXR-associated heterodimers, ERα is generally thought to form homodimers bound to either agonist or antagonist, depending upon the relative ligand concentrations. Thus, antiestrogens (e.g., tamoxifen) block E2 binding to ERα and antagonize estrogen-stimulated gene expression, which is highly desirable relative to breast cancer prevention and treatment. However, recent MCF-7 breast cancer cell microarray experiments revealed a group of novel genes cooperatively regulated by ERα agonist and antagonist (Chang et al., 2010; Wardell et al., 2012). This is difficult to reconcile with competitive antagonism, and argues for an additional model for combined agonist/antagonist regulation of ER activity. Based on RXR heterodimer models, it was hypothesized that an antagonist within an ERα heteroligand dimer (ER-HLD) complex, consisting of antagonist-bound and agonist-bound ERα subunits, could stimulate, rather than inhibit, gene expression. To date, this possibility has not been addressed experimentally, particularly because regulating the binding of agonist and antagonist to homodimers is considerably more difficult than controlling the interaction of two different ligands with RXR-containing heterodimers. Nonetheless, this is an important question that has implications for the pharmacology of SERMs when used to inhibit ERα function in breast tissues where systemic and even locally produced estrogens may be present (Yaghjyan and Colditz, 2011).
To evaluate whether antagonists could positively regulate the transcriptional activity of an ER-HLD complex, a chimeric luciferase reporter system was developed in conjunction with receptor mutations that regulate the specificity of ligand binding as well as DNA interaction. This model system demonstrates that ERα agonist and antagonist can cooperatively activate gene expression through an ER-HLD complex, and has implications for understanding the molecular pharmacology of clinically important estrogen receptor antagonists.
Materials and Methods
Cell Culture and Reagents.
The HeLa human cervical carcinoma and HepG2 human hepatoma cell lines were obtained from American Type Culture Collection (Manassas, VA). HeLa cells were cultured in Dulbecco’s modified Eagles medium (DMEM) media containing 10% fetal bovine serum (FBS). HepG2 cells were maintained in minimum essential medium (MEM) supplemented with 10% FBS. 17β-Estradiol, 4-hydroxytamoxifen, and raloxifene were purchased from Sigma-Aldrich (St. Louis, MO). Bazedoxifene was provided by Pfizer Inc. (New York, NY). The pure antiestrogen ICI 182,780 was obtained from Tocris (Ellisville, MO). The mixture of the unconjugated forms of the 10 most abundant conjugated estrogen (CE) components of Premarin (conjugated estrogens; Pfizer Inc.) was prepared in the same relative proportions as is present in the Premarin formulation (Chang et al., 2010). Thus, a 1 mM CE stock solution was prepared from 1 mM solutions of each of the 10 components mixed proportionally according to their percentage ratios.
Plasmids.
The expression plasmid pCR3.1-ERα and corresponding reporter gene ERE-e1b-Luc have been described previously (Nawaz et al., 1999a). The pCR3.1-ERα(GSCKV) plasmid was generated by polymerase chain reaction (PCR) mutagenesis, in which primers containing the DNA-binding domain mutation region (5′-GGAAGCTGTAAAGTT-3′) were used in two PCR reactions, generating ER cDNA fragments between the SmaI site and the mutation and between the mutation and the BglII site. The two PCR products were then used as a template in a third PCR reaction to create a fragment that was then digested with SmaI and BglII and subcloned back into the pCR3.1-ERα vector. The pCR3.1-ERα-G521R-Flag was generated by PCR with primers (5′-GGGGTACCCGGTCTGCACCCTGC-3′ and 5′-CCGCTCGAGCGGACCGTGGCAGGGAA-3′) using pCMV5-ERα-G521R as a template [provided by Dr. Benita Katzenellenbogen (Schodin et al., 1995)] and subcloned into pCR3.1 vector upstream of the sequence for a Flag epitope. The pcDNA3-ERα(GSCKV)-(HA) plasmid was generated by PCR with primers (5′-CGGAATTCCGGTCTGCACCCTGC-3′ and 5′-CCGCTCGAGGACCGTGGCAGGGAA-3′) using pCR3.1-ERα(GSCKV) as a template, and subcloned into the pcDNA3-HA vector [provided by Dr. Hank H. Qi from Harvard Medical School, Boston, MA (Qi et al., 2010)]. Glucocorticoid response element (GRE)-Luc and pC3-Luc have been described previously (Tzukerman et al., 1994; Nawaz et al., 1999b). The GGGTCAcagTGACCT estrogen response element of ERE-e1b-Luc was mutated to GGGTCAcagTGTTCT or TGTACAcagTGACCT for the EGRE-e1b-Luc and the GERE-e1b-Luc reporter genes, respectively, by site-directed mutagenesis. The 1/2ERE-Luc reporter was generated by mutating the GRE half-site in the EGRE-Luc construct from TGTTCT to TTTTTT using mutagenesis primers (sense 5′-CTGCGATCTAAGTAAGCTTGGGTCACAGTTTTTTGATCAAAGTTAATGT-3′ and antisense 5′-ACATTAACTTTGATCAAAAAACTGTGACCCAAGCTTACTTAGATCGCAG-3′) following the instruction of the Stratagene QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). The AF-1 mutant constructs 179C-ERα-G521R and 179C-ERα-GSCKV were generated by PCR, amplifying the fragments of 179-595AA of the pCR3.1-ERα-G521R and pCR3.1-ERα(GSCKV) vectors, respectively, and subcloning them into pCR3.1 vectors, respectively. All constructs were verified by DNA sequencing.
Luciferase Assay.
For the HeLa cell line, 200,000 cells/well were seeded in 6-well plates in phenol red-free DMEM containing 5% charcoal-stripped FBS (sFBS) one day before transfection. Cells were transfected with 1 μg of reporter gene and indicated amounts of expression vectors using Lipofectamine reagent (Invitrogen, Grand Island, NY) in Opti-MEM (Invitrogen) following the manufacturer’s instructions. Four to 6 hours later, Opti-MEM was replaced with phenol red-free DMEM containing 5% sFBS. For the HepG2 cell line, 750,000 cells/well were seeded in 6-well plates in phenol red-free MEM supplemented with 10% sFBS. The next day, the medium was removed. Cells were transfected with 1 μg of reporter gene and indicated amounts of expression vectors using Lipofectamine 2000 in Opti-MEM following the manufacturer’s instructions. Four to 6 hours later, Opti-MEM was replaced with phenol red-free MEM supplemented with 10% sFBS. The following day, cells were treated with the indicated ligands at the specified concentrations, and 24 hours thereafter were harvested using TEN buffer (40 mM Tris, pH 7.5, 1 mM EDTA, and 150 nM NaCl). Cell pellets were resuspended in Reporter Lysis Buffer (Promega, Madison, WI) and lysed according to the manufacturer’s instructions. Luciferase activity was determined using a Luminoskan Ascent Microplate Luminometer (Thermo Fisher Scientific, Waltham, MA) with Luciferase Assay Reagent (Promega) and normalized to protein concentration determined by Bio-Rad Protein Assay reagent (Bio-Rad Laboratories, Hercules, CA). All experiments were performed in duplicate and repeated three times.
Western Blot.
Cells were collected using TEN buffer and resuspended in ER extraction buffer (50 mM Tris, pH 8.0, 400 mM NaCl, 5 mM EDTA, 1% NP40, 0.2% Sarkosyl) supplemented with an EDTA-free Complete Mini protease inhibitor (Roche Applied Science, Indianapolis, IN). Cells were lysed on ice for 30 minutes, after which protein concentrations were determined as described earlier. Twenty micrograms of protein was mixed with LDS Sample Buffer containing Sample Reducing Agent (both Invitrogen), then heated to 75°C for 10 minutes. Proteins were separated using a 10% Bis-Tris gel and transferred in NuPAGE Transfer Buffer (Invitrogen) to a nitrocellulose membrane (GE Water & Process Technologies, Trevose, PA). The membrane was blocked in 1 × phosphate-buffered saline plus 0.1% Tween-20 (Sigma-Aldrich) containing 5% dry milk powder (w/v). Primary antibodies were diluted in 1 × phosphate-buffered saline plus 0.1% Tween-20 with 1% milk powder and detected using horseradish peroxidase–conjugated secondary antibodies and the ECL Plus Western Blotting Detection System (both from GE Healthcare, Waukesha, WI). Primary antibodies include ERα (HC-20; Santa Cruz Biotechnology, Santa Cruz, CA), actin (clone C4; EMD Millipore, Billerica, MA), anti-Flag (F7425; Sigma-Aldrich), and HA (Y-11; Santa Cruz Biotechnology).
Coimmunoprecipitation.
Following transfection, HeLa cells were cultured in phenol red-free DMEM containing 5% sFBS for 48 hours before being treated with the indicated ligands for 1 hour. Cells were harvested on ice in 1 × phosphate-buffered saline containing Complete Mini protease inhibitor (Roche, Basel, Switzerland). Cell pellets were lysed at 4°C for 30 minutes in immunoprecipitation lysis buffer [50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 (Sigma-Aldrich), and Complete Mini protease inhibitor], and lysate protein concentrations were determined as described earlier. One milligram of lysate was precleared for 2 hours at 4°C with 4 μg of mouse IgG and prewashed protein G PLUS agarose beads (both from Santa Cruz Biotechnology) under constant rotation. Thereafter, lysates were incubated with 40 μl of anti-Flag M2 affinity gel suspension overnight at 4°C. Immune complexes were washed four times with inositol phosphate lysis buffer, then eluted using LDS sample buffer, and analyzed by Western blot analysis as described earlier.
Results
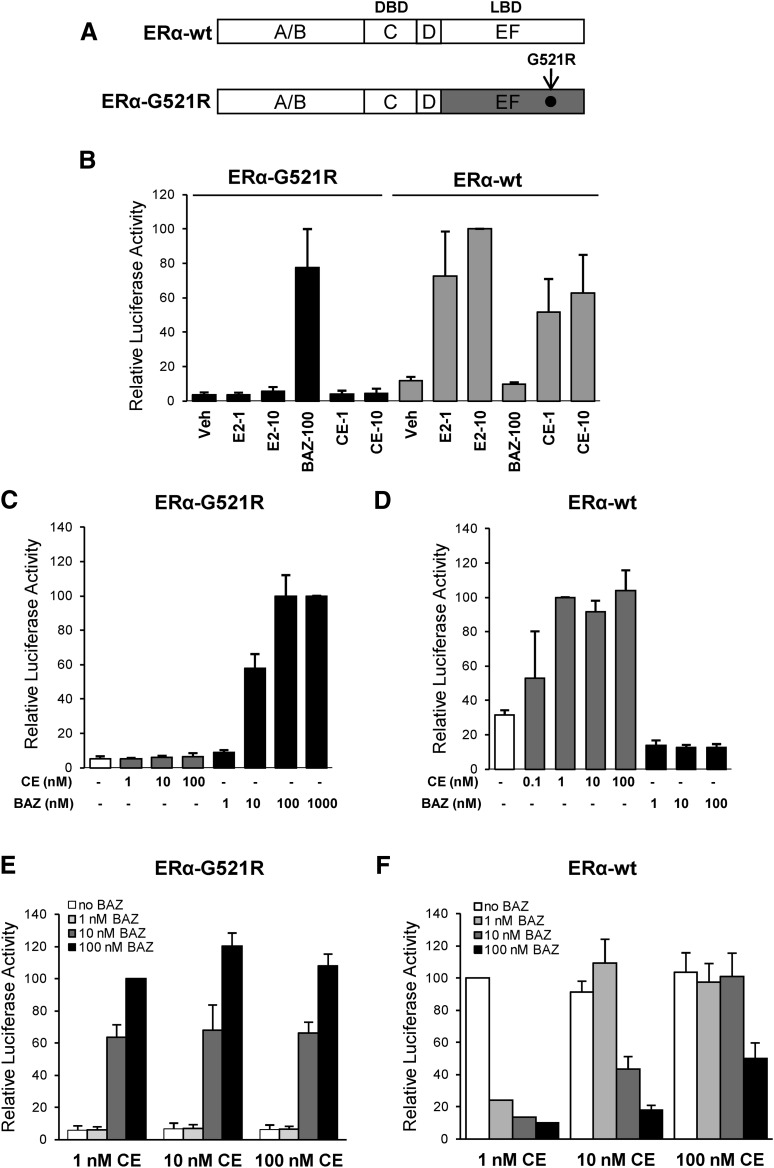
To test whether ER dimers in which each subunit binds to a different ligand can regulate gene expression in response to combined agonist/antagonist treatments, a chimeric receptor and reporter system was used. First, a mutant form of ERα, ERα-G521R (Schodin et al., 1995), that possesses a ligand-binding domain mutation permitting high-affinity antagonist but not agonist binding was generated (Fig. 1A). As shown in Fig. 1B, E2 and CEs, but not BAZ, activated wild-type ERα transcriptional activity measured by pC3-Luc reporter in HepG2 cells. In contrast, the ERα-G521R mutant was strongly activated by BAZ but not E2 or CE, demonstrating a complete change in the ligand specificity for inducing gene expression. Bazedoxifene was unable to increase pC3-Luc activity in cells cotransfected with pCR3.1 empty vector, indicating that the BAZ-induced luciferase expression was ERα-G521R–dependent (unpublished data). The relative ability of CE and BAZ to regulate the transcriptional activity of ERα-G521R was further evaluated in dose-response studies. Increasing concentrations of CE from 1 to 100 nM were unable to stimulate ERα-G521R activity, whereas BAZ stimulated the activity of ERα-G521R in a dose-responsive manner (Fig. 1C).
Fig. 1.
Activation of ERα-G521R with bazedoxifene. (A) Schematic diagram showing wild-type ERα (ERα-wt) and mutant ERα (ERα-G521R) used in these experiments. ERα consists of six domains (A–F). The A/B domains contain the ligand-independent AF-1 domain, the C domain is the DBD, D is the hinge, and the EF domain encompasses the LBD. The arrow indicates the position of the G521R point mutation. (B) HepG2 cells were cotransfected with 1 μg of pC3-Luc reporter and 50 ng of expression vectors for either ERα-G521R or ERα-wt. Cells were treated with the indicated concentrations of E2, BAZ, or CE for 24 hours, and then harvested for luciferase assay. The activity of 10 nM E2-treated ERα-wt transfected cells was set to 100. (C and E) HepG2 cells cotransfected with 1 μg of pC3-Luc reporter and 50 ng of ERα-G521R were treated with the indicated concentrations of ligands for 24 hours. The luciferase activity of cells treated with 1000 nM BAZ (C) or 100 nM BAZ + 1 nM CE (E) was set to 100. (D and F) HepG2 cells cotransfected with 1 μg of pC3-Luc reporter and 50 ng of ERα-wt were treated with the indicated concentrations of CE or BAZ alone or in combination for 24 hours. The luciferase activity of 1 nM CE treated cells was set to a value of 100 (D and F). Values represent the average ± S.E.M. of 3 independent experiments.
The second objective was to establish conditions in which wild-type ERα would be activated by CE in the presence of a concentration of BAZ sufficient to activate ERα-G521R. A dose-response study in cells transfected with wild-type ERα shows comparable activation of the pC3-Luc reporter gene by 1–100 nM CE, whereas BAZ failed to activate luciferase activity (Fig. 1D). As expected, BAZ activation of ERα-G521R was not influenced by cotreatment with CE, consistent with the inability of this receptor to be activated by estrogens (Fig. 1E). In contrast, antagonism studies demonstrated that BAZ, at a concentration that is equal to or greater than the concentration of CE, inhibited wild-type ERα transcriptional activity (Fig. 1F). Thus, BAZ at a concentration of 1 or 10 nM does not antagonize 10 nM CE- or 100 nM CE–induced wild-type ERα activity, respectively. Since 10 nM but not 1 nM BAZ was sufficient to activate ERα-G521R activity alone (Fig. 1C) or in combination with 1–100 nM CE (Fig. 1E), a combined treatment dosage of 100 nM CE and 10 nM BAZ was selected based on the ability of these treatments to effectively activate wild-type ERα and ERα-G521R, respectively.
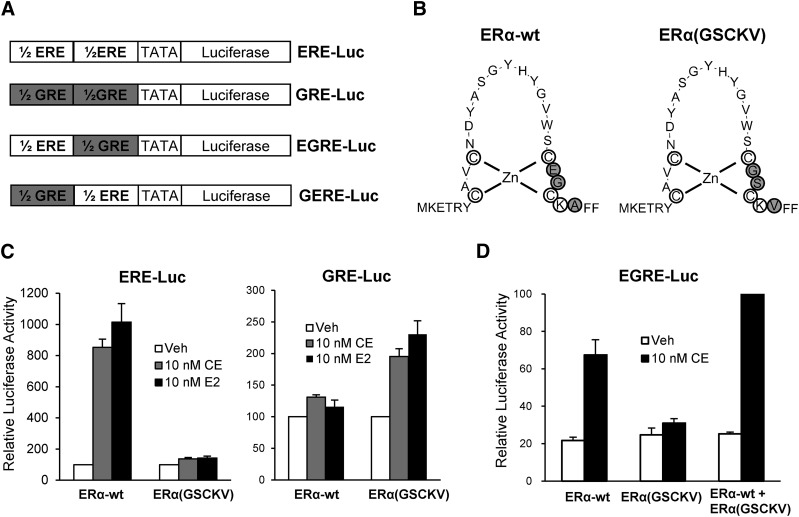
Third, a DNA-binding specificity mutation was introduced into wild-type ERα, and a modified ER responsive reporter was generated. These chimeric reporter genes, GERE-Luc or EGRE-Luc, consist of a luciferase cDNA downstream of a composite response element consisting of a GRE half-site fused 5′ to an ERE half-site and vice versa (Fig. 2A). To generate a receptor mutant that is able to bind to the GRE half-site, three amino acid residues located at the P-box, implicated in specific DNA interaction, in the first zinc finger module of the wild-type ERα DBD were mutated to the corresponding amino acids in the glucocorticoid receptor DBD (Fig. 2B), thereby generating ERα(GSCKV), which had been shown previously to enable binding to GREs (Mader et al., 1989). Luciferase assays were used to evaluate the function of ERα(GSCKV). As shown in Fig. 2C, ERα(GSCKV) was unable to activate ERE-Luc reporter activity in response to E2 or CE treatment, whereas expression of the GRE-Luc reporter was strongly stimulated. This was consistent with prior testing reports (Tremblay et al., 1999). In contrast, wild-type ERα strongly stimulated expression of luciferase from the ERE-Luc but not the GRE-Luc reporter. To test whether wild-type ERα and ERα(GSCKV) can work together to regulate the activity of the chimeric EGRE-Luc reporter, HepG2 cells were transfected with EGRE-Luc as well as wild-type ERα and ERα(GSCKV), alone or in combination. As shown in Fig. 2D, 10 nM CE induced EGRE-Luc activity in the presence of either wild-type ERα or ERα(GSCKV) single receptor to a variable extent. However, greater activity was observed when both receptors were coexpressed, indicating that they work together to activate expression of the EGRE-Luc reporter.
Fig. 2.
The DNA-binding mutant ERα(GSCKV) activates expression of a GRE-containing reporter gene. (A) Schematic diagram showing the ERE-Luc, GRE-Luc, and two chimeric reporter genes. (B) The amino acid sequence of the DBD of wild-type ERα (ERα−wt) and DNA-binding mutant ERα(GSCKV). The gray circles indicate the three amino acids that were mutated to enable binding to a GRE. (C) HepG2 cells cotransfected with 1 μg of the indicated reporter genes and 50 ng of expression vectors as indicated were treated with vehicle (Veh; 0.1% ethanol), 10 nM E2, or 10 nM CE for 24 hours. Data are presented as luciferase activity relative to their respective vehicle-treated control. (D) HepG2 cells were cotransfected with 1 μg of EGRE-Luc and ERα-wt (25 ng), ERα(GSCKV) (25 ng), or both [25 ng of ERα-wt + 25 ng ERα(GSCKV)] plasmids, and treated with vehicle or 10 nM CE for 24 hours. The luciferase activity of 10 nM CE-treated ERα-wt + ERα(GSCKV) cotransfected cells was set to 100. Values represent the average ± S.E.M.; (n = 3) for all experiments.
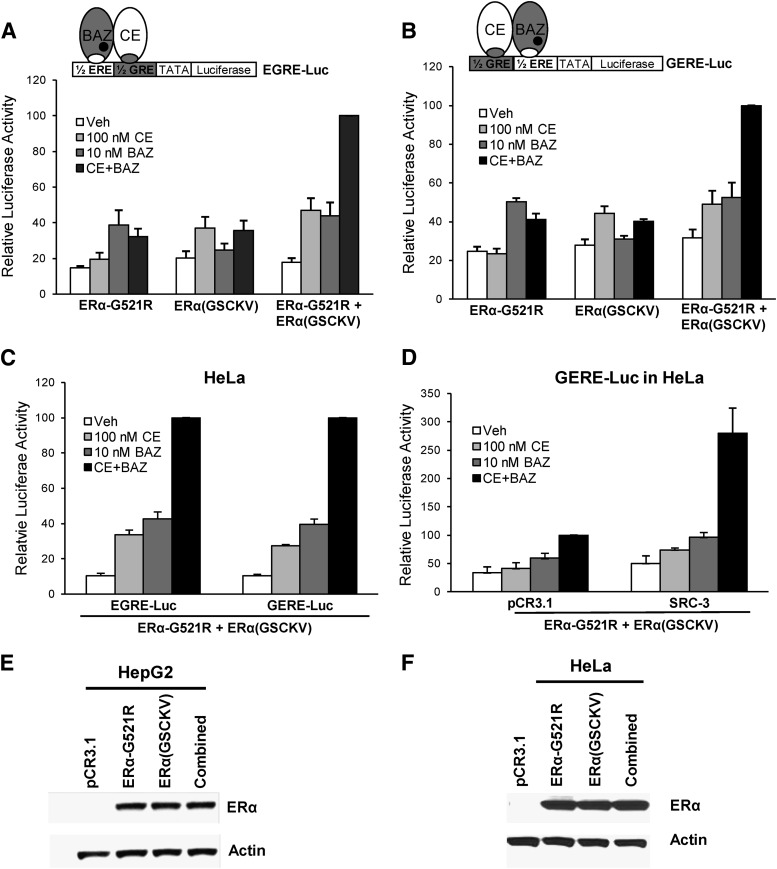
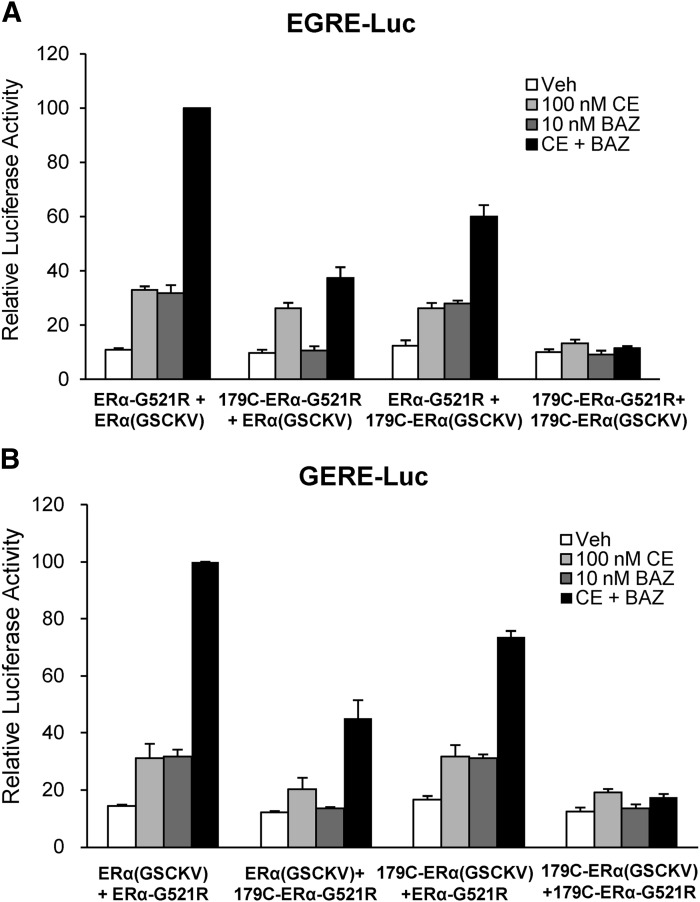
With the successful generation of the chimeric reporter genes and receptor mutations, the ability of ERα dimers bound to agonist and antagonist to activate gene expression was tested. First, HepG2 cells were cotransfected with EGRE-Luc as well as ERα-G521R and ERα(GSCKV), alone or in combination (Fig. 3A). In cells expressing ERα-G521R, luciferase activity was stimulated by BAZ ,but not CE treatment, and CE+BAZ did not further increase luciferase. In cells expressing ERα(GSCKV), CE, but not BAZ, induced reporter activity, whereas the combination of CE and BAZ showed no further induction. When both forms of ERα were expressed in the cells, the combined CE+BAZ treatment induced maximal reporter gene expression, whereas exposure to only agonist or antagonist yielded partial luciferase activity. The cooperative effect of CE+BAZ was also observed on the GERE-Luc reporter (Fig. 3B). To test whether this phenomenon is cell-type specific, similar luciferase assays were conducted in HeLa cells, which demonstrated that combined CE and BAZ treatment induced greater activity of the chimeric reporters than either agent alone (Fig. 3C).
Fig. 3.
Cooperative activation of chimeric reporter gene expression by combined ERα agonist and antagonist treatment. (A and B) HepG2 cells cotransfected with 1 μg of EGRE-Luc or GERE-Luc reporter genes with 50 ng of ERα-G521R, 50 ng ERα(GSCKV), or a combination [25 ng of ERα- G521R + 25 ng of ERα(GSCKV)] of the two were treated with vehicle (0.1% ethanol), 100 nM CE, 10 nM BAZ, or 100 nM CE + 10 nM BAZ for 24 hours. The luciferase activity of 100 nM CE + 10 nM BAZ–treated ERα-wt + ERα(GSCKV) cotransfected cells was set to 100. The illustration above each graph is a schematic representation of the respective chimeric reporter gene (EGRE-Luc or GERE-Luc) and transfected forms of ERα. (C) HeLa cells were cotransfected with 1 μg of EGRE-Luc or GERE-Luc along with 5 ng of ERα-G521R + 5 ng of ERα(GSCKV) and treated as indicated for 24 hours. The luciferase activity of 100 nM CE + 10 nM BAZ–treated cells was set as a value of 100. (D) HeLa cells were cotransfected with 1 μg of GERE-Luc, 5 ng of ERα-G521R, and 5 ng of ERα(GSCKV) along with either 100 ng of pCR3.1 or SRC-3 expression vector, and treated with agonist and antagonist as described earlier. The luciferase activity of 100 nM CE + 10 nM BAZ–treated pCR3.1 + ERα-wt + ERα(GSCKV) transfected cells was set to 100. Values represent the average ± S.E.M. of three independent experiments for all luciferase assays. (E and F) Representative Western blots show the corresponding expression level of transiently transfected ERα-G521R and ERα(GSCKV), alone or in combination, in HepG2 and HeLa cells. HepG2 cells were transfected with 50 ng of ERα-G521R, 50 ng of ERα (GSCKV), or 25 ng of ERα-G521R + 25 ng of ERα(GSCKV). HeLa cells were transfected with 10 ng of ERα-G521R, 10 ng of ERα(GSCKV), or 5 ng of ERα-G521R + 5 ng of ERα(GSCKV). Actin was used as a loading control.
The previous results indicate that ERα agonist and antagonist can work cooperatively to induce maximal transcriptional activity. To test whether coactivators such as steroid receptor coactivator (SRC)-3, which are known to enhance ERα transcriptional activity (Suen et al., 1998), could enhance CE+BAZ–stimulated expression of the chimeric reporter, HeLa cells were transfected with GERE-Luc, ERα-G521R, and ERα(GSCKV) in the absence or presence of SRC-3 expression vector. Exogenous SRC-3 significantly enhanced CE+BAZ–induced luciferase activity, indicating that this coactivator can functionally interact with these receptors following combined agonist/antagonist treatment, and suggesting a role for SRC-3 in mediating the gene expression induced by this combined ligand treatment (Fig. 3D). Additionally, transfection of SRC-1or SRC-2 can also boost the cooperative activation of CE+BAZ (unpublished data). Last, to exclude the effect of differential receptor expression levels on reporter activity, Western blots were performed. HepG2 or HeLa cells transfected with single receptors [ERα-G521R or ERα(GSCKV)] expressed similar levels of receptor in comparison with ERα-G521R and ERα(GSCKV) cotransfected cells, indicating that the greater activity observed in the latter condition is not due to increased receptor expression (Fig. 3, E and F). Taken together, the previous data demonstrate the ability of an antagonist treatment to work together with agonist to activate gene expression in the chimeric model system. This cooperative effect requires the presence of both forms of receptor, and can be coactivated by SRC-3.
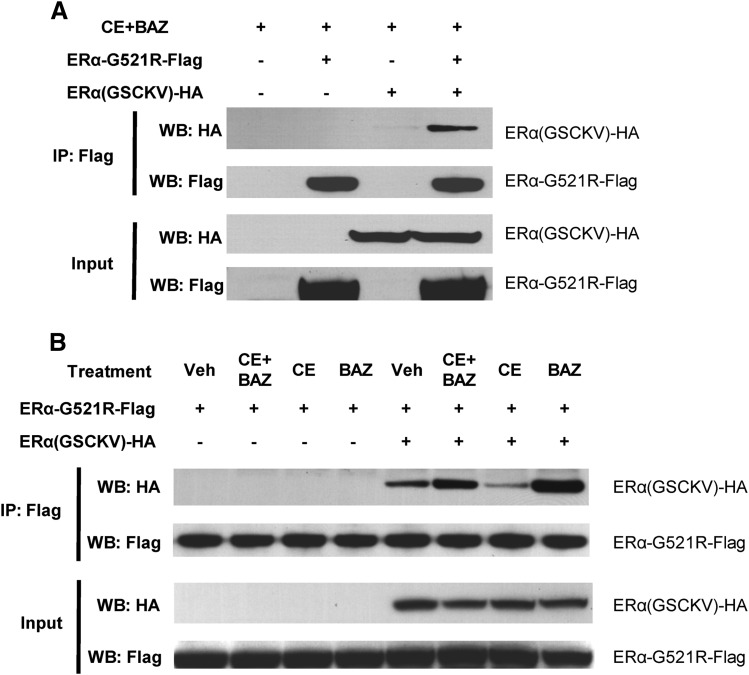
To further characterize the ability of these receptors to mediate the cooperative effect of combined agonist/antagonist treatment, the ability of epitope-tagged ERα-G521R-Flag and ERα(GSCKV)-HA receptors to interact with one another was tested in a coimmunoprecipitation assay. In cells treated with CE+BAZ, Flag antibody is able to immunoprecipitate ERα-G521R-Flag and ERα(GSCKV)-HA, indicating an interaction between the receptors (Fig. 4A). A low level of interaction between the two forms of receptor is detected in the absence of any ligand, and is enhanced upon treatment with CE+BAZ (Fig. 4B, compare lanes 5 and 6), indicating that a complex can be formed between CE-bound ERα(GSCKV)-HA and BAZ-bound ERα-G521R-Flag in vivo. Receptor-receptor interaction was also promoted by BAZ alone (compare lanes 5 and 8), consistent with the ability of this ligand to bind to both receptors. In contrast, the interaction between ERα-G521R-Flag and ERα(GSCKV)-HA was inhibited by CE (compare lanes 5 and 7), presumably due to the ability of CE to promote formation of ERα(GSCKV)-HA homodimers, and consequently reducing the amount of this receptor available for heterodimer formation with ERα-G521R-Flag. To test whether the epitope-tagged receptors are functional, HeLa cells transfected with GERE-Luc, ERα-G521R-Flag, and ERα(GSCKV)-HA were treated with CE and BAZ, alone or in combination. The epitope-tagged vectors were similar to ERα-G521R and ERα(GSCKV) in their ability to cooperatively activate luciferase activity in response to CE+BAZ treatment (unpublished data). These data demonstrate the possibility that agonist-bound ERα(GSCKV)-HA and antagonist-bound ERα-G521R can form an ERα-HLD complex in cells that can cooperatively stimulate gene expression.
Fig. 4.
Ligand-regulated interaction of ERα-G521R and ERα(GSCKV). HeLa cells were transfected with 5 ng of ERα-G521R-Flag and 5 ng of ERα(GSCKV)-HA, either alone or in combination; pCR3.1 empty vector was added to make equal amounts of DNA for each transfection. Forty-eight hours thereafter, cells were treated with 100 nM CE + 10 nM BAZ (A) or vehicle, 100 nM CE, 10 nM BAZ, or 100 nM CE + 10 nM BAZ (B) for 1 hour. Complexes were immunoprecipitated (IP) with anti-Flag antibody, and Western blots (WB) were performed with either Flag or HA antibody, as indicated.
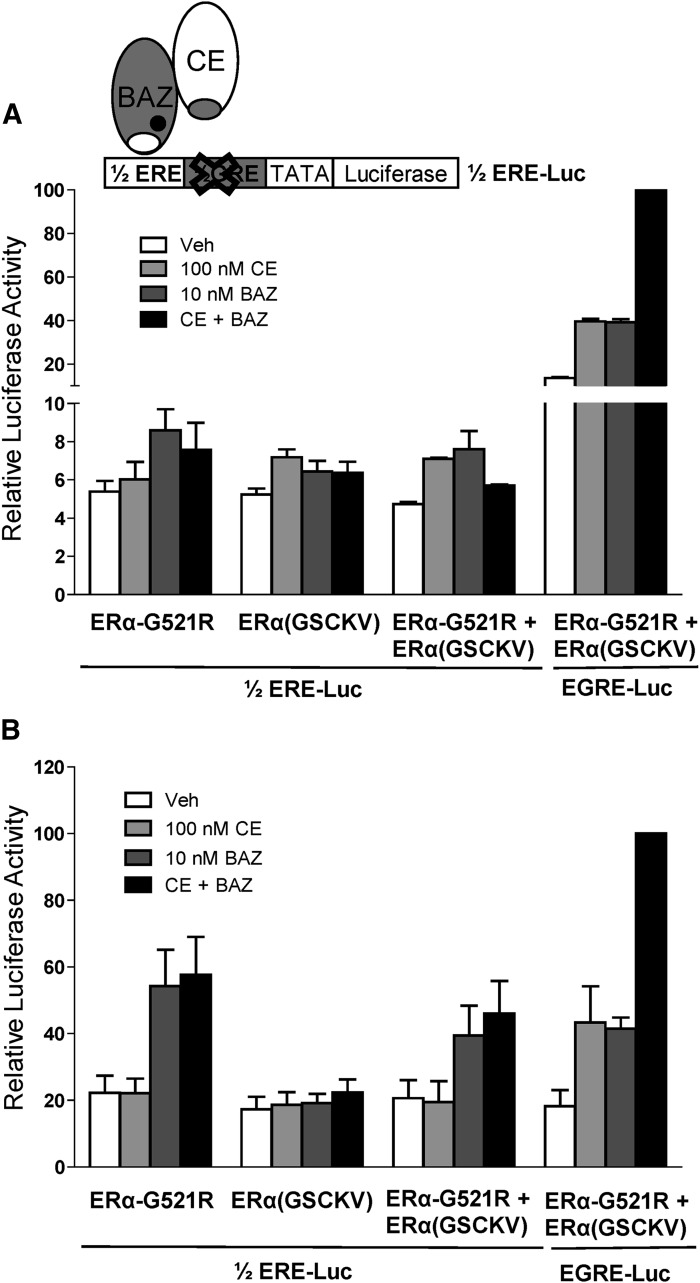
To test whether the CE+BAZ cooperative activation of chimeric reporter activity requires each monomer within the ERα-HLD to bind to DNA, a 1/2ERE-Luc reporter was generated in which the GRE half-site of the chimeric reporter was mutated. Activities induced by CE+BAZ treatment in cells transfected with ERα-G521R and ERα(GSCKV) were compared for the 1/2ERE-Luc versus the EGRE-Luc reporter (Fig. 5, A and B). No cooperative effect of CE+BAZ treatment was observed for the 1/2ERE-Luc reporter in either HepG2 or HeLa cells. This indicates that CE-bound ERα(GSCKV) and BAZ-bound ERα-G521R must both bind to DNA to cooperatively activate gene expression, rather than a single receptor binding to the ERE half-site and interacting with a non-DNA binding receptor partner solely via protein-protein interactions.
Fig. 5.
Cooperative activation by agonist and antagonist requires receptor dimer to bind to DNA. HepG2 (A) and HeLa (B) cells cotransfected with 1/2ERE-Luc or EGRE-Luc reporter genes as well as the indicated ER expression vectors were treated with vehicle (0.1% ethanol), 100 nM CE, 10 nM BAZ, or 100 nM CE + 10 nM BAZ for 24 hours. Data are presented as relative luciferase activity compared to EGRE-Luc plus ERα-G521R + ERα(GSCKV) cotransfected samples treated with 100 nM CE + 10 nM BAZ. Values represent the average ± S.E.M. of three independent experiments.
As the agonist activity of SERMs is dependent on AF-1 activity (Berry et al., 1990), the role of each receptor’s AF-1 domain on the activity achieved by the ERα-HLD complex was evaluated on the EGRE-Luc and GERE-Luc reporters. Deletion of the AF-1 domain of either of the ERα receptors attenuated the cooperative effect of CE+BAZ, regardless of the order in which the receptors bound to the target gene (Fig. 6, A and B). Moreover, removal of both AF-1 domains from the heteroligand dimer complex completely blocked the ability of CE+BAZ to stimulate luciferase gene expression, indicating that the AF-1 domains from each ERα subunit make important contributions to the transcriptional activity of the ERα heteroligand dimer.
Fig. 6.
AF-1 domain is required for maximal activation by combined ERα agonist and antagonist treatment. HeLa cells were cotransfected with EGRE-Luc (A) or GERE-Luc (B) and 5 ng each of the indicated plasmids. One day after transfection, cells were treated with vehicle, 100 nM CE, 10 nM BAZ, or 100 nM CE + 10 nM BAZ for 24 hours and harvested for luciferase assay. Data are presented as relative luciferase activity compared to EGRE-Luc plus ERα-G521R + ERα(GSCKV) cotransfected samples treated with 100 nM CE + 10 nM BAZ. Values represent the average ± S.E.M. of three independent experiments.
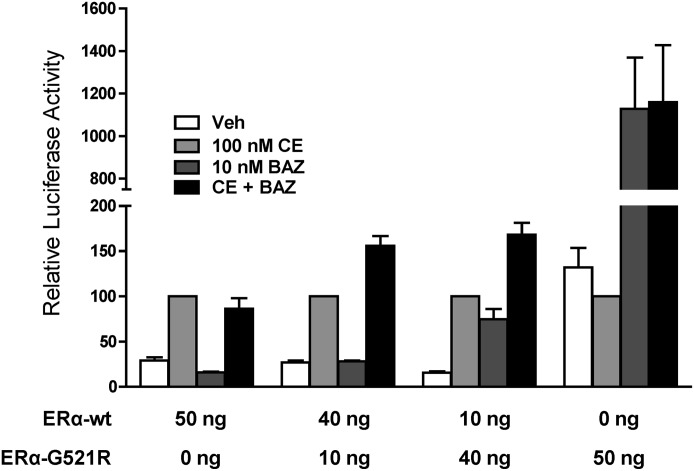
To extend these observations beyond the chimeric reporter model system, the ability of CE+BAZ to cooperatively activate a reporter gene regulated by a natural ERα target promoter was investigated. HepG2 cells were cotransfected with the pC3-Luc reporter, consisting of the complement 3 promoter linked to luciferase (Tzukerman et al., 1994) along with wild-type ERα and ERα-G521R, alone or in combination. For cells expressing only wild-type ERα or ERα-G521R, the activity from CE or BAZ single treatment was equivalent to the CE+BAZ combination treatment (Fig. 7). However, when both receptors are coexpressed, luciferase activity is greater in cells treated with CE+BAZ than either of the single ligands alone. These data clearly indicate that CE-bound wild-type ERα and BAZ-bound ERα-G521R can cooperatively work to promote a transcriptional response.
Fig. 7.
Cooperative activation of pC3-Luc by combined ERα agonist and antagonist treatment. HepG2 cells were cotransfected with the pC3-Luc reporter gene and the indicated amounts of expression vectors for wild-type ERα (ERα-wt) and ERα-G521R, alone or in combination. Treatments were with vehicle, 100 nM CE, 10 nM BAZ, or 100 nM CE + 10 nM BAZ for 24 hours. The level of activity achieved with 100 nM CE alone in each transfection was set to 100. Values represent the average ± S.E.M. of three independent experiments.
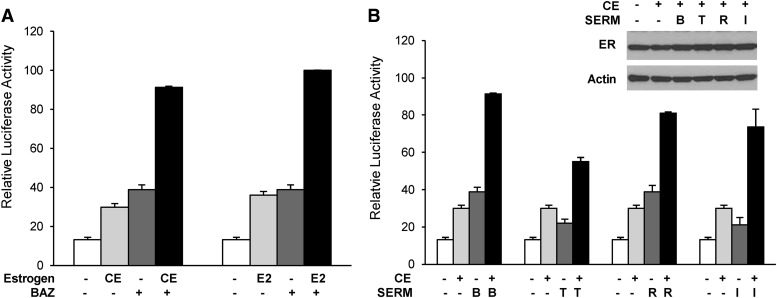
In addition to the combination of CE and BAZ, the ability of different combinations of estrogens and SERMs to cooperatively activate expression of the chimeric reporter was tested. As shown in Fig. 8A, E2 cooperated with BAZ to stimulate the activity of the GERE-Luc reporters in HeLa cells at a level comparable to CE+BAZ combined treatment, indicating that estradiol, the most potent naturally occurring estrogen, can work in combination with antagonists to induce gene expression. This effect is not limited to BAZ, as CE could cooperate with a variety of SERMs, including 4-hydroxytamoxifen, raloxifene, and ICI 182,780 to activate gene expression (Fig. 8B). Similar amounts of receptor expression were detected for all treatment groups, indicating that the greater activities in the combination conditions are not due to a corresponding change in receptor levels (Fig. 8B, inset). Thus, these data indicate that the agonist/antagonist cooperative transcriptional effects mediated by an ERα heteroligand dimer complex can be achieved by a variety of estrogen and antiestrogen combinations.
Fig. 8.
Different estrogens and SERMs synergistically activate chimeric reporter genes. HeLa cells were cotransfected with 1 μg of GERE-Luc reporter gene and 5 ng each of the expression vectors for ERα-G521R and ERα(GSCKV). One day after transfection, cells were treated with ERα agonist (either 100 nM CE or 100 nM E2), alone or in combination with 10 nM BAZ (A), or 100 nM CE, alone or combined with 10 nM of the indicated SERMs [bazedoxifene (B), 4-hydroxytamoxifen (T), raloxifene, and ICI 182,780 (I)] (B) for 24 hours. Data are presented as relative luciferase activity compared to 100 nM E2 + 10 nM BAZ–treated samples. Values represent the average ± S.E.M. of three independent experiments. The inset in panel (B) is a Western blot showing ER expression levels after 24-hour treatment with the indicated ligands.
Discussion
ER antagonists are generally thought to inhibit estrogen action through a competitive mechanism. However, microarray analyses reveal a group of genes for which ER agonist and antagonist cooperatively regulate expression, suggesting additional models of combined agonist/antagonist action must exist (Chang et al., 2010; Wardell et al., 2012). We demonstrate that ER agonist/antagonist combined treatment can cooperatively activate gene expression through an ER-HLD complex consisting of one receptor monomer bound to agonist and another occupied by antagonist. This cooperative activation of gene expression can be enhanced by an SRC-3 coactivator, and requires both ligand-bound subunits to bind to DNA and both AF-1 domains within the ER-HLD for maximal transcriptional activity. Moreover, ER-HLD complexes can activate transcription in the context of a natural promoter, and taken together, these findings demonstrate that ERs bound to different classes of ligands can form dimers that promote gene expression.
The ability of the CE-bound ERα(GSCKV)-HA and BAZ-bound ERα-G521R-Flag to form a dimer complex in cells, as demonstrated by coimmunoprecipitation experiments, revealed a weak level of basal interaction that was significantly enhanced upon treatment with CE+BAZ. This is analogous to the ability of wild-type ERα to exist as a dimer in the absence of ligand, and for E2 to promote dimer interaction (Tamrazi et al., 2002). Individual ER subunits come together to form dimers through two domains, one in the DBD and a second in the LBD (Bai and Gust, 2009). The weak dimerization interface located in the DBD is ligand-independent and responsible for the selection of the spacing distance between the two ERE half-sites (Kuntz and Shapiro, 1997; Bai and Gust, 2009). The latter constitutes the principal ERα dimerization interface with head-to-head contacts between the two subunits mediated primarily through helix 11 (H11) to H11 interactions (Brzozowski et al., 1997; Shiau et al., 1998). Crystal structures of the ERα LBDs bound to either E2 or raloxifene reveal that both ligands bind to the same pocket, and that the overall homodimeric arrangement is the same regardless of whether the LBD is agonist- or antagonist-bound (Brzozowski et al., 1997; Shiau et al., 1998). Indeed, with the possible exception of ICI-bound ERα, the relative monomer orientation at the H11 dimer interface does not vary substantially for receptors bound to agonist or antagonist (Pike et al., 2001), and it is therefore reasonable that these interaction surfaces can mediate dimer formation even when each subunit is occupied by a different ligand.
Through the use of chimeric reporters which directed CE-bound ERα(GSCKV) to bind to DNA either upstream or downstream of the BAZ-bound ERα-G521R, we were able to demonstrate that the relative position of the receptors does not impact cooperative activation by combined agonist/antagonist treatment. This is different from RXR-associated heterodimers in which the polarity of receptor binding to DNA impacts transcriptional activity (Jimenez-Lara and Aranda, 1999; Chandra et al., 2008; Orlov et al., 2012). For direct-repeat response elements, RXR is generally located on the upstream half-site, and this arrangement permits the ligand-bound NR partner to attain an active conformation that facilitates coactivator recruitment (Mangelsdorf and Evans, 1995; Orlov et al., 2012). However, the RAR/RXR heterodimer binds to DNA with a reversed polarity on direct-repeat spacing with 1bp response elements, and in most contexts this heterodimer constitutively represses transcription because the nuclear receptor corepressor remains associated with the RAR/RXR heterodimer, even in the presence of RAR or RXR ligands (Kurokawa et al., 1994, 1995). Thus, RAR/RXR experiments suggest that the relative orientation of half-sites within the response element can allosterically regulate receptor interaction with coregulators leading to either activation or repression of gene expression, but this is not the case for ER-HLDs since they interacted with the SRC-3 coactivator and induced gene expression in both DNA-binding orientations.
In the classic model for ER dimers binding to EREs, each receptor partner binds to one of the ERE half-sites, with the 3-base pair spacer influencing stability of the dimer interface and DNA binding specificity (Schwabe et al., 1993a,b). In contrast, evidence from multiple whole genome studies indicates that a majority of ERα binding sites do not correspond to consensus EREs, but rather encompass one or more half-site EREs (Carroll et al., 2005; Lin et al., 2007). This was somewhat surprising as individual half EREs were considered to be nonfunctional because early in vitro binding experiments, including electrophoretic mobility shift assays, did not detect binding to these half-sites (Klinge, 2001). However, a recent report demonstrates ERα binding to half-site EREs, particularly in conjunction with elevated high-mobility group protein B1 expression, and stimulation of luciferase reporter activity (Joshi et al., 2011). Our data also indicate that ERα can weakly activate gene transcription through a half-site ERE site in response to ligand treatment, but without a cooperative effect of combined agonist/antagonist treatment, indicating that effective ER-HLD promotion of gene expression requires both ER subunits to bind to DNA rather than one receptor tethering a second ER molecule to the gene. Whether ER-HLD can mediate transcriptional effects via multiple half-site EREs, as have been found in numerous genes, remains to be determined.
Activation of gene expression by ERs is dependent upon coactivators, such as SRC family coactivators, which make their primary contact through interaction of one or more LXXLL motifs with the AF-2 domain of agonist-bound receptors (Heery et al., 1997). Conversely, antagonist-bound ERα adapts a conformation in which the LXXLL-like sequence within helix H12 binds against the coactivator docking surface, and thereby prevents coactivators from effectively binding to the receptor (Shiau et al., 1998). It has been reported that SRC-1 and SRC-2 bind to the liganded ERα LBD with a stoichiometry of one coactivator per homodimer (Margeat et al., 2001; Osz et al., 2012), and a similar mode of binding was observed for SRC-1 and SRC-2 binding to RAR/RXR heterodimers (Osz et al., 2012). Moreover, 1,25-dihydroxyitamin D3 and 9-cis-retinoic acid synergistically activate VDR/RXR heterodimers by facilitating a concerted interaction between both receptors with distinct NR boxes of one molecule of SRC-1 (Zhang et al., 2011). Enhancement of agonist/antagonist-stimulated ER-HLD activity by SRC-3 indicates that this coactivator can interact with the heteroliganded dimer to active gene expression. Although it is highly likely that SRC-3 binds to the agonist-bound LBD, it is less certain that this coactivator can bind to antagonist-bound ER-G521R, and whether one molecule of SRC-3 can bind to the LBD of each member of the ER-HLD is therefore unclear.
Relative to costimulation by agonist/antagonist, the AF-1 domain is important for the partial agonist activity of SERMs such as tamoxifen (McInerney et al., 1996), and SRC-1 can bind to both the AF-1 and AF-2 domains of ERα through the coactivator’s Q-rich region and LXXLL motifs, respectively (Mérot et al., 2004). This raises the possibility that SRC-3 interacts with ER-HLDs through different AF domains on distinct subunits [e.g., AF-2 on agonist-bound ER(GSCKV) and AF-1 on antagonist-bound ER-G521R] to induce maximal transcriptional activity. Indeed, although our data revealed that AF-1 deletion for either ERα-G521R or ERα(GSCKV) attenuated transcriptional activity, loss of a single AF-1 domain had a greater impact on the ERα-G521R than the ERα(GSCKV) receptor, suggesting a greater dependence on AF-1 activity by the former component of the ER-HLD. The near complete loss of transcriptional activity when both AF-1 domains were deleted further demonstrates the importance of the AF-1 region to ER-HLD transcriptional activity, and suggests that ER-HLD activity may be more prominent in cell and tissue environments favorable to AF-1 activity (e.g., during elevated growth factor signaling).
Agonist and antagonist combinations can cooperatively activate gene expression mediated by ER-HLDs on chimeric reporter genes as well as on a target gene regulated by a natural promoter, indicating the potential of this novel mode of ERα function to regulate endogenous gene expression. Two recent microarray experiments demonstrated that, in MCF-7 cells treated with estrogen and SERMs, alone or in combination, there is a subset of genes induced to a greater extent by combined estrogen and SERM treatment than by either single agent (Chang et al., 2010; Wardell et al., 2012). The ability of ER-HLD to cooperatively regulate gene expression induced by combined agonist/antagonist treatments provides a possible mechanistic explanation for the induction, rather than the inhibition, of the expression of these genes. On a global level, the features of these potential ER-HLD–regulated genes that enable antagonists to cooperate with, rather than antagonize, agonists in the stimulation of their expression are unknown. Of potential significance, the existing gene microarray data also reveal unique gene sets induced by different antagonist/estrogen combinations, suggesting that the regulatory (e.g., promoter) regions of endogenous genes encode information that enables differential responses to distinct ER-HLD complexes (e.g., following bazedoxifene/estrogen versus raloxifene/estrogen exposure), perhaps through alterations in ER-HLD conformation and resultant recruitment of distinct coactivator complexes.
Our novel finding of ER-HLD mediating the cooperative effect of ER agonist and antagonist within a dimer context not only broadens our understanding of the complex mechanisms of action of ER agonists and antagonists, but is also of significant clinical interest. Combined administration of a SERM and an estrogen has been evaluated clinically, and these tissue-selective estrogen complexes alleviate postmenopausal symptoms without the increased risks associated with estrogen monotherapy (Lobo et al., 2009; Pinkerton et al., 2009; Archer, 2010; Lindsay, 2011). In addition, the well-established use of ER antagonists such as tamoxifen for breast cancer prevention and treatment is based on their demonstrated ability to inhibit E2-induced effects as well as their clinical efficacy (Osborne, 1998). Thus, antagonists are routinely used in settings where endogenous estrogens, be they systemic or locally produced by breast and/or tumor cells (Yaghjyan and Colditz, 2011), are available to generate ER-HLD complexes. The contribution of these types of complexes to the in vivo biologic activities of ER antagonists remains to be fully elucidated, but these data suggest a potential role for ER-HLDs in the gene-, cell-, and tissue-specificity of SERMs and tissue-selective estrogen complexes, as well as the ability of breast tumors to become resistant to SERM therapies.
Acknowledgments
The authors thank Patricia Dillard and Joe Roethele for technical assistance, and Dr. Benita Katzenellenbogen for providing materials.
Abbreviations
- AF-1
activation function 1
- AF-2
activation function 2
- BAZ
bazedoxifene
- CE
conjugated estrogen
- DBD
DNA-binding domain
- DMEM
Dulbecco’s modified Eagles medium
- E2
estradiol
- ER
estrogen receptor
- ERE
estrogen response element
- ER-HLD
ERα heteroligand dimer
- FBS
fetal bovine serum
- GRE
glucocorticoid response element
- H11
helix 11
- HA
hemagglutinin
- LBD
ligand-binding domain
- MEM
minimum essential medium
- NR
nuclear receptor
- pC3
promoter region of the human complement C3 gene
- PCR
polymerase chain reaction
- PPAR
peroxisome proliferator–activated receptor
- RAR
retinoic acid receptor
- RXR
retinoid X receptor
- SERM
selective estrogen receptor modulator
- sFBS
charcoal-stripped FBS
- SRC
steroid receptor coactivator
- VDR
vitamin D receptor
Authorship Contributions
Participated in research design: Liu, Smith.
Conducted experiments: Liu.
Contributed new agents or analytic tools: Han.
Performed data analysis: Liu, Smith.
Wrote or contributed to the writing of the manuscript: Liu, Smith.
Footnotes
This work was supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant DK53002]; and by Pfizer Pharmaceuticals.
References
- Archer DF. (2010) Tissue-selective estrogen complexes: a promising option for the comprehensive management of menopausal symptoms. Drugs Aging 27:533–544 [DOI] [PubMed] [Google Scholar]
- Bai Z, Gust R. (2009) Breast cancer, estrogen receptor and ligands. Arch Pharm (Weinheim) 342:133–149 [DOI] [PubMed] [Google Scholar]
- Berry M, Metzger D, Chambon P. (1990) Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J 9:2811–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engström O, Ohman L, Greene GL, Gustafsson J-Å, Carlquist M. (1997) Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389:753–758 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33–43 [DOI] [PubMed] [Google Scholar]
- Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. (2008) Structure of the intact PPAR-γ-RXR- nuclear receptor complex on DNA. Nature 456:350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KCN, Wang Y, Bodine PVN, Nagpal S, Komm BS. (2010) Gene expression profiling studies of three SERMs and their conjugated estrogen combinations in human breast cancer cells: insights into the unique antagonistic effects of bazedoxifene on conjugated estrogens. J Steroid Biochem Mol Biol 118:117–124 [DOI] [PubMed] [Google Scholar]
- Forman BM, Umesono K, Chen J, Evans RM. (1995) Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell 81:541–550 [DOI] [PubMed] [Google Scholar]
- Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H. (2006) International Union of Pharmacology. LXIII. Retinoid X receptors. Pharmacol Rev 58:760–772 [DOI] [PubMed] [Google Scholar]
- Germain P, Iyer J, Zechel C, Gronemeyer H. (2002) Co-regulator recruitment and the mechanism of retinoic acid receptor synergy. Nature 415:187–192 [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733–736 [DOI] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Ström A, Treuter E, Warner M, et al. (2007) Estrogen receptors: how do they signal and what are their targets. Physiol Rev 87:905–931 [DOI] [PubMed] [Google Scholar]
- Huang P, Chandra V, Rastinejad F. (2010) Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu Rev Physiol 72:247–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Lara AM, Aranda A. (1999) The vitamin D receptor binds in a transcriptionally inactive form and without a defined polarity on a retinoic acid response element. FASEB J 13:1073–1081 [DOI] [PubMed] [Google Scholar]
- Joshi SR, Ghattamaneni RB, Scovell WM. (2011) Expanding the paradigm for estrogen receptor binding and transcriptional activation. Mol Endocrinol 25:980–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. (1992) Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 358:771–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge CM. (2001) Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res 29:2905–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz MA, Shapiro DJ. (1997) Dimerizing the estrogen receptor DNA binding domain enhances binding to estrogen response elements. J Biol Chem 272:27949–27956 [DOI] [PubMed] [Google Scholar]
- Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld MG, Heyman RA, Glass CK. (1994) Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature 371:528–531 [DOI] [PubMed] [Google Scholar]
- Kurokawa R, Söderström M, Hörlein A, Halachmi S, Brown M, Rosenfeld MG, Glass CK. (1995) Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature 377:451–454 [DOI] [PubMed] [Google Scholar]
- Lala DS, Mukherjee R, Schulman IG, Koch SSC, Dardashti LJ, Nadzan AM, Croston GE, Evans RM, Heyman RA. (1996) Activation of specific RXR heterodimers by an antagonist of RXR homodimers. Nature 383:450–453 [DOI] [PubMed] [Google Scholar]
- Leblanc BP, Stunnenberg HG. (1995) 9-cis retinoic acid signaling: changing partners causes some excitement. Genes Dev 9:1811–1816 [DOI] [PubMed] [Google Scholar]
- Lin C-Y, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, et al. (2007) Whole-genome cartography of estrogen receptor α binding sites. PLoS Genet 3:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay R. (2011) Preventing osteoporosis with a tissue selective estrogen complex (TSEC) containing bazedoxifene/conjugated estrogens (BZA/CE). Osteoporos Int 22:447–451 [DOI] [PubMed] [Google Scholar]
- Lobo RA, Pinkerton JV, Gass MLS, Dorin MH, Ronkin S, Pickar JH, Constantine G. (2009) Evaluation of bazedoxifene/conjugated estrogens for the treatment of menopausal symptoms and effects on metabolic parameters and overall safety profile. Fertil Steril 92:1025–1038 [DOI] [PubMed] [Google Scholar]
- Mader S, Kumar V, de Verneuil H, Chambon P. (1989) Three amino acids of the oestrogen receptor are essential to its ability to distinguish an oestrogen from a glucocorticoid-responsive element. Nature 338:271–274 [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. (1995) The RXR heterodimers and orphan receptors. Cell 83:841–850 [DOI] [PubMed] [Google Scholar]
- Margeat E, Poujol N, Boulahtouf A, Chen Y, Müller JD, Gratton E, Cavailles V, Royer CA. (2001) The human estrogen receptor α dimer binds a single SRC-1 coactivator molecule with an affinity dictated by agonist structure. J Mol Biol 306:433–442 [DOI] [PubMed] [Google Scholar]
- McInerney EM, Tsai M-J, O’Malley BW, Katzenellenbogen BS. (1996) Analysis of estrogen receptor transcriptional enhancement by a nuclear hormone receptor coactivator. Proc Natl Acad Sci USA 93:10069–10073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérot Y, Métivier R, Penot G, Manu D, Saligaut C, Gannon F, Pakdel F, Kah O, Flouriot G. (2004) The relative contribution exerted by AF-1 and AF-2 transactivation functions in estrogen receptor α transcriptional activity depends upon the differentiation stage of the cell. J Biol Chem 279:26184–26191 [DOI] [PubMed] [Google Scholar]
- Nawaz Z, Lonard DM, Dennis AP, Smith CL, O’Malley BW. (1999a) Proteasome-dependent degradation of the human estrogen receptor. Proc Natl Acad Sci USA 96:1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz Z, Lonard DM, Smith CL, Lev-Lehman E, Tsai SY, Tsai M-J, O’Malley BW. (1999b) The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol 19:1182–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlov I, Rochel N, Moras D, Klaholz BP. (2012) Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA. EMBO J 31:291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CK. (1998) Tamoxifen in the treatment of breast cancer. N Engl J Med 339:1609–1618 [DOI] [PubMed] [Google Scholar]
- Osz J, Brélivet Y, Peluso-Iltis C, Cura V, Eiler S, Ruff M, Bourguet W, Rochel N, Moras D. (2012) Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors. Proc Natl Acad Sci USA 109:E588–E594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez E, Bourguet W, Gronemeyer H, de Lera AR. (2012) Modulation of RXR function through ligand design. Biochim Biophys Acta 1821:57–69 [DOI] [PubMed] [Google Scholar]
- Pike AC, Brzozowski AM, Walton J, Hubbard RE, Thorsell AG, Li Y-L, Gustafsson J-Å, Carlquist M. (2001) Structural insights into the mode of action of a pure antiestrogen. Structure 9:145–153 [DOI] [PubMed] [Google Scholar]
- Pinkerton JV, Utian WH, Constantine GD, Olivier S, Pickar JH. (2009) Relief of vasomotor symptoms with the tissue-selective estrogen complex containing bazedoxifene/conjugated estrogens: a randomized, controlled trial. Menopause 16:1116–1124 [DOI] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, et al. (2010) Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466:503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Taneja R, Chambon P. (1995) Synergistic activation of retinoic acid (RA)-responsive genes and induction of embryonal carcinoma cell differentiation by an RA receptor α (RAR α)-, RAR β-, or RAR γ-selective ligand in combination with a retinoid X receptor-specific ligand. Mol Cell Biol 15:6481–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schodin DJ, Zhuang Y, Shapiro DJ, Katzenellenbogen BS. (1995) Analysis of mechanisms that determine dominant negative estrogen receptor effectiveness. J Biol Chem 270:31163–31171 [DOI] [PubMed] [Google Scholar]
- Schwabe JW, Chapman L, Finch JT, Rhodes D, Neuhaus D. (1993a) DNA recognition by the oestrogen receptor: from solution to the crystal. Structure 1:187–204 [DOI] [PubMed] [Google Scholar]
- Schwabe JW, Chapman L, Finch JT, Rhodes D. (1993b) The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell 75:567–578 [DOI] [PubMed] [Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95:927–937 [DOI] [PubMed] [Google Scholar]
- Shulman AI, Larson C, Mangelsdorf DJ, Ranganathan R. (2004) Structural determinants of allosteric ligand activation in RXR heterodimers. Cell 116:417–429 [DOI] [PubMed] [Google Scholar]
- Suen C-S, Berrodin TJ, Mastroeni R, Cheskis BJ, Lyttle CR, Frail DE. (1998) A transcriptional coactivator, steroid receptor coactivator-3, selectively augments steroid receptor transcriptional activity. J Biol Chem 273:27645–27653 [DOI] [PubMed] [Google Scholar]
- Tamrazi A, Carlson KE, Daniels JR, Hurth KM, Katzenellenbogen JA. (2002) Estrogen receptor dimerization: ligand binding regulates dimer affinity and dimer dissociation rate. Mol Endocrinol 16:2706–2719 [DOI] [PubMed] [Google Scholar]
- Thomas C, Gustafsson J-Å. (2011) The different roles of ER subtypes in cancer biology and therapy. Nat Rev Cancer 11:597–608 [DOI] [PubMed] [Google Scholar]
- Tremblay GB, Tremblay A, Labrie F, Giguère V. (1999) Dominant activity of activation function 1 (AF-1) and differential stoichiometric requirements for AF-1 and -2 in the estrogen receptor α-β heterodimeric complex. Mol Cell Biol 19:1919–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzukerman MT, Esty A, Santiso-Mere D, Danielian P, Parker MG, Stein RB, Pike JW, McDonnell DP. (1994) Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol 8:21–30 [DOI] [PubMed] [Google Scholar]
- Wardell SE, Kazmin D, McDonnell DP. (2012) Research resource: Transcriptional profiling in a cellular model of breast cancer reveals functional and mechanistic differences between clinically relevant SERM and between SERM/estrogen complexes. Mol Endocrinol 26:1235–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin S, Kurokawa R, Nolte RT, Wisely GB, McInerney EM, Rose DW, Milburn MV, Rosenfeld MG, Glass CK. (1998) Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature 395:199–202 [DOI] [PubMed] [Google Scholar]
- Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. (1995) LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev 9:1033–1045 [DOI] [PubMed] [Google Scholar]
- Yaghjyan L, Colditz GA. (2011) Estrogens in the breast tissue: a systematic review. Cancer Causes Control 22:529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Wang Y, Busby SA, Pascal BD, Garcia-Ordonez RD, Bruning JB, Istrate MA, et al. (2011) DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat Struct Mol Biol 18:556–563 [DOI] [PMC free article] [PubMed] [Google Scholar]