Abstract
N-methyl-d-aspartate (NMDA) receptors are glutamate-gated ion channels whose function is critical for normal excitatory synaptic transmission in the brain and whose dysfunction has been implicated in several neurologic conditions. NMDA receptor function is subject to extensive allosteric regulation both by endogenous compounds and by exogenous small molecules. Elucidating the structural determinants and mechanism of action by which allosteric regulators control gating will enhance our understanding of NMDA receptor activation and facilitate the development of novel therapeutics. Here, we investigated the structural determinants for (3-chlorophenyl)(6,7-dimethoxy-1-((4-methoxyphenoxy)methyl)-3,4-dihydroisoquinolin-2(1H)-yl)methanone (CIQ), a GluN2C/2D-selective positive allosteric modulator. We show that CIQ does not bind to the amino-terminal domain of the NMDA receptor and does not share structural determinants with modulators acting at the agonist-binding domain dimer interface or ion channel pore. Rather, we identified critical determinants of CIQ modulation in the region near the first transmembrane helix of GluN2D, including in a putative pre-M1 cuff helix that may influence channel gating. We also show that mutations within the GluN2D pre-M1 region alter open probability of the NMDA receptor. These results suggest a novel site of action for potentiation of NMDA receptors by small molecules and implicate the pre-M1 region in NMDA receptor gating.
Introduction
N-methyl-d-aspartate (NMDA) receptors are ligand-gated ion channels that mediate excitatory synaptic transmission in the central nervous system (Traynelis et al., 2010). These nonselective cation channels are tetrameric complexes comprising GluN1, GluN2, and GluN3 subunits with typical NMDA receptors containing two GluN1 subunits and two GluN2 subunits (Ulbrich and Isacoff, 2008). The GluN1 subunit binds the coagonist glycine or d-serine and the GluN2 subunit binds the coagonist glutamate. There are four GluN2 subunits, GluN2A, 2B, 2C, and 2D, each of which is encoded by a separate gene (Hollmann and Heinemann, 1994). NMDA receptors composed of different GluN2 subunits exhibit markedly different biophysical and pharmacological properties (Vicini et al., 1998), which enables NMDA receptor subtypes to play distinct roles in brain physiology and development (Cull-Candy and Leszkiewicz, 2004).
NMDA receptor subunits are composed of three semi-autonomous domains: an amino-terminal domain (ATD), an agonist-binding domain (ABD), and a transmembrane domain. In addition, they contain a large intracellular region consisting of 100–600 amino acids. The transmembrane domain consists of three transmembrane helices—M1, M3, and M4—and a re-entrant pore loop, called M2. NMDA receptors have several allosteric sites, including the side-to-side GluN1/GluN2 dimer interface of the ATD (Karakas et al., 2011; Mony et al., 2011), the back-to-back GluN1/GluN2 dimer interface of the ABD (Gielen et al., 2008; Hansen et al., 2012) and the ion channel pore (Antonov and Johnson, 1996; Kashiwagi et al., 2002; Blanpied et al., 2005).
NMDA receptors are potentiated by several endogenous molecules, including arachidonic acid (Miller et al., 1992), dynorphin A (Zhang et al., 1997), sulfated neurosteroids (Wu et al., 1991), and polyamines (Ransom and Stec, 1988; McGurk et al., 1990; Reynolds, 1990; Williams et al., 1990). In addition, aminoglycosides potentiate NMDA receptors in a manner similar to potentiation by polyamines (Masuko et al., 1999). These compounds show varying subunit-selectivity and structural determinants of action. Additionally, the first class of positive allosteric modulators selective for GluN2C- and GluN2D-containing NMDA receptors was recently reported (Mullasseril et al., 2010). This class of potentiators doubles the current response to maximally effective concentrations of agonist for NMDA receptors containing GluN2C or GluN2D. These modulators are not agonists and do not affect the potency of either glutamate or glycine. Moreover, two regions of the GluN2 subunit were previously found to be critical for the selectivity of this class of potentiators: a 16-amino acid stretch linking the ATD with the ABD and a point mutation in the M1 transmembrane helix.
To gain a better understanding of allosteric potentiation of NMDA receptors, which could lead to therapeutics with novel selectivity and mechanisms of action, we sought to determine which regions of the receptor might contribute to the binding site and thereby control the actions of tetrahydroisoquinoline allosteric potentiators typified by CIQ.
Materials and Methods
DNA Constructs and Mutagenesis.
cDNAs for rat GluN1-1a (GenBank accession numbers U11418 and U08261; hereafter GluN1) and rat GluN2D (L31611) were used. The GluN2D amino-terminal domain deletion construct (2DΔATD) was generated by removing approximately 400 bp from the 5′ untranslated region of a previously described GluN2D ATD deletion construct (Yuan et al., 2009). The GluN1 ATD deletion construct was generated using a modified QuikChange reaction as described (Makarova et al., 2000). Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer’s protocol. All mutations were verified by dideoxy DNA sequencing (Eurofins MWG Operon, Huntsville, AL). The amino acids are numbered according to the full-length protein, including the signal peptide, with the initiating methionine as 1. For expression in Xenopus laevis oocytes, cDNA constructs were linearized by restriction enzymes and used as a template to produce cRNAs using the mMessage mMachine kit [Ambion (Life Technologies), Grand Island, NY] according to the manufacturer’s protocol.
Two-Electrode Voltage-Clamp Recordings.
Defolliculated stage VI Xenopus laevis oocytes (EcoCyte Bioscience, Austin, TX) were injected with cRNA encoding GluN1 and GluN2D. After cRNA injection, the oocytes were stored at 15–19°C in culture media containing (in mM) 88 NaCl, 2.4 NaHCO3, 1 KCl, 0.33 Ca(NO3)2, 0.41 CaCl2, 0.82 MgSO4, 5 Tris-HCl (pH 7.4 with NaOH), 1 U/ml penicillin, 0.1 mg/ml gentamicin sulfate, and 1 μg/ml streptomycin. Two-electrode voltage-clamp recordings were performed 2–7 days postinjection at room temperature (23°C). The extracellular oocyte recording solution contained (in mM) 90 NaCl, 1 KCl, 10 HEPES, 0.5 BaCl2, and 0.01 EDTA (pH 7.4 with NaOH). Solutions were applied by gravity and solution exchange was controlled by custom software operating an 8-port modular valve positioner (Hamilton Company, Reno, NV). Voltage and current electrodes were filled with 3.0 M KCl, and currents were recorded at a holding potential of −40 mV. Voltage control was accomplished with a two-electrode voltage-clamp amplifier (OC-725; Warner Instruments, Hamden, CT). For (3-chlorophenyl)(6,7-dimethoxy-1-((4-methoxyphenoxy)methyl)-3,4-dihydroisoquinolin-2(1H)-yl)methanone (CIQ) concentration-response curves, the recording solutions also included 5 mM 2-(hydroxypropyl)-β-cyclodextrin, which was found to improve the solubility of CIQ, and receptors were activated by 100 μM glutamate and 30 μM glycine. For glutamate concentration-response curves, 30 μM glycine was present in all solutions. For glycine concentration-response curves, 100 μM glutamate was present in all solutions.
Whole-Cell and Outside-Out Patch-Clamp Electrophysiology.
Human embryonic kidney 293 cells (CRL 1573, sex unknown; American Type Culture Collection, Rockville, MD) were seeded on glass coverslips coated with poly(d-lysine) (0.1 mg/ml) approximately 48 h before the experiments. The culture medium was Dulbecco’s modified Eagle’s medium with GlutaMAX-I [catalog #10569; Gibco (Life Technologies), Grand Island, NY] supplemented with 10% dialyzed fetal bovine serum (catalog #26400; Gibco), 10 U/ml penicillin, and 10 µg/ml streptomycin. Cells were transiently transfected using the calcium phosphate precipitation method (Chen and Okayama, 1987) with plasmid cDNAs encoding GluN1, GluN2D, and green fluorescent protein. Immediately after transfection, 200 µM d,l-2-amino-5-phosphonovalerate and 200 µM 7-chlorokynurenic acid were added to the culture medium. The cells were used for experiments approximately 18–24 h after transfection.
Whole-cell voltage-clamp recordings were performed at −60 mV using an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA) at room temperature (23°C). Recording electrodes (3–4 MΩ) were made from thin-wall glass micropipettes (World Precision Instruments, Sarasota, FL) using a Flaming-Brown puller (Model P-1000; Sutter Instrument Company, Novato, CA). The electrodes were filled with internal solution containing (in mM) 110 d-gluconic acid, 110 CsOH, 30 CsCl, 5 HEPES, 4 NaCl, 0.5 CaCl2, 2 MgCl2, 5 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA), 2 NaATP, and 0.3 NaGTP (pH 7.35 with CsOH). The extracellular recording solution was composed of (in mM) 150 NaCl, 10 HEPES, 3 KCl, 0.5 CaCl2, and 0.01 EDTA (pH 7.4 with NaOH). Currents were low-pass filtered at 8 kHz (-3 dB, 8-pole Bessel filter; Frequency Devices, Ottawa, IL) and digitized at 20 kHz using Clampex (Molecular Devices). Outside-out channel recordings were performed at −80 mV using recording electrodes (5–9 MΩ) made from thick-wall borosilicate glass (Warner Instruments). Internal and extracellular solutions were the same as above. Currents were filtered at 8 kHz as above and digitized at 40 kHz.
Data Analysis.
CIQ concentration-response curves were individually fit to the Hill equation using a variable slope:
where Max is the maximum CIQ current as a percentage of the glutamate/glycine current, EC50 is the half-maximally effective concentration of CIQ, x is the experimental concentration of CIQ, and nH is the Hill slope. Glutamate and glycine concentration-response curves from individual oocytes were also fit by the Hill equation:
where Max is the maximum current elicited by agonist, Min is the current elicited by the lowest concentration of agonist and except for 2D(Y578A) was not significantly different from baseline current, x is the experimental concentration of agonist, and nH is the Hill slope. Glutamate and glycine curves from individual oocytes were then normalized to the fitted minimum and maximum current and averaged for display.
The time constants describing the onset of 5H-dibenzo[a,d]cyclohepten-5,10-imine (dizocilpine maleate; MK-801) inhibition (τMK-801 block) were measured by fitting a single exponential function to the current decay from 90 to 10% of the peak current after MK-801 application:
where I is the current at time t, τ is the time constant of MK-801 inhibition, Ipeak is the peak agonist-evoked current, and C is an offset. Relative open probabilities (Popen) were calculated as 1/τMK-801 block and normalized to the average value for GluN1/GluN2D (Blanke and VanDongen, 2008; Gielen et al., 2009; Vance et al., 2011).
For the alanine scanning mutagenesis, statistical significance was assessed using a one-way analysis of variance with Dunnett’s post hoc test versus GluN1/GluN2D with α set to 0.05. For statistical testing of EC50 values, a one-way analysis of variance with Dunnett’s post hoc test was calculated using the logarithm of the EC50 values with α set to 0.05.
Reagents.
CIQ was purchased from Life Chemicals (catalog number F0535-0139; Burlington, ON, Canada). (+)-MK-801 was purchased from Tocris Bioscience (Bristol, UK). Ketamine was a gift from Dr. Stephen Holtzman (Emory University). Glutamate and glycine were purchased from Sigma-Aldrich (St. Louis, MO). 2-(hydroxypropyl)-β-cyclodextrin was purchased from Acros Organics (West Chester, PA). All other chemicals were purchased from Fisher Scientific.
Results
CIQ Does Not Act at Known Modulatory Sites.
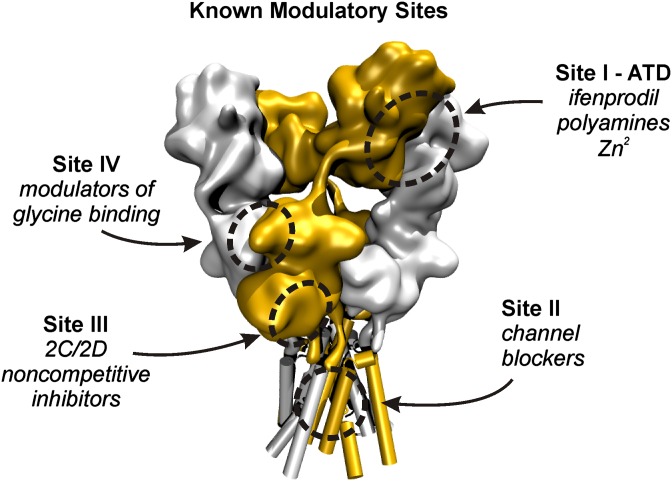
Previous work identified two distinct regions of GluN1/GluN2D that were critical for potentiation by CIQ and analogs: the ATD-ABD linker and the first transmembrane helix, M1 (Mullasseril et al., 2010). These regions are seemingly independent of each other based on homology to the GluA2 tetrameric crystal structure, with the ATD-ABD linker being about 65–70 Å extracellular to the M1 helix (Sobolevsky et al., 2009). Thus, it is unlikely that positive allosteric modulators could directly interact with both regions simultaneously. Moreover, it is unclear whether these regions might be allosterically coupled, as is the case for the ATD and the ABD in which ATD ligands such as Zn2+ and ifenprodil allosterically regulate glutamate binding (Kew et al., 1996; Paoletti et al., 1997; Zheng et al., 2001; Erreger and Traynelis, 2005). Therefore, we sought to reconcile the contribution of these apparently discrete regions to potentiation by CIQ. To do so, we systematically explored the importance of both well-established and emerging regulatory sites on the NMDA receptor (Fig. 1) for positive allosteric modulation of GluN1/GluN2D receptors. These sites, shown in Fig. 1, are the interface between the GluN1 and GluN2 ATDs (site I), the ion channel pore (site II), the lower lobe of the ABD (site III), and the agonist-binding domain dimer interface (site IV). These four modulatory sites are critical for the actions of GluN2B-selective antagonists (e.g., ifenprodil) and positive modulators (e.g., spermine), NMDA receptor channel blockers (e.g., ketamine), GluN2C/2D-selective antagonists [e.g., (E)-4-(6-methoxy-2-(3-nitrostyryl)-4-oxoquinazolin-3(4H)-yl)-benzoic acid (QNZ46)], and GluN2A-selective antagonists [e.g., 3-chloro-4-fluoro-N-[(4-[(2-(phenylcarbonyl)hydrazino)carbonyl]phenyl)methyl]-benzenesulfonamide (TCN-201)], respectively.
Fig. 1.
Known modulatory sites on the NMDA receptor are outlined on a volume representation of the GluA2 AMPA receptor with subunits corresponding to GluN1 shown in silver and those corresponding to GluN2 shown in orange. Site I, the ATD, is involved in modulation by GluN2B-selective inhibitors such as ifenprodil, which bind to the GluN1-GluN2 ATD dimer interface. In addition, Zn2+ ions bind the ATD at the cleft of the GluN2 clamshell-like domain. Site II resides deep within the transmembrane pore of the receptor and is critical for binding of channel blockers such as Mg2+, MK-801, ketamine, and memantine. Site III, located proximal to the membrane at the lower lobe of the GluN2 ABD, is critical for block by noncompetitive, glutamate-dependent inhibitors such as QNZ46 and 5-(4-Bromophenyl)-3-(1,2-dihydro-6-methyl-2-oxo-4-phenyl-3-quinolinyl)-4,5-dihydro-g-oxo-1H-pyrazole-1-butanoic acid (DQP-1105). Site IV is located at the GluN1-GluN2 ABD dimer interface and is critical for allosteric inhibition of glycine binding by TCN-201 and TCN-213.
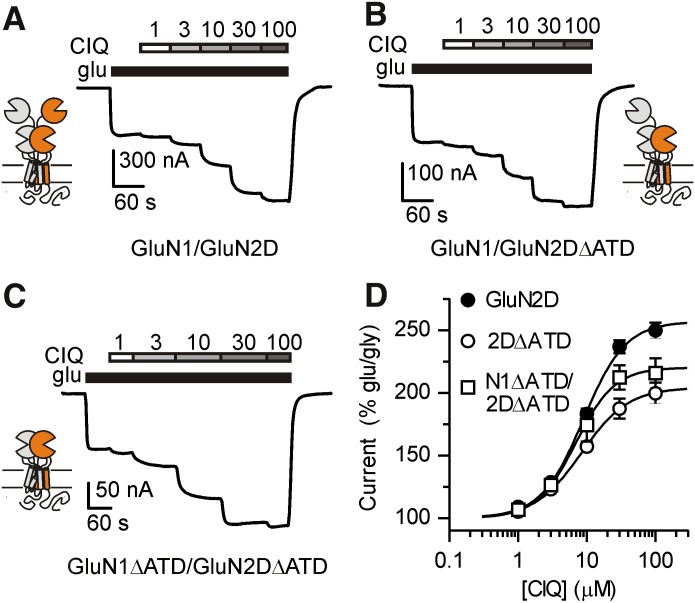
To evaluate the importance of site I in positive allosteric modulation of GluN1/GluN2D receptors, we recorded CIQ potentiation of receptors lacking the GluN2D ATD and the ATD-ABD linker (GluN2DΔATD) and found that CIQ potentiation was unaffected when then ATD was deleted from GluN2D (Fig. 2B). These results suggest that CIQ does not bind solely to the ATD of GluN2D. However, the GluN2B-selective antagonist ifenprodil was recently shown to bind NMDA receptors at the interface of the GluN1/GluN2 ATD dimer (Karakas et al., 2011), raising the possibility that residues from the GluN1 ATD could contribute to the CIQ binding site. Contrary to this hypothesis, removal of the ATD and ATD-ABD linker from both GluN1 and GluN2D (GluN1ΔATD/GluN2DΔATD) did not reduce potentiation by CIQ (Fig. 2C). These data eliminate the possibility that CIQ potentiates GluN2C- and GluN2D-containing NMDA receptors by binding to the ATD (site I).
Fig. 2.
GluN1/GluN2D (A), GluN1/GluN2DΔATD (B), or GluN1ΔATD/GluN2DΔATD (C) receptors were expressed in Xenopus laevis oocytes and current responses recorded using a two-electrode voltage-clamp. Currents were activated by 100 µM glutamate and 30 μM glycine and CIQ (1, 3, 10, 30, and 100 μM) was subsequently coapplied with agonists. (D) CIQ potentiated GluN1/GluN2D, GluN1/GluN2DΔATD (2DΔATD), and GluN1ΔATD/GluN2DΔATD (N1ΔATD/2DΔATD) with EC50 values of 8.9, 8.4, and 7.0 μM, respectively. Currents were normalized to the glutamate- and glycine-elicited currents. Data are shown as mean ± S.E.M. and are from 3–32 oocytes.
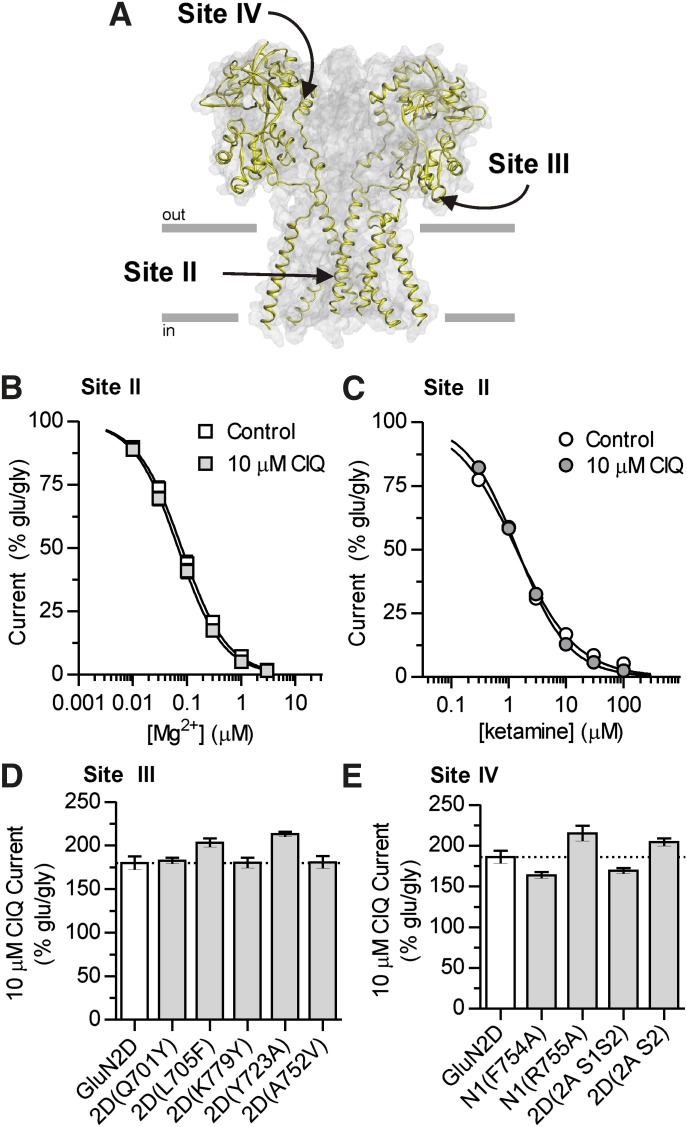
We next assessed whether CIQ acts at the channel blocker site (site II; Fig. 3A) on GluN2C- and GluN2D-containing receptors by measuring concentration-response curves for two structurally distinct channel blockers, Mg2+ (Fig. 3B) and ketamine (Fig. 3C), at a holding potential of −80 mV in the absence or presence of 10 μM CIQ. We predicted that if CIQ binds to or in some way perturbs the channel blocker site, then the potency of channel blockers, the extent of inhibition, or both would be altered in the presence of CIQ. Yet, CIQ affected neither the degree of inhibition nor the potency for both Mg2+ and ketamine at GluN1/GluN2D (Table 1). These data suggest that CIQ does not compete for binding with channel blockers or dramatically alter the nature of the conduction pathway. These data are consistent with no effect of CIQ on the stability of the open channel, that is, mean open time (Mullasseril et al., 2010), which is a key determinant of channel block.
Fig. 3.
(A) Modulatory sites II, III, and IV are depicted on a homology model of GluN1/GluN2D (Acker et al., 2011). The GluN2 subunits are highlighted in yellow and the predicted location of the plasma membrane is represented by gray lines with the orientation indicated. For clarity, the ATD is not shown. (B and C) Interaction of CIQ with site II was assessed by measuring the potencies of two channel blockers, Mg2+ and ketamine, in the absence (control) or presence of 10 μM CIQ. Concentration-response curves were evaluated from current responses of GluN1/GluN2D receptors expressed in oocytes. Currents were normalized to a percentage of the initial glutamate (100 μM)- and glycine (30 μM)-activated currents in the absence of inhibitor. Data are depicted as mean ± S.E.M. and are from 6 oocytes for each condition. (D) CIQ positive modulation was assessed at GluN1/GluN2D receptors containing point mutations at site III that critically affected inhibition of GluN2D by QNZ46. (E) CIQ potentiation was not affected by point mutations or chimeric NMDA receptors that significantly affected inhibition by TCN-201 at site IV in GluN1/GluN2A receptors.
TABLE 1.
CIQ does not interact with site II modulators
Mg2+ and ketamine concentration-response curves were measured at GluN1/GluN2D receptors expressed in oocytes and recorded using two-electrode voltage-clamp in the absence (control) or presence of CIQ (10 μM). Receptors were activated by 100 μM glutamate and 30 μM glycine at −80 mV. Mg2+ data are from 10 oocytes for controls and 7 oocytes for CIQ, whereas ketamine data are from 6 oocytes for both controls and CIQ. The log IC50 values were not significantly different between control and CIQ for both Mg2+ and ketamine (P > 0.05, unpaired t test).
| IC50
|
Minimum Response |
|||
|---|---|---|---|---|
| Control | CIQ | Control | CIQ | |
| μM | % Glutamate | |||
| Mg2+ | 80 ± 10 | 66 ± 6 | 1.8 ± 0.8 | 1.6 ± 0.6 |
| Ketamine | 1.58 ± 0.08 | 1.41 ± 0.12 | 5.4 ± 0.8 | 2.4 ± 0.6 |
The ABD harbors several ligand binding sites; thus, we explored the potential interaction of CIQ with each of these sites. Agonists bind in the cleft of the clamshell-shaped ABD; however, CIQ is unlikely to bind within the agonist-binding pockets for two reasons. First, CIQ neither activates nor inhibits the receptor, in contrast to other molecules binding at these sites, which are either agonists or competitive antagonists. Second, CIQ does not detectably alter glutamate or glycine potency (Mullasseril et al., 2010), suggesting that the nature of the agonist-binding pockets is unchanged by CIQ. In addition to the agonist-binding sites, the ABD contains two regulatory sites that are critical for the actions of subunit-selective inhibitors. One site is located at the membrane-proximal lower lobe of the ABD (site III in Fig. 1) and is important for noncompetitive inhibition by quinazoline-4-ones (Hansen and Traynelis, 2011) and dihydroquinoline-pyrazolines (Acker et al., 2011). We tested whether this region may also be critical for positive allosteric modulation of GluN1/GluN2D by measuring CIQ potentiation of GluN2D receptors containing point mutations that markedly affected inhibition by quinazoline-4-ones (Hansen and Traynelis, 2011) and dihydroquinoline-pyrazolines (Acker et al., 2011). CIQ potentiation, however, was unaffected by mutations in this region that alter inhibition by both quinazoline-4-ones and dihydroquinoline-pyrazolines (Fig. 3D), suggesting that the lower portion of the ABD clamshell proximal to the membrane helices of the receptor does not contribute to the molecular determinants for positive allosteric modulation of GluN1/GluN2D receptors.
A second modulatory site for subunit-selective inhibitors resides at the dimer interface between GluN1 and GluN2 ABDs (site IV in Fig. 1) and is critical for glycine-dependent inhibition by TCN-201 and N-(cyclohexylmethyl)-2-({5-[(phenylmethyl)amino]-1,3,4-thiadiazol-2-yl}thio)acetamide (TCN-213) (Hansen et al., 2012; McKay et al., 2012). Given the role of the ABD dimer interface in mediating allosteric coupling between the ATD and the ion channel gate (Gielen et al., 2008, 2009), and the importance of the ABD dimer interface of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors for the activity of positive modulators such as cyclothiazide (Sun et al., 2002), we asked whether the ABD dimer interface of NMDA receptors might contribute to potentiation by CIQ. Two residues in GluN1 contributing to the dimer interface, Phe754 and Arg755, were critical for inhibition of GluN1/GluN2A receptors by TCN-201 (Hansen et al., 2012), and mutation of these residues to alanine altered the binding of TCN-201. By contrast, CIQ potentiation was not altered at GluN1(F754A)/GluN2D and GluN1(R755A)/GluN2D receptors (Fig. 3E). The S2 segment of the GluN2 ABD also contributes residues to the ABD dimer interface and introducing this region of GluN2A into GluN2D conferred inhibition by TCN-201 to GluN2D (Hansen et al., 2012). Yet, CIQ potentiation was not diminished in GluN2D chimeric receptors containing the entire ABD from GluN2A [i.e., 2D(2A S1S2)] or just the S2 segment, as in the chimera 2D(2A S2) (Fig. 3E). These data suggest that positive allosteric modulation of GluN2C- or GluN2D-containing receptors is not mediated through the ABD dimer interface. These results are consistent with the molecular determinants for CIQ residing outside the ATD and being more membrane-proximal than the ABD dimer interface.
Residues in the M1 Helix Affect CIQ Potentiation.
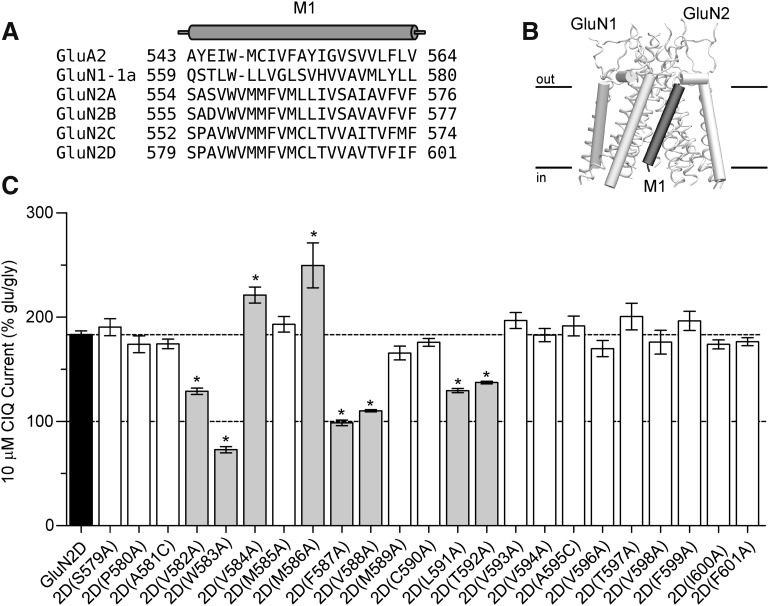
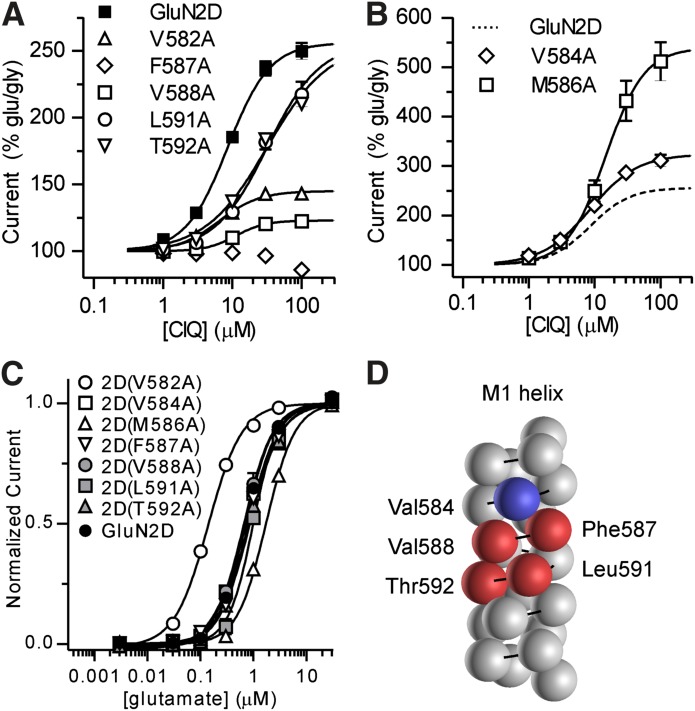
Previous studies identified a single amino acid residue in the M1 transmembrane helix of GluN2D, Thr592, that when mutated to the corresponding GluN2A residue (isoleucine) eliminated potentiation by CIQ (Mullasseril et al., 2010). To further explore the importance of the M1 helix in mediating potentiation by CIQ, we used alanine scanning mutagenesis of this region. A sequence alignment of the GluN1 and GluN2 NMDA receptor subunits together with the GluA2 AMPA receptor (Fig. 4A) was used as a guide to individually mutate 23 residues in GluN2D to alanine (or cysteine if the wild-type residue was alanine). These residues correspond to the residues comprising the M1 transmembrane helix in the GluA2 membrane-spanning crystal structure (Sobolevsky et al., 2009) and are shown in a homology model of GluN1/GluN2D in Fig. 4B. We assessed the effects of 10 μM CIQ on these mutants and found six residues in GluN2D that when mutated to alanine reduced potentiation by CIQ: Val582, Trp583, Phe587, Val588, Leu591, and Thr592 (Fig. 4C). In addition, we found two residues that increased the potentiation by CIQ: Val584 and Met586 (Fig. 4C). CIQ concentration-response curves (Fig. 5, A and B) revealed varying effects of these mutations on CIQ potency and efficacy. CIQ potency, but not efficacy, was reduced at 2D(L591A) and 2D(T592A). By contrast, the efficacy of CIQ at 2D(V582A) and 2D(V588A) was significantly attenuated. CIQ caused no detectable potentiation at 2D(F587A). CIQ efficacy increased at 2D(V584A) and 2D(M586A) with little to no decrease in CIQ potency (Table 2). Currents elicited from 2D(W583A), however, were small (<50 nA) and showed linear rundown with agonist application, preventing reliable estimation of CIQ potency, consistent with previous studies (Kashiwagi et al., 2002; Thomas et al., 2006). In addition, mutation of this tryptophan to phenylalanine, 2D(W583F), did not affect CIQ modulation [10 μM CIQ response (% glu/gly) was 179 ± 6 for 2D(W583F) versus 201 ± 7 for GluN2D; P > 0.05 unpaired t test; n = 4]. Hence, it is likely that this tryptophan residue is a critical structural element for the M1 helix and not necessarily involved with modulation by CIQ.
Fig. 4.
(A) A sequence alignment of NMDA receptor subunits with GluA2 is shown. The M1 helix of GluA2 (Sobolevsky et al., 2009) is indicated as a cylinder above the alignment. (B) The position of the GluN2D M1 helix is highlighted in a side-on view of the transmembrane region of a homology model of GluN1/GluN2D (Acker et al., 2011). (C) GluN2D subunits with point mutations in the M1 transmembrane helix were coexpressed with GluN1 in Xenopus laevis oocytes and current responses recorded using two-electrode voltage clamp. Currents were first activated by 100 μM glutamate and 30 μM glycine and then 10 μM CIQ was coapplied with glutamate and glycine. *Statistically significant change from wild-type GluN1/GluN2D (P < 0.05, one-way analysis of variance with Dunnett’s post-test) and those mutants are highlighted in gray. Responses were normalized to the glutamate- and glycine-induced current. Data are from 4–52 oocytes.
Fig. 5.
Currents were recorded under two-electrode voltage clamp in response to increasing concentrations of CIQ coapplied with glutamate (100 μM) and glycine (30 μM) to oocytes expressing GluN2D point mutants that attenuated CIQ potentiation (A) or enhanced CIQ potentiation (B). See Table 2 for CIQ EC50 values. Responses were normalized to the currents elicited by glutamate and glycine in the absence of CIQ. Data are from 4–32 oocytes. (C) Glutamate concentration-response curves were measured using two-electrode voltage-clamp recordings of oocytes expressing the GluN2D point mutants from (A) and (B). We coapplied 30 μM glycine with all glutamate concentrations. Glutamate EC50 values (Table 2) measured for 2D(V582A) and 2D(M586A) were significantly different from GluN2D. Data are shown as mean ± S.E.M. from 6–9 oocytes. (D) Residues comprising the M1 helix are depicted as spheres on a generic protein α helix. Residues with diminished CIQ potentiation are colored red and those with increased CIQ potentiation are colored blue. The residues affecting modulation by CIQ but not glutamate potency cluster on one side of the α helix.
TABLE 2.
CIQ, glutamate, and glycine EC50 values for GluN1/GluN2D point mutants
EC50 values were determined from two-electrode voltage-clamp recordings of Xenopus laevis oocytes expressing the indicated GluN1/GluN2D receptor. For CIQ EC50 determination, receptors were activated by 100 μM glutamate and 30 μM glycine at −40 mV; 5 mM 2-(hydroxypropyl)-β-cyclodextrin was present in all solutions. For glutamate EC50 measurements, glycine was 30 μM. For glycine EC50 measurements, glutamate was 100 μM. Data are from 4–36 oocytes.
| Mutant | Region | CIQ EC50 | 100 μM CIQ Response | Glutamate EC50 | Glycine EC50 |
|---|---|---|---|---|---|
| µM | % glu/gly | μM | μM | ||
| GluN2D | – | 9.3 ± 0.3 | 253 ± 5 | 0.82 ± 0.07 | 0.14 ± 0.02 |
| 2D(F574A) | Pre-M1 | 13.0 ± 1.5* | 150 ± 1 | 0.057 ± 0.003* | 0.060 ± 0.001* |
| 2D(L575A) | Pre-M1 | 38 ± 18 | 53 ± 4 | 0.24 ± 0.03* | 0.15 ± 0.02 |
| 2D(E576A) | Pre-M1 | 10.7 ± 0.4 | 321 ± 12 | 0.178 ± 0.009* | 0.106 ± 0.002 |
| 2D(P577A) | Pre-M1 | 7.2 ± 0.6* | 127 ± 2 | 0.398 ± 0.014 | 0.147 ± 0.002 |
| 2D(Y578A) | Pre-M1 | 4.5 ± 0.4* | 320 ± 30 | 0.0027 ± 0.0002a* | 0.0011 ± 0.0001a* |
| 2D(V582A) | M1 | 6.6 ± 0.6* | 143 ± 3 | 0.17 ± 0.01* | 0.04 ± 0.01* |
| 2D(V584A) | M1 | 8.7 ± 0.7 | 311 ± 12 | 0.78 ± 0.04 | 0.14 ± 0.02 |
| 2D(M586A) | M1 | 15.3 ± 1.1* | 510 ± 40 | 1.90 ± 0.12* | 0.26 ± 0.01* |
| 2D(F587A) | M1 | NE | 86 ± 3 | 0.80 ± 0.05 | 0.25 ± 0.02* |
| 2D(V588A) | M1 | 11.3 ± 1.2 | 123 ± 3 | 0.93 ± 0.09 | 0.12 ± 0.02 |
| 2D(L591A) | M1 | 34.6 ± 5.9* | 218 ± 9 | 0.99 ± 0.06 | 0.18 ± 0.01 |
| 2D(T592A) | M1 | 32.5 ± 2.9* | 210. ± 4 | 1.06 ± 0.08 | 0.12 ± 0.02 |
| N1(M813A) | M4 | 11.0 ± 0.7 | 337 ± 19 | 0.168 ± 0.008* | 0.082 ± 0.003 |
| N1(F817A) | M4 | 12.1 ± 0.6 | 430 ± 30 | ND | ND |
ND, not determined; NE, no effect.
The lowest concentration of glutamate tested (3 nM) elicited currents greater than 100 nA from GluN1/GluN2D(Y578A) receptors. Therefore, the EC50 of glutamate was calculated by fixing the minimum current to be 0 pA.
P < 0.05 versus GluN2D, one-way analysis of variance with Dunnett’s post hoc test.
To assess the effects of these GluN2 M1 mutations on channel function, we recorded glutamate and glycine concentration-response curves (Fig. 5C; Supplemental Fig. 1). We found glutamate and glycine potencies were significantly increased at 2D(V582A) (Table 2). By contrast, glutamate potency was reduced at 2D(M586A) (Table 2). No other M1 mutations affected glutamate or glycine potency. Although agonist potency is determined by both the affinity and efficacy of the agonist at the receptor, these residues do not comprise the agonist-binding pocket (Furukawa et al., 2005; Vance et al., 2011); therefore, we expect that the affinity of glutamate and glycine remain unchanged at these mutants. Thus, we interpret the change in glutamate potency at 2D(V582A) and 2D(M586A) to reflect a change in the efficacy of glutamate (i.e., the ability of glutamate binding to cause the ion channel pore to open) and suggest that these mutations alter gating of the receptor.
We next asked whether the residues in the M1 helix that when mutated affect CIQ potentiation, but not glutamate potency, clustered in three-dimensional space. We plotted the residues comprising the M1 helix as spheres on a generic protein α helix, having 3.6 amino acids per turn and a 5.4 Å translation per turn, and highlighted the residues affecting CIQ potentiation (Fig. 5D). Strikingly, the residues seemed to reside on only one side of the helix, suggesting that these residues could all potentially interact directly with CIQ. It is also noteworthy that the equivalent residues in the GluA2 tetrameric crystal structure line “gaps” between the transmembrane domains that were hypothesized to be occupied by amino acid residues from transmembrane AMPA receptor regulatory proteins (Sobolevsky et al., 2009).
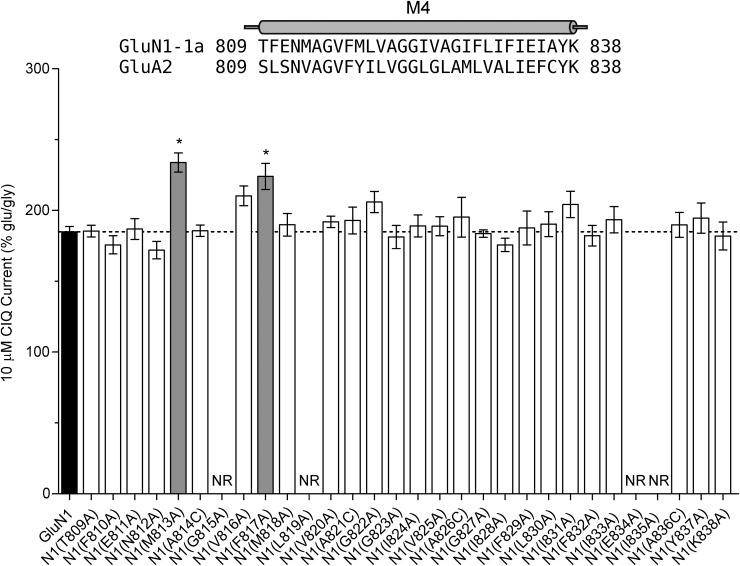
If CIQ interacts with residues in the M1 transmembrane helix, there may be other residues in this region of the receptor on either GluN1 or GluN2 that mediate potentiation by CIQ. To explore this idea, we used a homology model of GluN1/GluN2D (Acker et al., 2011) to identify residues located within 5 Å of the GluN2 M1 helix. The residues we identified resided in the M4 transmembrane helix of GluN1 and a short stretch of amino acids immediately extracellular to the GluN2 M1 helix. We mutated these residues and assessed potentiation by 10 μM CIQ. Only two residues in the M4 helix of GluN1 affected CIQ potentiation: GluN1(M813A) and GluN1(F817A) (Fig. 6). Although these residues do not cluster in three-dimensional space with the amino acids from the GluN2 M1 helix that affected CIQ modulation, they do occupy a provocative location in a homology model of GluN1/GluN2D. They are positioned on the GluN1 M4 helix such that their side chains protrude into a region between the M1 helix and the GluN2 M3 gate helix. For example, Met813 of GluN1 is about 4.5 Å from the serine in the SYTANLAAF motif of GluN2D. Thus, Met813 and Phe817 of the GluN1 M4 helix may be positioned to mediate interactions between the GluN2D M1 helix and the GluN2D M3 gate helix.
Fig. 6.
Residues comprising the M4 helix of GluN1 were individually mutated to alanine and coexpressed with GluN2D in oocytes. CIQ potentiation of glutamate (100 μM)- and glycine (30 μM)-stimulated currents was then measured using two-electrode voltage-clamp recordings. *Residues displaying significantly altered CIQ potentiation, which are also highlighted in gray (P < 0.05, one-way analysis of variance with Dunnett’s post hoc test). NR signifies that oocytes expressing the corresponding GluN1 point mutant failed to generate agonist-evoked currents larger than 5 nA in at least three separate injections of cRNA. Shown above the graph is an amino acid sequence alignment of the M4 region of GluN1 and GluA2; the M4 helix from the GluA2 crystal structure (Sobolevsky et al., 2009) is depicted as a cylinder on top of the alignment. Amino acids are numbered with the initiating methionine as 1.
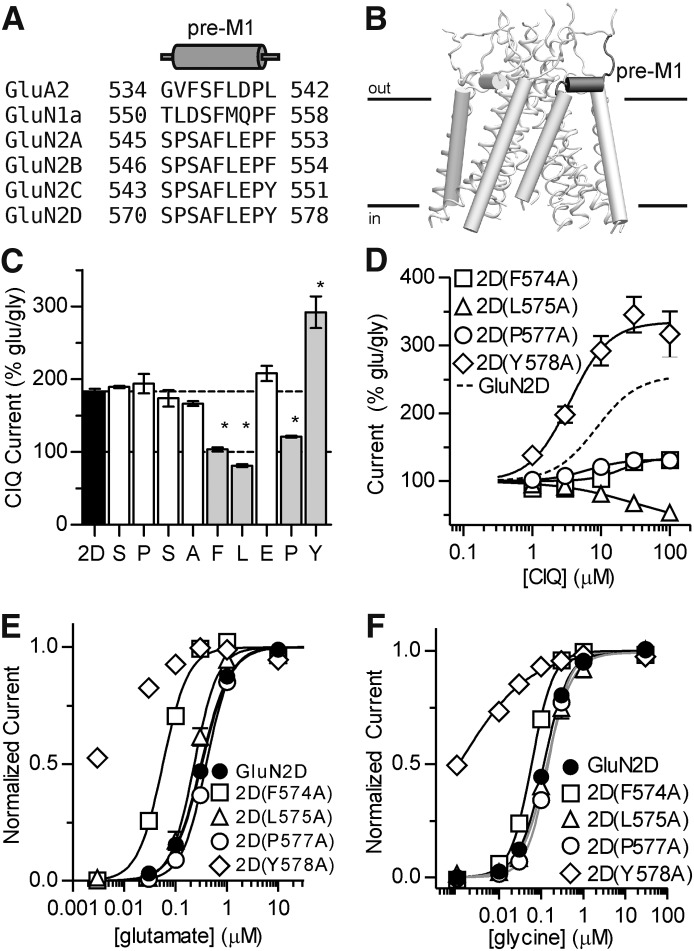
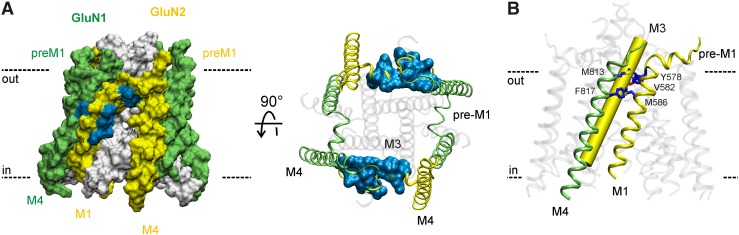
We also identified four residues in the GluN2D S1-M1 linker that influence positive modulation by CIQ: Phe574, Leu575, Pro577, and Tyr578 (Fig. 7). These residues, which are immediately extracellular to the GluN2 M1 helix, are of interest because they form a purportedly crucial gating element for glutamate receptor ion channels. The corresponding amino acids in the tetrameric crystal structure of a GluA2 AMPA receptor form a cuff helix that is parallel to the membrane and makes van der Waals contacts with the M3 helix that forms the gate (Sobolevsky et al., 2009). Moreover, the pre-M1 cuff helices in AMPA receptors have been proposed to be key determinants of receptor gating by restricting movement of the M3 helices in the closed state of the ion channel, but mediating channel opening upon agonist binding (Sobolevsky et al., 2009). The proposed role of the pre-M1 region in receptor gating is quite interesting given that CIQ increases the channel opening frequency in a gating step that precedes channel opening (Mullasseril et al., 2010). Consistent with the proposed role of these pre-M1 residues in receptor gating, we observed profound changes in both glutamate (Fig. 7; Table 2) and glycine (Table 2; Supplemental Fig. 1) potencies for 2D(F574A) and 2D(Y578A).
Fig. 7.
(A) An amino acid sequence alignment of the pre-M1 region of NMDA receptor subunits and the GluA2 subunit is shown with the pre-M1 helix of the GluA2 crystal structure depicted above as a cylinder. (B) The position of the GluN2D pre-M1 helix is illustrated in a homology model of GluN1/GluN2D viewed parallel to the membrane. The ATD and ABD are omitted for clarity. (C) Residues comprising the GluN2D pre-M1 region were mutated to alanine and potentiation by 10 μM CIQ was measured using two-electrode voltage-clamp recordings of oocytes. Residues highlighted in gray and marked with an asterisk exhibited CIQ potentiation that was significantly different from GluN2D (P < 0.05, one-way analysis of variance with Dunnett's post hoc test). (D) CIQ concentration-response curves were evaluated on point mutants from (C) with altered CIQ modulation. CIQ EC50 values are given in Table 2. The data from GluN2D are replicated from Fig. 5A for comparison. Data are depicted as mean ± S.E.M. and are from 3–4 oocytes. (E and F) Glutamate and glycine concentration-response curves for the GluN2D mutants in (D) were measured using two-electrode voltage-clamp recordings (see Table 2 for EC50 values). Individual curves were normalized to fitted minimum and maximum currents for that curve except for 2D(Y578A). The lowest concentration of glutamate (3 nM) or glycine (1 nM) tested at 2D(Y578A) produced currents that were larger than 100 nA and were at least half of the maximal agonist-evoked response. Hence, curves for 2D(Y578A) were fixed to a minimum of zero. Data are presented as mean 6 S.E.M. and are from 3–6 oocytes.
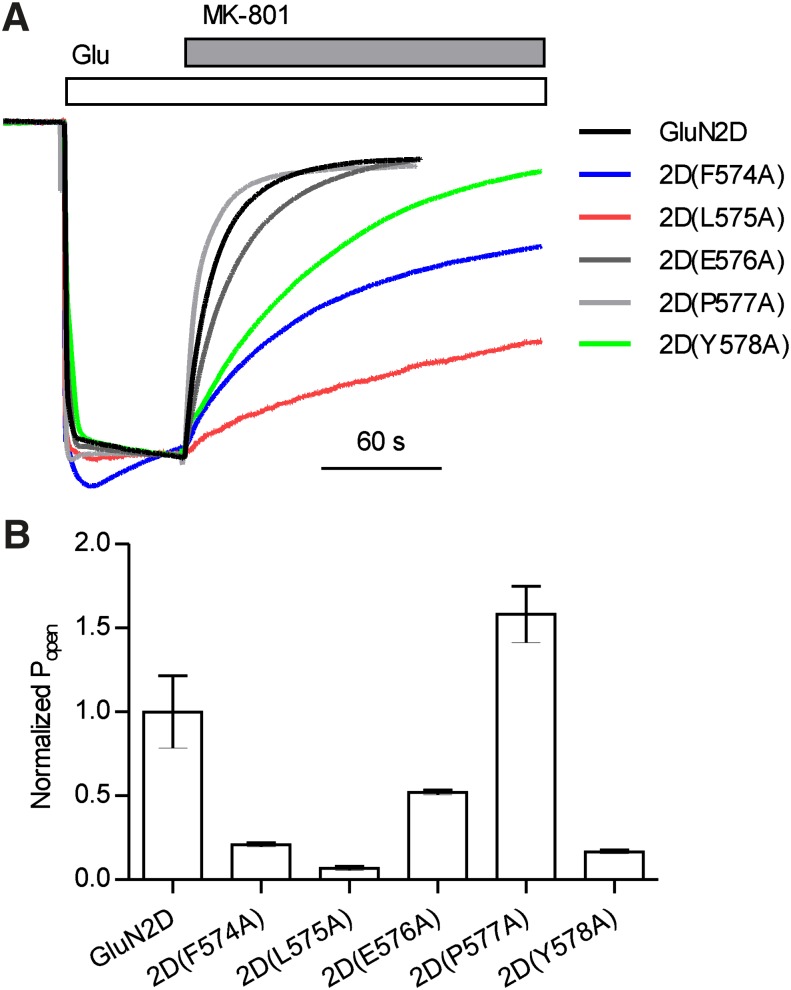
Pre-M1 Residues Control Channel Open Probability.
We further explored the potential of these GluN2D pre-M1 residues to contribute to channel gating by estimating the open probability of the 2D pre-M1 alanine mutants using the onset of MK-801 inhibition (Blanke and VanDongen, 2008; Gielen et al., 2009; Vance et al., 2011). We expected that mutations at residues influencing gating would accelerate or decelerate MK-801 binding depending on whether the mutations increased or decreased gating efficiency, respectively. The time course of MK-801 inhibition was dramatically slowed for 2D(F574A), 2D(L575A), 2D(E576A), and 2D(Y578A) (Fig. 8), suggesting that these residues are involved in mediating ion channel opening after agonist binding. Surprisingly, mutation of Pro577, which corresponds to an “elbow” in the pre-M1 helix of GluA2 and is highly conserved across glutamate receptor ion channels but absent from K+ channels (Sobolevsky et al., 2009), caused only a modest increase in the rate of onset of MK-801 inhibition. These results suggest that molecular determinants of CIQ potentiation converge on key determinants of receptor gating and imply an interesting mechanism of action whereby CIQ binding to the M1 helix of GluN2D increases the efficiency by which the pre-M1 region can promote channel opening.
Fig. 8.
(A) Shown are two-electrode voltage-clamp recordings illustrating the rate of block by MK-801 of GluN1/GluN2D receptors containing point mutations in the GluN2D pre-M1 helix. Currents were activated by 100 μM glutamate and 30 μM glycine at a holding potential of −60 mV followed by coapplication of 200 nM (+)-MK-801. Traces were normalized to the glutamate/glycine current. (B) Open probability (Popen) of GluN2D point mutations, calculated as the reciprocal of the time constant of onset of MK-801 block (τMK-801 block) and normalized to the values for GluN2D, are shown. Bars represent mean ± S.E.M. for 3–10 oocytes.
CIQ Cannot Reach Its Modulatory Site by Diffusion through the Membrane.
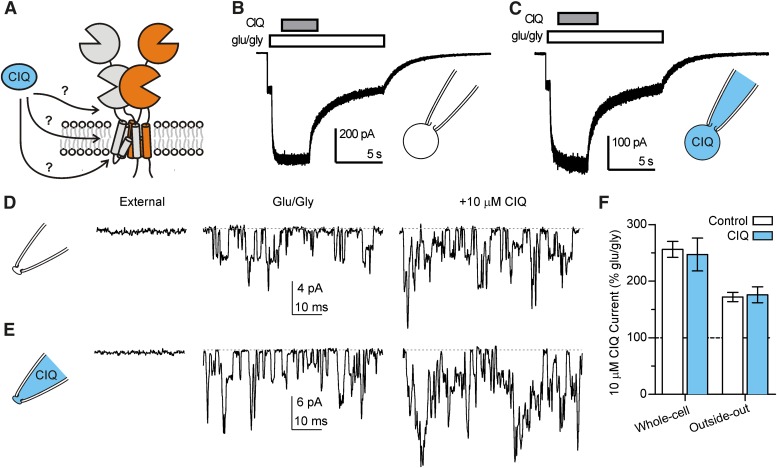
The location of multiple residues that affect the action of CIQ clustered in the transmembrane region raised the possibility that CIQ was required to partition into the plasma membrane to exert its effects. To determine whether this was the case, we recorded GluN1/GluN2D currents in the whole-cell configuration and included 10 μM CIQ in the recording pipette. We waited for 10 min after achieving the whole-cell configuration to allow for dialysis of the cell and then coapplied 10 μM CIQ with glutamate and glycine to the exterior of the cell. We reasoned that if CIQ must partition into the membrane, then it could do so equally well from the intracellular or extracellular face. If CIQ included in the patch pipette entered the plasma membrane to access its site, then the receptors should prebind CIQ and no further potentiation would be observed when CIQ was applied extracellularly. However, when CIQ was included in the pipette solution, GluN1/GluN2D receptors were still potentiated by extracellular CIQ to the same extent as control cells with normal internal solution (Fig. 9; P > 0.05, versus control pipette solution).
Fig. 9.
(A) Potential routes by which CIQ can access residues in the M1 helix are illustrated in a schematic representation of GluN1/GluN2D receptors. For simplicity, only one pair of GluN1/GluN2 subunits is shown. CIQ could access its site directly from the extracellular solution, by first partitioning into the plasma membrane and then laterally diffusing to its site, or by first crossing the plasma membrane and then accessing the M1 helix from the cytosolic face of the receptor. (B) A whole-cell voltage-clamp recording of an human embryonic kidney cell expressing GluN1/GluN2D receptors is shown. Currents were activated by 100 μM glutamate and 50 μM glycine and then 10 μM CIQ was rapidly coapplied with glutamate and glycine. The pipette tip contained control internal solution and currents were recorded at least 10 minutes after breaking through the cell membrane. (C) A whole-cell recording similar to (B) is shown, except the pipette tip contained internal solution that included 10 μM CIQ. (D) An outside-out patch-clamp recording of GluN1/GluN2D receptors is shown at different times with the applied extracellular solution indicated above. Channels were activated by 100 μM glutamate and 50 μM glycine. Coapplication of 10 μM CIQ with glutamate and glycine increased the current response to 170% that of glutamate/glycine. (E) An outsideout patch-clamp recording similar to (D) is shown except the pipette tip contained internal solution plus 10 μM CIQ. CIQ applied in the extracellular solution increased the current response to 220% that of glutamate/glycine. In both (D) and (E), channel openings were recorded at least 5 minutes after pulling the patch. (F) The results from experiments in (B–E) are summarized. Potentiation of GluN2D receptors by externally applied CIQ was not affected by pre-incubating the cytosolic face of the receptor with CIQ. Bars represent mean ± S.E.M. from 5 cells or 3 patches.
One possible caveat to this experiment is that CIQ could be a substrate for transporters in the cell, degradative enzymes, or otherwise be moved into organelles with a consequent decrease in its effective intracellular concentration. We therefore repeated this experiment in excised outside-out patches that lack all organelles and contain only about 1 μm2 of membrane and associated cytoskeletal components. We selected patches containing multiple GluN1/GluN2D channels so as to avoid potential complications of variable activity of a single channel throughout the duration of the experiment and to maximize our ability to measure an increase in the average current response of the patch. The potentiation of GluN1/GluN2D receptors by CIQ applied to the exterior of the patch was comparable when the internal pipette solution contained no CIQ (control) or contained 10 μM CIQ (Fig. 9; P > 0.05, CIQ pipette solution versus control). These results are similar to those obtained in the whole-cell configuration, and together suggest that CIQ cannot access its modulatory site from the intracellular side of the receptor nor by diffusion into the plasma membrane. Rather, direct extracellular aqueous access to the receptor appears necessary for positive modulation by CIQ.
Discussion
The most important conclusion of this study is that CIQ, a positive allosteric modulator of GluN2C- and GluN2D-containing NMDA receptors, does not bind the ATD. Rather, our data suggest that CIQ interacts with residues in the M1 transmembrane helix, and that CIQ potentiation is mediated by residues in the GluN2 pre-M1 region and the GluN1 M4 transmembrane helix. Moreover, we show for the first time that the GluN2 pre-M1 region may be a critical determinant of NMDA receptor gating. Mutations in this region not only influence allosteric regulation by CIQ, but also alter receptor open probability, assessed by the rate of onset of MK-801 channel block.
Structural Determinants of CIQ Potentiation Reside in the Transmembrane Region.
Although the linker between the ATD and ABD was previously identified as a molecular determinant of CIQ action (Mullasseril et al., 2010), CIQ does not bind this region of the receptor because removal of the ATD and the ATD-ABD linker from both GluN1 and GluN2D did not affect CIQ potentiation. The actions of CIQ are in contrast to positive modulation of GluN1/GluN2B receptors by polyamines such as spermine, which seems to involve positive charges located on the lower lobe of the GluN2B ATD (Mony et al., 2011) and alternatively spliced GluN1 ATD (Traynelis et al., 1995). Hence, the structural determinants of positive allosteric modulation by CIQ are distinct from those of polyamines. It remains to be determined whether the ATDs of GluN2C and GluN2D harbor binding sites for allosteric modulators and whether the downstream mechanisms of GluN2B potentiation by polyamines are conserved at GluN2C and GluN2D receptors.
The majority of residues in the GluN2D M1 helix critical for potentiation by CIQ cluster on one side of the helix (Fig. 5D). However, two of those residues (Val582 and Met586) are located on the opposite side of the helix. Mutation of both of these residues to alanine also changes glutamate potency, which likely reflects changes in gating of these mutants because these residues are situated far outside the agonist-binding pocket. Perhaps mutation of these residues alters the conformation of the M1 helix, thereby preventing CIQ from binding. Alternatively, these mutations change the manner in which the M1 helix moves upon agonist binding and by extension disrupt the changes that occur in gating when CIQ is present. In this context, it is interesting to note that in a homology model of GluN1/GluN2D, Val582 and Met586 of GluN2D are positioned within approximately 3 Å of Met813 and Phe817 in the GluN1 M4 helix, raising the possibility that these residues interact during channel gating. Moreover, these residues are also located adjacent to the M3 gate helix, in particular the serine in the conserved SYTANLAAF gating motif and two phenylalanines that are one and two helical turns below SYTANLAAF (Fig. 10). Although further experiments would be needed to confirm interaction of these residues, it is tempting to speculate that Val582 and Met586 of GluN2D together with Met813 and Phe817 of GluN1 couple movement of the M1 helix upon agonist binding to movement of the M3 gate helix.
Fig. 10.
(A) Amino acid residues at which mutations altered CIQ potentiation are depicted in a homology model of GluN1/GluN2D. For clarity, only the transmembrane region is shown. (Left) The outside of the receptor viewed parallel with the membrane is shown as a surface representation. GluN1 is colored in green and GluN2D is colored in yellow. Residues affecting positive modulation by CIQ are highlighted in blue. (Right) A view from the extracellular side of the receptor down the pore axis is shown. The pre-M1, M1, and M4 helices from all four subunits are colored (GluN1 in green and GluN2D in yellow). Residues affecting CIQ activity are highlighted in blue. (B) The proximity of the GluN2D pre-M1 region and M1 helix, GluN1 M4 helix, and M3 gate helix is shown in a side-on view of the receptor. Residues Tyr578, Val582, and Met586 in GluN2D and Met813 and Phe817 in GluN1 are highlighted. These residues affected both CIQ potentiation and glutamate potency when mutated to alanine. GluN1 is colored in green and GluN2D is colored in yellow.
Of the five residues we identified in the GluN2 M1 helix that appear critical specifically to potentiation by CIQ (Val584, Phe587, Val588, Leu591, and Thr592; Fig. 5D), only one of those residues, Thr592, differs between GluN2A/2B and GluN2C/2D. Indeed, CIQ did not potentiate GluN2D(T592I) receptors, in which this residue had been mutated to the homologous residue in GluN2A/2B (Mullasseril et al., 2010). We further observed that mutation of this residue to valine also eliminates CIQ potentiation, whereas mutation to serine has no effect on CIQ potentiation (unpublished data). Hence, hydrogen bond capabilities at this residue might be critical for the actions of CIQ and may help explain the selectivity of CIQ for GluN2C/2D over 2A/2B. For example, CIQ may directly interact with the side chain of Thr592 and loss of the hydroxyl group, which occurs with the isoleucine residue at this position in GluN2A/2B, may prevent CIQ from binding. Alternatively, the side chain of Thr592 may be critical for conformational changes that occur downstream of CIQ binding and lead to increased channel openings.
Selectivity of CIQ for GluN2C/2D over GluN2A/2B could also arise from differences in the arrangement of the transmembrane helices in GluN2A/2B versus GluN2C/2D. Perhaps the M1 helix of 2A/2B is rotated compared with the M1 helix in 2C/2D and thus the residues on the outside of the transmembrane region that could potentially interact with modulators are different. It is also likely that movements of the transmembrane helices upon agonist binding differ between 2A/2B and 2C/2D given the markedly different open and closed times of these receptors (Traynelis et al., 2010). Hence, functional rearrangements of the transmembrane helices may be differentially sensitive to modulation by CIQ.
Role of Pre-M1 Region in Gating.
Several lines of evidence implicate the pre-M1 region in gating of glutamate receptors. In AMPA receptors, changes in receptor leak currents occur when amino acids on the M3 helix facing the pre-M1 helix are mutated to cysteine and reacted with methanethiosulfonate (MTS) reagents, suggesting these residues are important for gating of AMPA receptors (Sobolevsky et al., 2003). Additionally, residues at the interface of the pre-M1 and M4 helices were critical for noncompetitive inhibition of AMPA receptors by 1-(4-Aminophenyl)-3-methylcarbamyl-4-methyl-3,4-dihydro-7,8-methylenedioxy-5H-2,3-benzodiazepine (GYKI-53655) and 3-(2-chloro-phenyl)-2-[2-(6-diethylaminomethyl-pyridin-2-yl)-vinyl]-6-fluoro-3H-quinazolin-4-one (CP-465022) (Balannik et al., 2005). In NMDA receptors, mutations in the pre-M1 region of GluN1 (Gln556 and Pro557; Kashiwagi et al., 2002) and GluN2C (Glu530 and Ser533; Sobolevsky et al., 2007) result in either spontaneously active channels or channels that become spontaneously active upon modification by MTS reagents. Moreover, introduction of cysteines at several residues in the pre-M1 region of GluN2A resulted in channels with small or abnormal glutamate-activated currents (Thomas et al., 2006). Mutations in the pre-M1 region giving rise to spontaneously active channels (either alone or after reaction with MTS reagents) may reflect a shift in the gating equilibrium toward the open state, that is, an increase in the gating efficiency, which has been shown for several residues in the S1-M1 linker (Talukder et al., 2010). In a complementary way, mutations yielding receptors with small glutamate-activated currents may be due to uncoupling of the ion channel pore from agonist binding.
In this study, mutations in the pre-M1 region of GluN2D at Phe574, Leu575, and Pro577, disrupted positive modulation by CIQ. CIQ potentiates the receptor by accelerating a pregating step, thereby increasing the opening frequency of the receptor (Mullasseril et al., 2010). Hence, mutations at these pre-M1 residues likely disrupt the gating steps accelerated by CIQ. By contrast, mutation at Tyr578 enhanced both the potency and maximum effect of CIQ potentiation. This effect may be explained by increased space for CIQ to interact with the receptor as the larger side chain of tyrosine was replaced with the smaller methyl group of alanine. However, the alanine also lacks the hydrogen bond capabilities of the tyrosine, which may interact with the thioether of Met813 on GluN1 (Fig. 10). It is worth noting that both 2D(Y578A) and N1(M813A) receptors displayed large leak currents in the absence of glutamate. These leak currents were blocked by 1 mM Mg2+ and 1 µM (+)-MK-801 and could be potentiated by 10 µM CIQ (unpublished data), suggesting that these currents were mediated by NMDA receptors.
Although residues in pre-M1 region seem critical for potentiation of NMDA receptors by CIQ, residues in a similar region of AMPA receptors mediate noncompetitive inhibition (Balannik et al., 2005) and it remains an open question whether noncompetitive inhibition of NMDA receptors may be achieved through the pre-M1 region. We hypothesize that compounds exist that can act at this site to bring about negative allosteric modulation, rendering it functionally analogous to the benzodiazepine site on γ-aminobutyric acid receptors at which ligands can have positive, neutral, or negative actions.
Supplementary Material
Acknowledgments
The authors thank Phuong Le and Jing Zhang for excellent technical assistance, and Drs. Kasper B. Hansen and Katie M. Vance for critical discussion of the manuscript.
Abbreviations
- ABD
agonist-binding domain
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- ATD
amino-terminal domain
- BAPTA
1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid
- CIQ
(3-chlorophenyl)(6,7-dimethoxy-1-((4-methoxyphenoxy)methyl)-3,4-dihydroisoquinolin-2(1H)-yl)methanone
- CP-465022
3-(2-chloro-phenyl)-2-[2-(6-diethylaminomethyl-pyridin-2-yl)-vinyl]-6-fluoro-3H-quinazolin-4-one
- DQP-1105
5-(4-Bromophenyl)-3-(1,2-dihydro-6-methyl-2-oxo-4-phenyl-3-quinolinyl)-4,5-dihydro-g-oxo-1H-pyrazole-1-butanoic acid
- GYKI-53655
1-(4-Aminophenyl)-3-methylcarbamyl-4-methyl-3,4-dihydro-7,8-methylenedioxy-5H-2,3-benzodiazepine
- MK-801
(5S,10R)-10,11-dihydro-5-methyl-5H-dibenzo[a,d]cyclohepten-5,10-imine (dizocilpine maleate)
- MTS
methanethiosulfonate
- NMDA
N-methyl-d-aspartate
- QNZ46
(E)-4-(6-methoxy-2-(3-nitrostyryl)-4-oxoquinazolin-3(4H)-yl)-benzoic acid
- TCN-201
3-chloro-4-fluoro-N-[(4-[(2-(phenylcarbonyl)hydrazino)carbonyl]phenyl)methyl]-benzenesulfonamide
- TCN-213
N-(cyclohexylmethyl)-2-({5-[(phenylmethyl)amino]-1,3,4-thiadiazol-2-yl}thio)acetamide
Authorship Contributions
Participated in research design: Ogden, Traynelis.
Conducted experiments: Ogden.
Performed data analysis: Ogden.
Wrote or contributed to the writing of the manuscript: Ogden, Traynelis.
Footnotes
This study was supported by the Michael J. Fox Foundation; the National Institutes of Health National Institute of Neurological Disorders and Stroke [Grants R01-NS036654, R01-NS065371, and F31-NS071802]; the National Institutes of Health National Institute of General Medical Sciences [Grant T32-GM008602]; and the National Institutes of Health National Institute on Drug Abuse [Grant T32-DA01504006]; as well as by research grants to Emory University from Pfizer Inc. and Lundbeck AS. S.T. is a co-inventor of Emory-owned intellectual property, has an equity position in NeurOp Inc., and is a paid consultant of NeurOp Inc., which is developing NMDA receptor allosteric modulators.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Acker TM, Yuan H, Hansen KB, Vance KM, Ogden KK, Jensen HS, Burger PB, Mullasseril P, Snyder JP, Liotta DC, et al. (2011) Mechanism for noncompetitive inhibition by novel GluN2C/D N-methyl-D-aspartate receptor subunit-selective modulators. Mol Pharmacol 80:782–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov SM, Johnson JW. (1996) Voltage-dependent interaction of open-channel blocking molecules with gating of NMDA receptors in rat cortical neurons. J Physiol 493:425–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balannik V, Menniti FS, Paternain AV, Lerma J, Stern-Bach Y. (2005) Molecular mechanism of AMPA receptor noncompetitive antagonism. Neuron 48:279–288 [DOI] [PubMed] [Google Scholar]
- Blanke ML, VanDongen AMJ. (2008) Constitutive activation of the N-methyl-D-aspartate receptor via cleft-spanning disulfide bonds. J Biol Chem 283:21519–21529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpied TA, Clarke RJ, Johnson JW. (2005) Amantadine inhibits NMDA receptors by accelerating channel closure during channel block. J Neurosci 25:3312–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Okayama H. (1987) High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol 7:2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cull-Candy SG, Leszkiewicz DN. (2004) Role of distinct NMDA receptor subtypes at central synapses. Sci STKE 2004:re16. [DOI] [PubMed] [Google Scholar]
- Erreger K, Traynelis SF. (2005) Allosteric interaction between zinc and glutamate binding domains on NR2A causes desensitization of NMDA receptors. J Physiol 569:381–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. (2005) Subunit arrangement and function in NMDA receptors. Nature 438:185–192 [DOI] [PubMed] [Google Scholar]
- Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P. (2008) Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron 57:80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. (2009) Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature 459:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Ogden KK, Traynelis SF. (2012) Subunit-selective allosteric inhibition of glycine binding to NMDA receptors. J Neurosci 32:6197–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Traynelis SF. (2011) Structural and mechanistic determinants of a novel site for noncompetitive inhibition of GluN2D-containing NMDA receptors. J Neurosci 31:3650–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann M, Heinemann S. (1994) Cloned glutamate receptors. Annu Rev Neurosci 17:31–108 [DOI] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. (2011) Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature 475:249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi K, Masuko T, Nguyen CD, Kuno T, Tanaka I, Igarashi K, Williams K. (2002) Channel blockers acting at N-methyl-D-aspartate receptors: differential effects of mutations in the vestibule and ion channel pore. Mol Pharmacol 61:533–545 [DOI] [PubMed] [Google Scholar]
- Kew JN, Trube G, Kemp JA. (1996) A novel mechanism of activity-dependent NMDA receptor antagonism describes the effect of ifenprodil in rat cultured cortical neurones. J Physiol 497:761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O, Kamberov E, Margolis B. (2000) Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques 29:970–972 [DOI] [PubMed] [Google Scholar]
- Masuko T, Kuno T, Kashiwagi K, Kusama T, Williams K, Igarashi K. (1999) Stimulatory and inhibitory properties of aminoglycoside antibiotics at N-methyl-D-aspartate receptors. J Pharmacol Exp Ther 290:1026–1033 [PubMed] [Google Scholar]
- McGurk JF, Bennett MV, Zukin RS. (1990) Polyamines potentiate responses of N-methyl-D-aspartate receptors expressed in xenopus oocytes. Proc Natl Acad Sci USA 87:9971–9974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay S, Griffiths NH, Butters PA, Thubron EB, Hardingham GE, Wyllie DJ. (2012) Direct pharmacological monitoring of the developmental switch in NMDA receptor subunit composition using TCN 213, a GluN2A-selective, glycine-dependent antagonist. Br J Pharmacol 166:924–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B, Sarantis M, Traynelis SF, Attwell D. (1992) Potentiation of NMDA receptor currents by arachidonic acid. Nature 355:722–725 [DOI] [PubMed] [Google Scholar]
- Mony L, Zhu S, Carvalho S, Paoletti P. (2011) Molecular basis of positive allosteric modulation of GluN2B NMDA receptors by polyamines. EMBO J 30:3134–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullasseril P, Hansen KB, Vance KM, Ogden KK, Yuan H, Kurtkaya NL, Santangelo R, Orr AG, Le P, Vellano KM, et al. (2010) A subunit-selective potentiator of NR2C- and NR2D-containing NMDA receptors. Nat Commun 1:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Ascher P, Neyton J. (1997) High-affinity zinc inhibition of NMDA NR1-NR2A receptors. J Neurosci 17:5711–5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom RW, Stec NL. (1988) Cooperative modulation of [3H]MK-801 binding to the N-methyl-D-aspartate receptor-ion channel complex by L-glutamate, glycine, and polyamines. J Neurochem 51:830–836 [DOI] [PubMed] [Google Scholar]
- Reynolds IJ. (1990) Arcaine uncovers dual interactions of polyamines with the N-methyl-D-aspartate receptor. J Pharmacol Exp Ther 255:1001–1007 [PubMed] [Google Scholar]
- Sobolevsky AI, Prodromou ML, Yelshansky MV, Wollmuth LP. (2007) Subunit-specific contribution of pore-forming domains to NMDA receptor channel structure and gating. J Gen Physiol 129:509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. (2009) X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462:745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Yelshansky MV, Wollmuth LP. (2003) Different gating mechanisms in glutamate receptor and K+ channels. J Neurosci 23:7559–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. (2002) Mechanism of glutamate receptor desensitization. Nature 417:245–253 [DOI] [PubMed] [Google Scholar]
- Talukder I, Borker P, Wollmuth LP. (2010) Specific sites within the ligand-binding domain and ion channel linkers modulate NMDA receptor gating. J Neurosci 30:11792–11804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CG, Krupp JJ, Bagley EE, Bauzon R, Heinemann SF, Vissel B, Westbrook GL. (2006) Probing N-methyl-D-aspartate receptor desensitization with the substituted-cysteine accessibility method. Mol Pharmacol 69:1296–1303 [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Hartley M, Heinemann SF. (1995) Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science 268:873–876 [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. (2010) Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62:405–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich MH, Isacoff EY. (2008) Rules of engagement for NMDA receptor subunits. Proc Natl Acad Sci USA 105:14163–14168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KM, Simorowski N, Traynelis SF, Furukawa H. (2011) Ligand-specific deactivation time course of GluN1/GluN2D NMDA receptors. Nat Commun 2:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicini S, Wang JF, Li JH, Zhu WJ, Wang YH, Luo JH, Wolfe BB, Grayson DR. (1998) Functional and pharmacological differences between recombinant N-methyl-D-aspartate receptors. J Neurophysiol 79:555–566 [DOI] [PubMed] [Google Scholar]
- Williams K, Dawson VL, Romano C, Dichter MA, Molinoff PB. (1990) Characterization of polyamines having agonist, antagonist, and inverse agonist effects at the polyamine recognition site of the NMDA receptor. Neuron 5:199–208 [DOI] [PubMed] [Google Scholar]
- Wu FS, Gibbs TT, Farb DH. (1991) Pregnenolone sulfate: a positive allosteric modulator at the N-methyl-D-aspartate receptor. Mol Pharmacol 40:333–336 [PubMed] [Google Scholar]
- Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. (2009) Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci 29:12045–12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Peoples RW, Oz M, Harvey-White J, Weight FF, Brauneis U. (1997) Potentiation of NMDA receptor-mediated responses by dynorphin at low extracellular glycine concentrations. J Neurophysiol 78:582–590 [DOI] [PubMed] [Google Scholar]
- Zheng F, Erreger K, Low CM, Banke T, Lee CJ, Conn PJ, Traynelis SF. (2001) Allosteric interaction between the amino terminal domain and the ligand binding domain of NR2A. Nat Neurosci 4:894–901 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.