Abstract
Background:
β-thalassaemia is one of the most common single-gene disorders worldwide. Each ethnic population has its own common mutations, accounting for the majority of cases, with a small number of mutations for the rarer alleles. Due to the heterogeneity of β-thalassaemia and the multi-ethnicity of Malaysians, molecular diagnostics may be expensive and time consuming.
Methods:
A simple polymerase chain reaction (PCR) approach involving a multiplex amplification refractory mutation system (MARMS) and one amplification refractory mutation system (ARMS), consisting of 20 β-globin gene mutations, were designed and employed to investigate β-thalassaemia patients and carriers.
Results:
Out of 169 carriers tested with the MARMS, Cd 41/42 (–TTCT), Cd 26 (A–G) HbE, IVS 1–1 (G–T), and IVS 1–5 (G–C) were the most common mutations, accounting for 78.1%. Among the Malays, Cd 26 (A–G) HbE, Cd 41/42 (–TTCT), IVS 1–1 (G–T), and IVS 1–5 (G–C) were the most common mutations, accounting for 81.4%, whereas Cd 41/42 (–TTCT) and IVS 2–654 (C–T) were most common among the Chinese (79.1%).
Conclusion:
We propose the use of this cheap, easy to interpret, and simple system for the molecular diagnostics of β-thalassaemia among Malaysians at the Institute for Medical Research (IMR).
Keywords: β-globin gene mutations, β-thalassaemia, MARMS, Molecular diagnostics
Introduction
β-thalassaemia is an autosomal haematological disorder resulting in a genetically deficient synthesis of the β-globin chain in haemoglobin (1). β-globin chain deficit leads to the intracellular precipitation of excess α-globin chains, causing ineffective erythropoiesis (2,3). Generally, a defective β-globin gene leads to a reduction (β+) or absence (β°) of the gene production.
To date, the database of haemoglobin variants and thalassaemia mutations, HbVar, has recorded more than 800 mutation entries involving the β-globin gene (http://globin.bx.psu.edu/cgi–bin/hbvar/query_vars3). The majority of the β-thalassaemia mutations are nucleotide substitutions, frameshifts or minor deletions (4), and rarely, large deletions are also reported (5). Blood transfusions and iron-chelator treatment for patients with β-thalassaemia major are expensive, and beyond the economic reach of many families, and therefore, the provision of genetic counselling even before marriage is important.
About 4.5% of the Malaysian population are carriers for β-thalassaemia (6,7). It is common among the Malaysian Malays and Chinese (8), with a structural haemoglobin variant, HbE found very commonly among the Malays (8,9). A molecular diagnostic approach to the Malaysian population has been reported previously (6,10) using ARMS, and MARMS to detect most of the common mutations, and with either PCR or genomic sequencing for the rarer mutations (9,11,12).
The aims of this study are to design and use a rapid, inexpensive, and simple PCR approach to update, and characterise the spectrum of β-globin gene mutations among Malaysians. Based on earlier reported studies of β-thalassaemia heterogeneity among Malaysians (11,13,14), we modified the MARMS developed by Bhardwaj et al., (15) by adding primers, suitable for the local mutation heterogeneity. Five sets of MARMS and one single ARMS were used to detect 20 different mutations of the β-globin gene. Direct sequencing and reverse dot blot hybridization analysis, using β-Globin StripAssay SEA™ (ViennaLab Diagnostics GmbH, Vienna, Austria) were used for mutation confirmation.
Materials and Methods
Subjects
A total of 208 patients from various parts of Peninsular Malaysia, Sabah, and Sarawak were screened for β-thalassaemia. The carrier state was diagnosed in samples in which the mean corpuscular volume (MCV) was below 83 fL, mean corpuscular haemoglobin (MCH) was below 27 pg, and HbA2 exceeded 3.4%, whereby HbA2 of more than 10% was screened for Cd 26 (G–A) HbE. The patient’s ethnicity was recorded to determine the sequence of the MARMS test. Representation by ethnic group was as follows: Malay 173, Chinese 27, Indian 3, with 3 from the indigenous population of East Malaysia, and 2 from the indigenous population of West Malaysia.
Genomic DNA Extraction
A total of 2 mL of a patient’s blood was collected with ethylenediaminetetraacetic acid (EDTA) as an anticoagulant. The DNA was extracted using QiAmp® DNA Blood Mini Kit (Qiagen GmbH, Hilden, Germany). The kit was used according to the manufacturer’s instructions. The extracted genomic DNA was used as a template and was kept at 4 °C until further use.
Mutation analysis
Multiplex Amplification Refractory Mutation System– Polymerase Chain Reaction (MARMS–PCR)
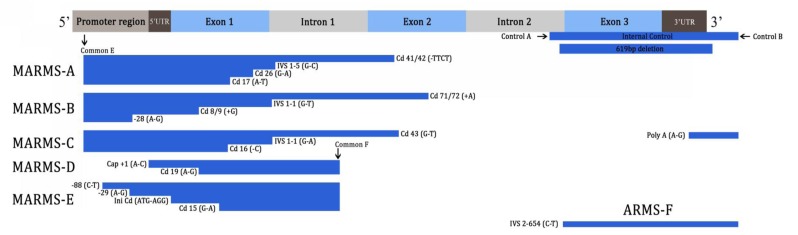
Five MARMS and one ARMS were used to detect 20 mutations: In MARMS-A, we screened for the IVS 1–5 (G–C), Cd 41/42 (–TTCT), Cd 17 (A–T), and Cd 26 (G–A) HbE mutations. In MARMS-B, we screened for the IVS 1–1 (G–T), Cd 8/9 (+G), –28 (A–G), and Cd 71/72 (+A) mutations. In MARMS-C, we screened for the IVS 1–1 (G–A), Cd 43 (G–T), Cd 16 (–C), and Poly A (A–G) mutations. In MARMS-D, we screened for the Cd 19 (A–G) and cap +1 (A–C) mutations. In MARMS-E, we screened for the –88 (C–T), Initiation Cd (ATG–AGG), Cd 15 (G–A), and –29 (A–G) mutations. Another mutation of IVS 2–654 (C–T) was detected in a single ARMS.
Most of the primer sequences were obtained from earlier publications (14–17). Primers to detect IVS 1–1 (G–A), Cd 43 (G–T), Cd 19 (A–G) and Initiation Cd (ATG–AGG) mutations were designed during the course of the project. A pair of primers was used as an internal control for each reaction. Primers A and B were used with MARMS, and primers C and D were used with the single ARMS. They amplify 861 bp and 493 bp amplicons, respectively. Sequences and concentrations of primers, and amplicon size are described in Table 1.
Table 1.
Sequence of primers and their concentration used in multiplex ARMS (MARMS)
| Mutation | Sequence | Used with primer | Primer concentration (nm) | Product size (bp) | |
|---|---|---|---|---|---|
| Normal | Mutant | ||||
| MARMS-A | |||||
| IVS 1–5 (G–C) | CTC CTT AAA CCT GTC TTG TAA CCT TGT TAG a | E | 500 | 500 | 319 |
| Cd 41/42 (–TTCT) | GAG TGG ACA GAT CCC CAA AGG ACT CAA CCT a | E | 50 | 50 | 476 |
| Cd 17 (A–T) | CTC ACC ACC AAC TTC ATC CAC GTT CAG CTA b | E | 50 | 50 | 275 |
| Cd 26 (G–A) | TAA CCT TGA TAC CAA CCT GCC CAG GGC GTT d | E | 50 | 50 | 301 |
| MARMS-B | |||||
| IVS 1–1 (G–T) | TTA AAC CTG TCT TGT AAC CTT GAT ACG AAA a | E | 300 | 600 | 315 |
| Cd 8/9 (+G) | CCT TGC CCC ACA CGG CAG TAA CGG CAC ACC a | E | 500 | 250 | 250 |
| –28 (A–G) | TAA GCA ATA GAT GGC TCT GCC CTG AGT TC b | E | 300 | 180 | 145 |
| Cd 71/72 (+A) | GGT TGT CCA GGT GAG CCA GGC CAT CAG TT b | E | 500 | 140 | 569 |
| MARMS-C | |||||
| IVS 1–1 (G–A) | TTA AAC CTG TCT TGT AAC CTT GAT ACG AAT e | E | 300 | 600 | 315 |
| Cd 43 (G–T) | ATC AGG GAG TGG ACA GAT CCC CAA GGA GTA e | E | 80 | 50 | 482 |
| Cd 16 (–C) | TCA CCA CCA ACT TCA TCC ACG TTC ACG TTC d | E | 80 | 160 | 273 |
| Poly A (A–G) | GGC CTT GAG CAT CTG GAT TCT GCC TAT TAG c | B | 160 | 50 | 393 |
| MARMS-D | |||||
| Cd 19 (A–G) | TGC CGT TAC TGC CCT GTG GGG CAA GGA GAG e | F | 50 | 30 | 173 |
| CAP +1 (A–C) | AAA AGT CAG GGC AGA GCC ATC TAT TGG TTC d | F | 100 | 80 | 281 |
| MARMS-E | |||||
| –88 (C–T) | TCA CTT AGA CCT CAC CCT GTG GAG CCT CAT a | F | 50 | 50 | 369 |
| Initiation Cd | TGT TCA CTA GCA ACC TCA AAC AGA CAG CAG e | F | 100 | 160 | 248 |
| Cd 15 (G–A) | TGA GGA GAA GTC TGC CGT TAC TGC CCA GTA d | F | 60 | 30 | 203 |
| –29 (A–G) | CAG GGA GGG CAG GAG CCA GGG CTG GGT ATG a | F | 50 | 20 | 310 |
| ARMS-F | |||||
| IVS 2–654 (C–T) | GAA TAA CAG TGA TAA TTT CTG GGT TAA CGT a | B | 160 | 400 | 826 |
| Control A | CAA TGT ATC ATG CCT CTT TGC ACC a | 120 | 120 | 861 | |
| Control B | GAG TCA AGG CTG AGA GAT GCA GGA a | 150 | 105 | ||
| Control C | CAA CTT GCT CAA GCA TAC ACT C b | 105 | 105 | 493 | |
| Control D | AAT AAT AGG CAT AGT GAC AAG TGC b | 105 | 105 | ||
| Common E | TGA AGT CCA ACT CCT AAG CCA GTG b | 160 | 160 | ||
| Common F | CAA TAG GCA GAG AGA GTC AGT GCC TAT CA b | 160 | 160 | ||
* Each normal primer was tested with internal control only. Primer sequences (superscripted a–e) in reference were obtained from earlier publications. Sequences tagged “a” were from (16), “b” from (15), “c” from (14), “d” from (17), and “e” were designed during the course of the project. Underlined text identifies mutation, bolded text identifies mismatch.
To determine the primer mix in each MARMS tube, consideration was given to the size of the amplicons and the prevalence rate of the mutations in the local population. Samples were tested with MARMS-A and B first, followed by the other MARMS and IVS 2–654 (C–T) if the patient was negative for any of the A and B mutations. Patients who had received regular or irregular blood transfusions, and who were not compound heterozygotes, were characterised using the corresponding normal primers to detect homozygosity. Each normal ARMS primer was used with the common primer, and the amplification was carried out only in the presence of the internal control. The presence of 861 bp amplicon in MARMS, and 493 bp amplicon in the single ARMS, showed successful amplification and homozygosity for that specific mutation.
Amplification was carried out in a 20 μL reaction containing HotStarTaq® Master Mix (1X PCR Buffer, 1.5 mm MgCl2, HotStarTaq® DNA Polymerase and 200 μm of each DNTP) (Qiagen GmbH, Hilden, Germany), various primers of different concentrations, as shown in Table 1, and 0.3–0.5 μg of genomic DNA. All oligonucleotide primers were synthesised by Sigma-Aldrich® (St. Louis, MO, USA).
Amplifications were carried out in a PTC-200 thermal cycler (MJ Research, Watertown, MA, USA). The thermal cycling consisted of an initial denaturation at 95 °C for 15 min, followed by 30 cycles at 94 °C for 45 s, annealing at 65 °C for 45 s, at 72 °C for 1 min 30 s, and final extension at 72 °C for 7 min. For IVS 2–654 (C–T), annealing temperature was set at 60 °C. The products were electrophoresed with 3.0% Bioline™ Multipurpose Agarose (London, UK) gel in 1× Tris-borate-EDTA buffer. The gel was stained with ethidium bromide and visualised under a transilluminator (Vilber Lourmat, Marne-la-Vallée, France).
Reverse dot blot hybridization
Reverse dot blot hybridization, using β-Globin StripAssay SEA™ (ViennaLab Diagnostics GmbH, Vienna, Austria) was used for mutation confirmation. Genomic DNA was used as a template for PCR amplification using biotinylated primers, followed by hybridization of the amplification products to a test strip containing allele-specific oligonucleotide probes, immobilised as an array of parallel lines. Bound biotinylated sequences were detected using streptavidin-alkaline phosphatase and colour substrates. The assay covered 22 mutations: –31 (A–G), –29 (A–G), –28 (A–G), cap+1 (A–C), Initiation Cd (ATG–AGG), Cd 8/9 (+G), Cd 15 (TGG–TAG), Cd 17 (A–T), Cd 19 (A–G), Cd 26 (G–A) HbE, Cd 27/28 (+C), IVS1–1 (G–T), IVS 1–5 (G–C), Cd 41/42 (–TTCT), Cd 43 (G–T), Cd 71/72 (+A), Cd 89/90 (–GT), Cd 90 (G–T), Cd 95 (+A), IVS2–1 (G–A), IVS 2–654 (C–T), and Cd 121 (G–T).
Direct Sequencing
Poly A (A–G), –86 (C–G), –88 (C–T), IVS 1–1 (G–A), and Cd 16 (–C) are not detectable using β-Globin StripAssay SEA™ (ViennaLab Diagnostics GmbH, Vienna, Austria). We therefore confirmed Poly A (A–G), and IVS 1–1 (G–A) by direct sequencing (18). Samples were amplified using two sets of primers. The resulting nucleotides were checked by gel electrophoresis. Purification and sequencing were performed by First Base Laboratories (Selangor, Malaysia). The sequencing data were analysed using GeneScreen software (19).
Results
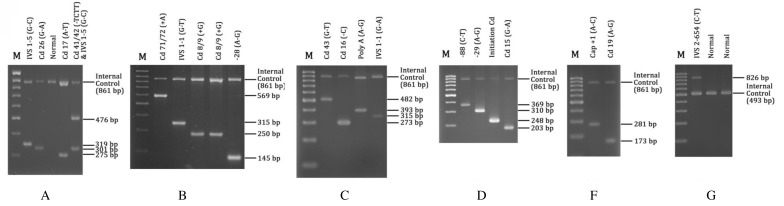
Nearly all mutations were amplified at an annealing temperature of 65 °C, except for IVS 2–654 (C–T) which was amplified at 60 °C. For IVS 2–654 (C–T), primers C and D were used as internal control primers, producing a control band of 493 bp, whereas primers A and B were used as internal controls, producing an 861-bp control band on the MARMS. All samples were analysed simultaneously with positive controls for a particular mutation (Figure 1b). A deletion of 619-bp was located within primers A and B (Figure 1a). Heterozygous 619 bp deletion was shown by the presence of a 242 bp band and an 861-bp control band. The presence of a 242 bp band without the 861 bp control band indicated homozygosity for the deletion. To differentiate the 619-bp deletion from Cd 8/9 (+G) and Initiation cd (ATG–AGG) mutations which produce 250- and 248-bp bands, respectively, PCR was repeated with internal control primers A and B only.
Figure 1b:
A representative agarose gel electrophoresis of PCR products using ARMS and MARMS-PCR. M is 100 bp molecular weight marker. A-E represents MARMS A, B, C, D, and E. F represents single ARMS-F.
Figure 1a:
Schematic illustration of β-globin gene (HBB) and relative positions of various primers for ARMS and MARMS-PCR and mutation sites within the HBB gene. Common C and D are not within HBB.
A total of 169 heterozygous β-thalassaemia carriers were detected (Table 2). The IVS 1–5 (G–C) mutation was the most common β-thalassaemia defect in this population. The mutations detected are presented in decreasing frequency: IVS 1–5 (G–C) (23.1%), Cd 26 (G–A) HbE (23.1%), Cd 41/42 (–TTCT) (16%), and IVS 1–1 (G–T) (16%). Seventy-five percent of the Malay β-thalassaemia carriers had either Cd 26 (G–A) HbE, IVS1–5 (G–C) or IVS1–1 (G–T) mutations. In the Chinese, Cd 41/42 (–TTCT) and IVS 2–654 (C–T) were the most common (79.1%).
Table 2.
Distribution of β-globin gene mutations identified in β-thalassemia carriers of different ethnic groups
| Mutations | Chinese | Malay | Indians | Others | Total |
|---|---|---|---|---|---|
| –88 (C>T) | 0 | 0 | 0 | 0 | 0 |
| Cap +1 (A>C) | 0 | 2 | 0 | 0 | 2 |
| –28 (A>G) | 2 | 0 | 0 | 0 | 2 |
| –29 (A>G) | 0 | 2 | 0 | 0 | 2 |
| Initiation Cd (T>G) | 0 | 0 | 0 | 0 | 0 |
| Cd 8/9 (+G) | 0 | 4 | 0 | 0 | 4 |
| Cd 15 (G>A) | 0 | 0 | 1 | 0 | 1 |
| Cd 16 (–C) | 0 | 0 | 0 | 0 | 0 |
| Cd 17 (A>T) | 0 | 3 | 0 | 0 | 3 |
| Cd 19 (A>G) | 0 | 8 | 0 | 0 | 8 |
| Cd 26 (G>A) Hb E | 2 | 36 | 0 | 1 | 39 |
| IVS1–1 (G>T) | 0 | 27 | 0 | 0 | 27 |
| IVS1–1 (G>A) | 0 | 1 | 0 | 0 | 1 |
| IVS1–5 (G>C) | 1 | 37 | 0 | 1 | 39 |
| Cd 43 (G>T) | 0 | 0 | 2 | 0 | 2 |
| Cd 41/42 (–TCTT) | 13 | 14 | 0 | 0 | 27 |
| Cd 71/72 (+A) | 0 | 0 | 0 | 0 | 0 |
| IVS 2–654 (C>T) | 6 | 1 | 0 | 0 | 7 |
| Poly A (A>G) | 0 | 5 | 0 | 0 | 5 |
| 619 Deletion | 0 | 0 | 0 | 0 | 0 |
| Total | 24 | 140 | 3 | 2 | 169 |
Sixteen homozygous and 23 compound heterozygous were found among 39 β-thalassaemia patients (Table 3 and 4). Homozygosity of the Cd 26 (G–A) HbE mutation was found in 8 Malays, 1 indigenous Sabahan, and 1 indigenous West Malaysian. Homozygosity for the Cd 41/42 (–TTCT), Cd 71/72 (+A), IVS 1–5 (G–C), Cd 19 (A–G), and IVS 1–1 (G–T) mutations was also detected among the Malays. Most compound heterozygous cases involved common mutations of Cd 26 (G–A) HbE, IVS 1–5 (G–C), IVS 1–1 (G–T), Cd 41/42 (–TTCT), IVS 2–654 (C–T), and –28 (A–G) (Table 3). In total, 16 mutations were discovered, whereas Cd 16 (–C), –88 (C–T), Initiation Cd (T–G), and 619 deletion were not detected in this study. The results from MARMS showed 100% concordance with those from the reverse dot blot analysis by β-Globin StripAssay SEA™ and direct sequencing.
Table 3.
Cases of Homozygous β-thalassaemia patients
| Mutations | Number of patients |
|---|---|
| Cd 41/42 (–TTCT) | 1 |
| Cd 26 (G>A) | 10 |
| Cd 19 (A>G) | 1 |
| IVS1–5 (G>C) | 2 |
| IVS 1–1 (G>T) | 1 |
| Cd 71/72 (+A) | 1 |
| Total | 16 |
Table 4.
Cases of Compound Heterozygous β-thalassaemia patients
| Mutations | Number of patients |
|---|---|
| Cd 26 (G–A) & Cd 41/42 (–TTCT) | 2 |
| Cd 26 (G>A) & IVS1–5 (G–C) | 4 |
| Cd 26 (G>A) & IVS1–1 (G>T) | 4 |
| Cd 26 (G–A) & –28 (A>G) | 2 |
| –28 (A>G) & Cd 41/42(–TTCT) | 1 |
| –28 (A>G) & IVS 2–654 (C–T) | 1 |
| –28 (A>G) & IVS1–1 (G>T) | 1 |
| –28 (A>G) & Cd 19 (A>G) | 2 |
| IVS 2–654 (C–T) & Cd 41/42 (–TTCT) | 1 |
| Cd 19 (A>G) & IVS1–1 (G>T) | 1 |
| CAP +1 (A>C) & Cd 19 (A>G) | 1 |
| CAP +1 (A>C) & IVS1–5 (G–C) | 1 |
| Poly A (A–G) & IVS1–5 (G–C) | 1 |
| Cd 8/9 (+G) & Poly A (A–G) | 1 |
| Total | 23 |
Discussion
Every molecular diagnostic technique has its limitations. Molecular analysis is highly accurate, but can be expensive and time-consuming without proper planning. Allele-specific oligonucleotide probes (ASO) dot blot hybridization is reliable and easy for populations predominated by only one or two mutations (20), but becomes time-consuming because it requires separate hybridization and washing steps to screen for multiple mutations (21). Direct DNA sequencing, denaturing gradient gel electrophoresis (DGGE) and single-strand conformation polymorphism (SSCP) can be employed to screen for unknown mutations. DGGE and SSCP require special apparatus and a skilled operator to interpret the heteroduplex patterns (22). The conventional reverse dot blot is cheap, but laborious and timeconsuming, whereas the commercial reverse dot blot kit is rapid but expensive. ARMS is laborious and expensive, therefore multiplexing the most common mutations in an ethnic group is more desirable. It can be employed if funds and access to more advanced technology are limited as it does not require extensive equipment (23).
Recently, more sophisticated techniques, such as real-time PCR, high resolution melting analysis (HRM), and oligonucleotide microarray analysis, have been reported. Allele-specific Q-primer real time-PCR (24) is cheap, rapid and high-throughput, but limited to the screening of several common mutations only. The bead-based biosensor described by Ng et al., (25) is rapid, high-throughput and more sensitive, but the fabrication process can be complicated and costly. HRM analysis is a powerful screening tool, being a closed-tube method where amplification and subsequent analyses are sequentially performed in one well. However, the presence of unexpected polymorphisms, close to mutations of interest, may interfere with genotyping (26). Furthermore, it is important that molecular techniques be aimed at directly interrogating the site of a mutation as opposed to determining the effect of a putative mutation on an amplicon, such as in melting profile identification (27).
Carrier screening in Southeast Asia revealed a high frequency of β-thalassaemia ranging from 3% to 10% in Indonesia, 0.5% to 1.5% in Myanmar, 0.93% in Singapore, 1.5% in Vietnam, and 3% to 9% in Thailand (28). We found a high frequency of carriers with Cd 26 (G–A), IVS1–5 (G–C), IVS 1–1 (G–T), and Cd 41/42 (–TTCT) among the Malays. Previously, only Cd 26 (G–A) and IVS1–5 (G–C) were reported as common among the Malays (6,12). The high frequencies of Cd 26 (G–A) HbE and IVS 1–5 (G–C) were similar to an earlier finding in Indonesia (29). We recorded five times higher frequencies of Cd 26 (G–A) HbE carriers (23%) as compared to previous micromapping studies that showed 4% among the Malays (30). This could be due to the larger sample size studied. In total, 29% (54/185) of patients were found with Cd 26 (G–A) HbE, including 11 compound heterozygous Cd 26 (G–A) HbE/β-thalassaemia and 73.3% (11/15) homozygous for Cd 26 (G–A) HbE. The prevalence of Cd 26 (G–A) HbE is relatively similar to that in neighbouring countries (10,31–33).
Among the Chinese, Cd 41/42 (–TTCT), IVS2–654 (C–T), and –28 (A–G) mutations accounted for 88% (24/27) of the mutations. Thong et al. (12) reported a high frequency of similar mutations among the Chinese. Interestingly, we found a 22-year-old Kedayan with Cd 8/9 (+G) and Poly A (A–G) who was diagnosed as β-thalassaemia major, and who had undergone regular blood transfusions since he was 8-months old; and a Chinese-Malay descendant patient with heterozygous IVS 2–654 (C–T) mutations. Other rare compound heterozygous cases were Cap+1 (A–C) with Cd 19 (A–G), Cap+1 (A–C) with IVS 1–5 (G–C), and Poly A (A–G) with IVS1–5 (G–C), which was detected in Malays.
Although five mutations, IVS–2–654 (C–T), CD41/42 (–TTCT), IVS1–5 (G–C), Cd 26 (G–A) HbE, and IVS 1–1 (G–T) accounted for more than 80% of the mutant alleles in carriers, another 10 different mutations were identified in our patients. This demonstrates the heterogeneity of β-thalassaemia mutations which may have been brought about by the ethnic diversity and interracial marriages among Malaysians.
Conclusion
In conclusion, we have reported the used of inexpensive and easily interpreted techniques to identify 6 common and 14 rare β-thalassemia mutations among Malaysians. The genotyping system uses commonly available reagents and equipment and is simple to perform, requiring only a basic understanding of molecular techniques. By employing these techniques under the National Thalassaemia Control and Prevention Program, we hope that screening will facilitate the genetic counselling of transfusion-dependent children, and pre-marital and pregnancy planning among Malaysians.
Acknowledgments
We would like to thank the Director General of Health, Dato’ Sri Dr Hasan bin Abdul Rahman for his permission to publish this paper. This project was funded by the Ministry of Health, Malaysia, under the National Thalassaemia Control and Prevention Program. We also thank the developers of GeneScreen software, lead by Dr Ian M Carr, of the Leeds Institute for Molecular Medicine, for technical assistance, and Dr Law Hai Yang for supplying the human genomic DNA from patients with a wide range of defined β-thalassaemia mutations.
Footnotes
Authors’ contributions
Conception and design, analysis and interpretation of the data: SH
Drafting of the article: SH, RA
Critical revision of the article: WZA, RA, ZEZ
Final approval of the article: WZA, RA, ZEZ, ZZ
Provision of study materials or patients, obtaining of funding, and administrative, technical or logistic support: RA
Conflict of interest
No conflict of interest.
Funds
Ministry of Health, Malaysia, under the National Thalassaemia Control and Prevention Program (B42/251/190000/032100/20000), Warrant no 32132.
References
- 1.Thein SL. Genetic modifiers of β-thalassemia. Haematologica. 2005;90(5):649–660. [PubMed] [Google Scholar]
- 2.Stamatoyannopoulos G, Nienhuis AW. In: The Molecular Basis of Blood Diseases. 2nd ed. Stamatoyannopoulos G, Nienhuis AW, Majerus PW, Varmus H, editors. Philadelphia: WB Saunders; 1994. Hemoglobin switching; pp. 107–136. [Google Scholar]
- 3.Steinberg MH, Forget BG, Higgs DR, Nagel RL. New York (NY): Cambridge University Press; 2001. Disorders of hemoglobin. [Google Scholar]
- 4.Huisman THJ, Carver MFH, Baysal E. Augusta, GA: The Sickle Cell Anemia Foundation; 1997. A syllabus of thalassaemia mutations. [Google Scholar]
- 5.Higgs DR, Thein SL, Woods WG. In: The thalassaemia syndromes. 4th ed. Weatherall DJ, Clegg JB, editors. Oxford: Blackwell Science; 2001. The molecular pathology of the thalassaemias; pp. 133–191. [Google Scholar]
- 6.George E, Li HJ, Fei YJ, Reese AL, Baysal E, Cepreganova B, et al. Types of thalassaemia among patients attending a large university clinic in Kuala Lumpur, Malaysia. Hemoglobin. 1992;16(1–2):51–66. doi: 10.3109/03630269209005676. [DOI] [PubMed] [Google Scholar]
- 7.Thong MK, Law HY, Ng ISL. Molecular heterogeneity of β-thalassaemia in Malaysia: a practical approach to diagnosis. Ann Acad Med Singapore. 1996;25(1):79–83. [PubMed] [Google Scholar]
- 8.George E. In: Thalassaemia carrier diagnosis in Malaysia. George E, editor. Kuala Lumpur (Malaysia): SP-Muda; 1998. Thalassemia: a public health problem in Malaysia. [Google Scholar]
- 9.Tan JAMA, George E, Tan KL, Chow T, Hassan J, Chia P, et al. Molecular defects in the β-globin gene identified in different ethnic groups/populations during prenatal diagnosis for β-thalassaemia: a Malaysian experience. Clin Exp Med. 2004;4(3):142–147. doi: 10.1007/s10238-004-0048-x. [DOI] [PubMed] [Google Scholar]
- 10.Abdullah WA, Jamaluddin NK, Kham SK, Tan JA. The spectrum of the β-thalassaemia mutations in the Malays in Singapore and Kelantan. Southeast Asian J Trop Med Public Health. 1996;27(1):164–168. [PubMed] [Google Scholar]
- 11.Tan KL, Tan JAMA, Wong YC, Wee YC, Thong MK, Yap SF. Combine-ARMS: a rapid and cost effective protocol for molecular characterization of β-thalassaemia in Malaysia. Genetic Testing. 2001;5(1):17–22. doi: 10.1089/109065701750168626. [DOI] [PubMed] [Google Scholar]
- 12.Thong MK, Tan JAMA, Tan KL, Yap SF. Characterisation of β-globin gene mutations in Malaysian children: A strategy for the control of β-thalassaemia in a developing country. J Trop Pediatr. 2005;51(6):328–333. doi: 10.1093/tropej/fmi052. [DOI] [PubMed] [Google Scholar]
- 13.Tan JAMA, Yap SF, Tan KL, Thong MK. The use of the Amplification Refractory Mutation System (ARMS) as an effective and economical tool for prenatal diagnosis of β-thalassaemia in Malaysian subjects. Int Med Res J. 1998;26(2):65–68. [Google Scholar]
- 14.Chan YF, Tan KL, Wong YC, Wee YC, Yap SF, Tan JAMA. The use of the Amplification Refractory Mutation System (ARMS) in the detection of rare β-thalassaemia in the Malays and Chinese in Malaysia. Southeast Asian J Trop Med Public Health. 2001;32(4):872–879. [PubMed] [Google Scholar]
- 15.Bhardwaj U, Zhang YH, Lorey F, McCabe LL, McCabe ER. Molecular genetic confirmatory testing from newborn screening samples for the common African-American, Asian Indian, Southeast Asian, and Chinese β-thalassaemia mutations. Am J Hematol. 2005;78(4):249–255. doi: 10.1002/ajh.20269. [DOI] [PubMed] [Google Scholar]
- 16.Varawalla NY, Old JM, Sarkar R, Venkatesan R, Weatherall DJ. The spectrum of β-thalassemia mutations on the Indian subcontinent: the basis of prenatal diagnosis. Br J Hematol. 1991;78(2):242–247. doi: 10.1111/j.1365-2141.1991.tb04423.x. [DOI] [PubMed] [Google Scholar]
- 17.Old JM. In: Methods Pediatric Hematology: Methods and Protocols. Voume 91. Goulden NJ, Steward CG, editors. New Jersey: Humana Press Inc; 2004. Antenatal Diagnosis of Hemoglobinopathies; pp. 33–62. [Google Scholar]
- 18.Mirasena S, Shimbhu D, Sanguansermsri M, Sanguansermsri T. The spectrum of β-thalassaemia mutations in Phitsanulok Province: Development of multiplex ARMS for mutation detection. Naresuan University J. 2007;15(1):43–53. [Google Scholar]
- 19.Carr IM, Camm N, Taylor GR, Charlton R, Ellard S, Sheridan EG, et al. GeneScreen: a program for highthroughput mutation detection in DNA sequence electropherograms. J Med Gens. 2011;48(2):123–130. doi: 10.1136/jmg.2010.082081. [DOI] [PubMed] [Google Scholar]
- 20.Ristaldi MS, Pirastu M, Rosatelli C, Cao A. Prenatal diagnosis of β-thalassaemia in Mediterranean populations by dot blot analysis with DNA amplification and allele specific oligonucleotide probes. Proc Natl Acad Sci USA. 1989;9(9):629–638. doi: 10.1002/pd.1970090906. [DOI] [PubMed] [Google Scholar]
- 21.Old JM. Screening and genetic diagnosis of hemoglobin disorders. Blood Reviews. 2003;17(1):43–45. doi: 10.1016/s0268-960x(02)00061-9. [DOI] [PubMed] [Google Scholar]
- 22.Clark BE, Thein SL. Molecular diagnosis of haemoglobin disorders. Clin Lab Haem. 2004;26(3):159–176. doi: 10.1111/j.1365-2257.2004.00607.x. [DOI] [PubMed] [Google Scholar]
- 23.Joshi VA, DiNardo DM, Funke BH. In: Current Protocol in Human Genetics. Joshi VA, DiNardo DM, Funke BH, editors. United States (US): John Wiley; 2008. Selection of a Platform for Mutation Detection. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Law HY, Tan YM, Hong Y. High-throughput β-thalassaemia carrier screening by allele-specific Q-primer real-time polymerase chain reaction. Analyt Biochem. 2010;404(1):97–99. doi: 10.1016/j.ab.2010.04.025. [DOI] [PubMed] [Google Scholar]
- 25.Ng JKK, Wang W, Liu W-T, Chong SS. Spatially addressable bead-based biosensor for rapid detection of β-thalassaemia mutations. Analatica Chimica Acta. 2010;658(2):193–196. doi: 10.1016/j.aca.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Shih HC, Er TK, Chang TJ, Chang YS, Liu TC, Chang JG. Rapid identification of HBB gene mutations by high-resolution melting analysis. Clin Biochem. 2009;42(16–17):1667–1676. doi: 10.1016/j.clinbiochem.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 27.Chehab FF. In: Molecular Diagnostics: Techniques and Applications for the Clinical Laboratory. 1st ed. Grody WW, Nakamura RM, Kiechle FL, Strom C, editors. New York (NY): Academic Press; 2010. Molecular Diagnostic Challenges of the Thalassaemia; pp. 441–452. [Google Scholar]
- 28.Fucharoen S, Winichagoon P. Prevention and control of thalassemia in Asia. Asian Biomed. 2007;1(1):1–6. [Google Scholar]
- 29.Setianingsih I, Williamson R, Marzuki S, Harahap A, Tamam M, Forrest S. Molecular basis of β-thalassemia in Indonesia: application to prenatal diagnosis. Mol Diag. 1998;3(1):11–20. doi: 10.154/MODI00300011. [DOI] [PubMed] [Google Scholar]
- 30.George E, Khuziah R. Malays with thalassaemia in west Malaysia. Trop Geogr Med. 1984;36(2):123–125. [PubMed] [Google Scholar]
- 31.Sicard D, Kaplan JC, Labie D. Haemoglobinopathies and G6PD deficiency in Laos. Lancet. 1978;2(8089):571–572. doi: 10.1016/s0140-6736(78)92899-4. [DOI] [PubMed] [Google Scholar]
- 32.Svasti MLS, Hieu TM, Munkongdee T, Winichagoon P, Be TV, Binh TV, et al. Molecular analysis of β-thalassemia in South Vietnam. Am J Hematol. 2002;71(2):85–88. doi: 10.1002/ajh.10193. [DOI] [PubMed] [Google Scholar]
- 33.Choopayak C, Mirasena S, Poodendaen C, Jiraviriyakul A, Sangarun K, Shimbhu D. Thalassemia mutations in lower northern part of Thailand. Naresuan University J. 2005;13(3):19–29. [Google Scholar]