Abstract
Purpose.
Innate immunity plays a role in many diseases, including glaucoma and AMD. We have used transcriptome profiling in the mouse to identify a network of genes involved in innate immunity that is present in the normal retina and that is activated by optic nerve crush (ONC).
Methods.
Using a recombinant inbred (RI) mouse strain set (BXD, C57BL/6 crossed with DBA/2J mice), we generate expression datasets (Illumina WG 6.2 arrays) in the normal mouse retina and 2 days after ONC. The normal dataset is constructed from retinas from 80 mouse strains and the ONC dataset is constructed from 62 strains. These large datasets are hosted by GeneNetwork.org, along with a series of powerful bioinformatic tools.
Results.
In the retina datasets, one intriguing network involves transcripts associated with the innate immunity. Using C4b to interrogate the normal dataset, we can identify a group of genes that are coregulated across the BXD RI strains. Many of the genes in this network are associated with the innate immune system, including Serping1, Casp1, C3, Icam1, Tgfbr2, Cfi, Clu, C1qg, Aif1, and Cd74. Following ONC, the expression of these genes is upregulated, along with an increase in coordinated expression across the BXD strains. Many of the genes in this network are risk factors for AMD, including C3, EFEMP1, MCDR2, CFB, TLR4, HTA1, and C1QTNF5.
Conclusions.
We found a retina-intrinsic innate immunity network that is activated by injury including ONC. Many of the genes in this network are risk factors for retinal disease.
Keywords: innate immunity, genetic networks, retinal injury
An intrinsic innate immune network was identified in the mouse retina. This network is activated by injury and mutations in members of this network result in disease, including AMD.
Introduction
The innate immune system plays many different roles in the central nervous system (CNS)1 during development2,3 and disease.4–7 The primary component of innate immunity is the complement system, and its involvement in CNS disorders is well documented. For example, members of the complement cascade are associated with the pathological changes occurring in Alzheimer disease8–11 and spinal cord injury.12,13 Within the past decade, the significance of the complement system was dramatically demonstrated by its role in AMD, where recent studies have shown that common variants of complement factor H (CFH) represent major risk factors for AMD.14–17 Many of the components of the complement system are also found in drusen that accumulate in AMD, examples of which include CFH,15 complement factor B (CFB),18 complement receptor 1,19 C3,19 C5,20 C6–C9,21 and members of the terminal pathway, vitronectin, and clustrin.22 As it turns out, these same genes are also risk factors for AMD (for review see Anderson et al.4), including CFB,18 C2,18 C3,23 and SERPING1.24
The complement components also play a significant role in animal models of glaucoma.25–29 One of the most accepted murine models of naturally occurring glaucoma is the DBA/2J mouse. This mouse carries two mutations that result in a progressive iris stromal atrophy and pigment dispersion.30,31 The dispersed pigment blocks the trabecular meshwork causing an elevation in IOP, and a phenotype similar to pigmentary dispersion glaucoma.30 Studies using the DBA/2J model26–28 examined the changes in gene expression during the progression of the disease.26,28 In this mouse model, many components of the complement system increased in expression as the disease process progressed, specifically including C1q, C3, and Cfi (see Fig. 5 in Howell et al.28). The upregulation of these components of the complement system suggests that the innate immune system plays an important role in the immune-privileged environment of the retina and specifically in the response of the retina to disease. The role of complement in glaucomatous damage was tested by Howell et al.28 by knocking out C1q on the DBA/2J background. These C1q knockout mice developed iris disease and elevated IOP similar to the normal DBA/2J mice; however, knocking out C1q reduced and delayed the loss of axons in the optic nerve,28 which suggests that interrupting the complement cascade in DBA/2J mice may alter disease severity and progression.
The present study investigates the regulation of the innate immune system in the retina, and identifies changes in global expression of the complement system after optic nerve crush (ONC). We created two comprehensive and complementary expression data sets for the retina of the BXD mouse strains. One dataset was generated using untreated control retinas and the other was generated using the same genotypes of mice but 2 days after ONC. By examining the changes that occur in the gene expression profiles, we have been able to define an innate immune network active in the normal retina that is upregulated by the ONC.
Materials and Methods
Animals: Strains, Sex, and Age
The full Hamilton Eye Institute (HEI) Retina Database contains the data analysis of 346 Illumina Sentrix Mouse WG-6 v2.0 arrays (Illumina, San Diego, CA).32 Eighty strains of mice are represented, including 75 BXD RI strains with 307 independent retinal samples (for details see GeneNetwork.org). In the ONC HEI Retina Database, there are 62 strains (57 BXD RI strains, C57BL/6J, DBA/2J, and the reciprocal F1 crosses) with 184 independent biological samples run on the Illumina Sentrix Mouse WG-6 v2.0 array (for detailed method of ONC see Templeton and Geisert 201233). Samples were collected for microarray analyses 2 days after ONC. Based on our findings from C57BL/6 and DBA/2J mice,34 this is the time point when we are able to see early response genes upregulated and the changes that reach maxima at later time points, such as the upregulation of Gfap. Mice were either purchased from Jackson Laboratory (Bar Harbor, ME)35,36 or were from the breeding colonies of Robert Williams and Lu Lu at the University of Tennessee Health Science Center.37 All of the animals were young adults between 60 and 120 days of age. The housing room was maintained on a 12 hour on, 12 hour off light cycle. Expression data were obtained from independent biological samples from both sexes. For details regarding the animals in each of the datasets see Freeman et al.32 or the Info file on GeneNetwork.org for each of the datasets. All protocols used in this study were approved by the Animal Care and Use Committee of the University of Tennessee Health Science Center and were in accordance with the Institute for Laboratory Animal Research and the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Sample Processing and RNA Isolation
All mice were deeply anesthetized with a mixture of 13 mg/kg of xylazine (Rompun) and 87 mg/kg of ketamine (Ketalar) and killed by rapid cervical dislocation. The retinas were removed immediately. Two retinas per mouse were immersed in RNALater (Qiagen, Germantown, MD) and stored in a single tube overnight at 4°C. Total RNA was prepared from the retinal tissue with RNA-Stat-60 (Tel-Test, Friendswood, TX). The quality and purity of RNA were assessed using an Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA) to assess the relative quantities of 18S and 28S RNA, as well as the RNA integrity.
Sample Preparation and Hybridization
Total RNA (150 ng) was processed with the Illumina TotalPrep RNA Amplification Kit (Applied Biosystems, Foster City, CA) to produce biotinylated cRNAs. The concentration of the cRNA solution was determined by measuring the absorbance at 260 nm and 280 nm using the NanoDrop 1000A Spectrophotometer (Thermo Scientific, West Palm Beach, FL). The biotinylated cRNAs (1.5 μg/sample) were hybridized to the Illumina Sentrix Mouse WG-6 v2.0 arrays for 19.5 hours at 58°C according to the manufacturer's instructions (Illumina).
Quality Control of Array Data
Quality control analysis of the raw image data was performed using the Illumina BeadStudio software. Rank invariant normalization with BeadStudio software was used to calculate the data. Once these data were collected, they were globally normalized in a four-step process: (1) Compute the log base 2 of each raw signal value. (2) Calculate the mean and SD of each Mouse WG-6 v2.0 array. (3) Normalize each array using the formula 2 (z score Log2 [intensity]) + 8. The result is to produce arrays that have a mean of 8 and an SD of 2. (4) Compute the mean of the values for the set of microarrays for each strain. This process produces a distribution of probe labeling that is z-scaled with a mean probe labeling set to 8 on a log base 2 scale. Probes with scores below 8 are either expressed at extremely low levels or are not detected (i.e., absent) from the tissue.
Results
The Innate Immune System in the Retina
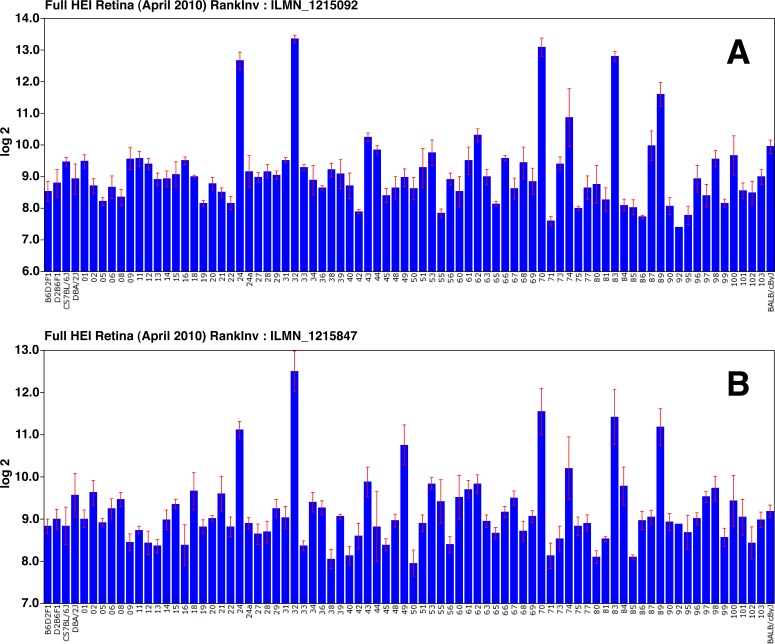
To begin our analysis of the innate immunity in the retina, we searched the Full HEI Retina Database in GeneNetwork to find mRNAs that correlate with the levels of C4b mRNA, a well-established marker of innate immunity. In the Illumina dataset, there are seven probes that target different exons in C4b. For this analysis, we chose the probe with the highest expression (ILMN_1215092 coding exon 39). All seven probes that recognize different exons within the gene covary (the lowest of 21 pairwise correlations among these probes was greater than 0.77). As a next step in the analysis, we examined genes within the Full HEI Retina Dataset that had a similar pattern of expression across the BXD RI strains (Fig. 1A). Using the correlation program within GeneNetwork, we found 100 genes that had a Pearson correlation greater than r = 0.73 (within this dataset six strains are present that display retinal degeneration and elevated glial fibrillary acidic protein [GFAP]). In this list of genes, many are associated with the innate immune network (see Table 1), including genes such as C4, C3, Cd1q, and Serping1.
Figure 1.
The expression of C4b (A) and Gfap (B) is shown across all of the BXD RI strains in the Full HEI Retina Datasets. The scale to the left is expression from the microarray studies expressed in log base 2 with the mean expression of mRNA across the microarray set to 8. Notice that the strains expressing high levels of C4b also express high levels of Gfap, suggesting that the elevation in C4b expression may be associated with reactive gliosis. These high Gfap strains were removed from this dataset to form the Normal HEI Retina Dataset.
Table 1. .
The Selected Genes in the Innate Immune System Are Presented Along With Their Illumina ID, Chromosomal Location, and Mean Expression Level in the Normal Retina Database
|
Symbol |
Description |
Record |
Location, Chr, Mb |
Mean Expression Level |
| C4b | Complement component 4B | ILMN_1215092 | Chr17: 34.866133 | 9.10 |
| Serping1 | Serine peptidase inhibitor, clade G, member 1 | ILMN_2913166 | Chr2: 84.605753 | 9.66 |
| C3 | Complement component 3 | ILMN_2759484 | Chr17: 57.343430 | 7.97 |
| Casp1 | Caspase 1 | ILMN_1247592 | Chr9: 5.306713 | 7.11 |
| lcam1 | Intercellular adhesion molecule 1 | ILMN_2896601 | Chr9: 20.832882 | 7.32 |
| Tgfbr2 | Transforming growth factor, beta receptor 2 | ILMN_2762979 | Chr9: 115.997139 | 8.86 |
| Cfi | Complement component factor i | ILMN_2631704 | Chr3: 129.577959 | 7.44 |
| Clu | Clusterin | ILMN_2727153 | Chr14: 66.600224 | 15.85 |
| C1qg | Complement component 1, q subcomponent, gamma | ILMN_2715840 | Chr4: 136.446132 | 8.18 |
| Aif1 | Allograft inflammatory factor 1 | ILMN_2804487 | Chr17: 35.308081 | 7.52 |
| Cd74 | CD74 antigen | ILMN_1221817 | Chr18: 60.971927 | 7.39 |
| Cp | Ceruloplasmin | ILMN_2520239 | Chr3: 19.90862 | 11.27 |
| Gfap | Glial fibrillary acidic protein | ILMN_1215847 | Chr11: 102.748858 | 9.15 |
| Stat3 | Signal tranducer and activator of transcription 3 | ILMN_2698046 | Chr11: 100.749555 | 11.22 |
All of these genes are expressed in the retina.
Reactive Gliosis as a Potential Confound
In the list of C4b correlates from the Full Retina Dataset were a group of genes associated with reactive gliosis, including Gfap, Stat3, Egrf, Cp, and Cd74. Examination of Gfap expression across all of the strains in the HEI Retinal Dataset (Fig. 1B) reveals that some of the strains express very high levels of Gfap. For example, BXD24 expresses high levels of Gfap and also expresses C4b at a 9-fold higher level than BXD22. During the inbreeding process, BXD24 acquired a mutation in Cep290 that results in early-onset photoreceptor degeneration.38 This degeneration results in reactive gliosis throughout the retina and clearly an elevation in C4b. When we checked the expression of Gfap and C4b across the BXD strains (Figs. 1A, 1B, respectively) we observed that some strains in the Full HEI Retinal Dataset (BXD24, BXD32, BXD49, BXD70, BXD83, and BXD 89) have extremely high levels of these two genes. Immunostaining for GFAP showed that all strains with elevated levels of Gfap message displayed reactive gliosis, with high levels of GFAP in both Müller cells and astrocytes (data not shown). We assume that either a mutation occurred during the breeding process necessary to produce an inbred strain (as is the case for BXD24) or the specific constellation of genes in these particular strains cause reactive gliosis. To eliminate the potential confounding influence due to reactive gliosis, we created a second “normal” retina dataset (Normal HEI Retina), eliminating strains with elevated levels of Gfap (i.e., BXD24, BXD32, BXD49, BXD70, BXD83, and BXD89). We also examined the effects of ONC using the ONC HEI Retina Database. All of these datasets are presented on our GeneNetwork Web site (genenetwork.org).
Network Maps of Innate Immunity
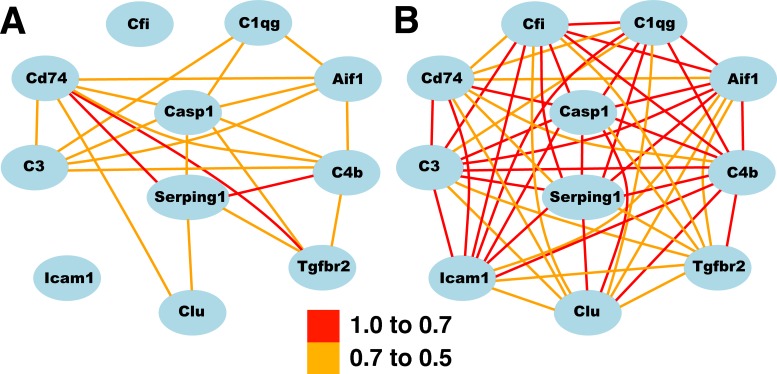
One way to illustrate the changes occurring in the network is to generate network maps using the Normal HEI Retinal Dataset (without the six animals with elevated Gfap) and comparing the correlations with a map generated from the ONC Retina Dataset (Fig. 2). When the top 100 correlates of C4b were analyzed using the Gene Set Analysis Toolkit V2 (Vanderbilt University, Nashville, TN; http://bioinfo.vanderbilt.edu/webgestalt), 30 of the 100 probes (25 of the 82 genes) were associated with the immune system process (see Supplementary Table S2). In the analysis of biological processes, there were no other significant enrichments of genes in any other functional category. Thus, immune system genes dominate this network, and examining the list of genes within the network reveals that most are associated with the innate immune system. We selected a small group of genes from this list (C4b, Serping1, Casp1, C3, Icam1, Tgfbr2, Cfi, Clu, C1qg, Aif1, Cd74) to illustrate effectively the relationships of the innate immune network in the retina. These genes were used to generate the two network maps in the normal retina and in the retina following ONC (Fig. 2).
Figure 2. .
The increased correlation after ONC among selected genes in the innate immune network. The correlation among genes in the Normal Retina Database is shown in (A). The increased correlation among these genes following ONC is clearly demonstrated in (B). The scale indicates the correlation between genes (Pearson). See Table 1 for a complete list of gene symbols and descriptions.
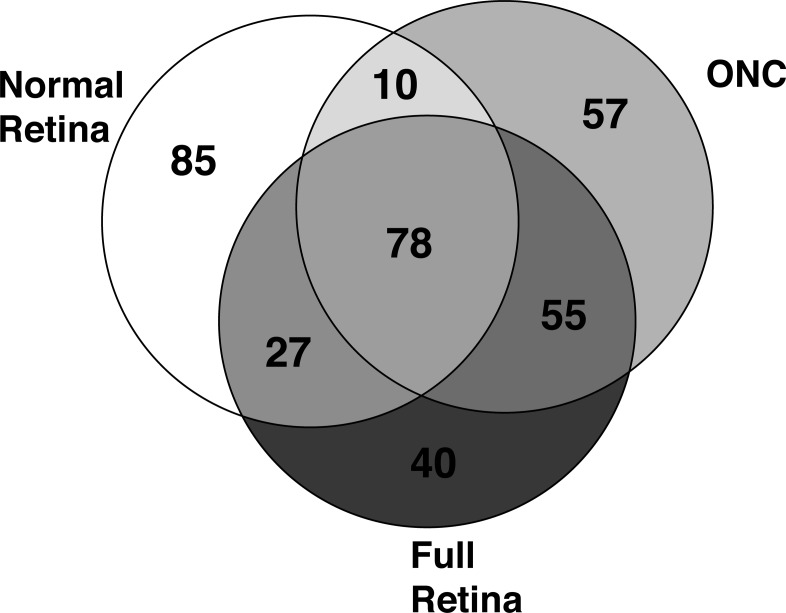
The innate immune system network is present within the normal retina (see Table 1). This small set of genes was selected to highlight genes associated with the innate immune system and to illustrate the changes occurring in a large group of genes within the retinal innate immune network (Supplementary Table S1 lists the top 100 genes in this network). The correlates of C4b in the Full Retina Dataset, the Normal Retina Dataset, and the ONC Dataset have a considerable overlap. All three correlate lists that contain the top 200 correlates share 66 genes (Fig. 3), represent 78 probes on the Illumina microarray. Among these 78 probes, the correlation values increase following ONC, with virtually all of the genes having a correlation above 0.7 (see Fig. 2B and Table 2). In addition to the increases in correlation values across the BXD RI strains, the expression of these genes was significantly upregulated following optic nerve injury (Table 2). Relative to the average expression of these genes in the normal retina, there was an average of a 3-fold increase in expression 2 days after ONC. Taken together, these data demonstrate that there is an existing innate immune network in the normal retina and that network is activated by crushing the axons within the optic nerve.
Figure 3.
The Venn diagram reveals the overlap in the top 200 correlates of C4b in the normal retina dataset (Normal Retina), the ONC Dataset (ONC), and the Full Retinal Dataset (Full Retina). Of the top 200 correlates from each dataset, 78 probes (representing 66 different genes) are in common among the three experimental groups.
Table 2. .
The Mean Expression Level of the Genes and Correlation to C4b for a Select Group of Genes in the Innate Immune Network Is Shown for the Normal HEI Retinal Database (Control Retina), the ONC HEI Retinal Database (ONC), and the Meta-Analysis of the Howell et al.28 DBA/2J Data (D2 Glaucoma)
|
Symbol |
Control Retina Expression |
ONC Expression |
D2 Glaucoma Expression |
Control Retina Correlation |
ONC Correlation |
D2 Glaucoma Correlation |
| C4b | 8.75 | 10.23 | 8.85 | 1 | 1 | 1 |
| Serping1 | 8.48 | 10.62 | 9.50 | 0.768 | 0.811 | 0.924 |
| C3 | 8.14 | 8.54 | 7.83 | 0.636 | 0.779 | 0.88 |
| Casp1 | 6.52 | 7.44 | 7.03 | 0.646 | 0.768 | 0.704 |
| Icam1 | 7.66 | 7.50 | 7.26 | 0.231 | 0.749 | 0.807 |
| Tgfbr2 | 8.21 | 8.99 | 8.83 | 0.644 | 0.723 | 0.69 |
| Cfi | 12.45 | 7.65 | 7.38 | 0.23 | 0.72 | 0.692 |
| Clu | 11.19 | 16.42 | 15.78 | 0.406 | 0.719 | 0.835 |
| C1qg | 6.91 | 9.02 | 8.08 | 0.481 | 0.708 | 0.818 |
| Aif1 | 6.40 | 7.93 | 7.47 | 0.432 | 0.667 | 0.723 |
| Cd74 | 6.22 | 7.66 | 7.29 | 0.721 | 0.657 | 0.628 |
| Cp | 8.96 | 11.93 | 11.59 | 0.169 | 0.603 | 0.741 |
| Gfap | 7.41 | 10.31 | 8.96 | 0.308 | 0.45 | 0.883 |
| Stat3 | 9.28 | 11.58 | 11.13 | −0.137 | 0.437 | 0.774 |
| Mean | 8.33 | 9.70 | 9.07 | 0.47 | 0.70 | 0.79 |
Notice that in the case of naturally occurring glaucoma and after ONC there is an increase in gene expression and an increased correlation among genes in this network.
QTL Mapping of the Innate Immune Network
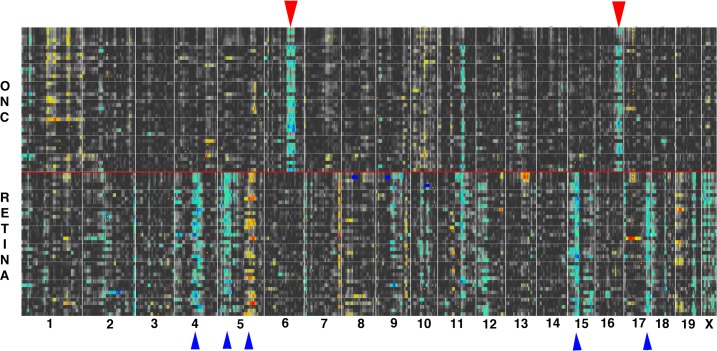
The finding that the innate immune system in the retina is so tightly correlated indicates that there are common regulatory mechanisms in the retina that are activated by the ONC injury. To define genomic loci that modulate the alterations in gene expression following ONC, we examined the changes occurring in the quantitative trait locus (QTL) map. The normal QTL Map was built using the top 40 correlates of C4b, and the same genes were used for the optic nerve injury heat map (Fig. 4). In the heat maps displayed in Figure 4, the regions of the genome that modulate the expression of the members of the innate immune network are highlighted (for a full description see Geisert et al.39). When the QTLs extend across the entire group of genes within the network (indicated by arrowheads in Fig. 4), we call these “signature QTLs” because they represent regions of the genome that affect the entire network. Within these signature QTLs are cis-acting loci (cis-QTLs) that are candidates for loci that modulate the innate immune network. When comparing the QTL maps from the normal retina database and ONC dataset, it is obvious that the signature QTL patterns change. The signature QTLs (marked with upward-pointing blue arrowheads in the lower part of Fig. 4) identify regions of the genome that modulate the innate immune network in the normal retina. Following ONC, the pattern of signature QTLs changes with most of the bands in the normal retina dataset diminishing in influence and two new prominent signature QTLs appearing in the ONC dataset (upper part of Fig. 4). The two major new signature bands (downward-pointing red arrowheads in Fig. 4) are on chromosome 6 at 116-127 Mb and chromosome 16 at 80-95 Mb. These two genome loci contain elements that potentially modulate the response of the innate immune network to ONC. Within these regions, cis-acting QTLs39 are putative candidate genes that modulate the innate immune network. In the chromosome 6 signature locus there are 9 genes with significant cis-QTLs: Adipor2, Cops7a, Dcp1b, Ing4, Lrrc23, Phc1, Slc6a12, Tmcc1, and Afp9. Within the signature band on chromosome 16, three genes are present with significant cis-QTLs: Cbr3, Sfrs15, and Tiam1. These genes represent putative upstream modulators in the activation of the innate immune network that occurs during optic nerve injury.
Figure 4. .
The genomic regions modulating the change in the innate immune network following ONC are shown by the differential QTL maps from the ONC HEI Database and the Normal Retinal Database (RETINA). The upper heat map was generated from the top 40 correlates of C4b found in the ONC HEI Database. The same genes (Illumina probes) were used to generate the lower heat map for the normal retina (RETINA). The upper map of the ONC data is separated from the lower normal retina data by a red line. The individual QTL maps are stacked on top of each other to create heat maps revealing signature QTLs for each dataset. Notice the two prominent new bands on Chr 6 and Chr 16 in the ONC heat map (red downward arrows). These represent regions of the genome that modulate the activation of the innate immune network following ONC. Note the loss of several cis-QTLs in the normal retina on Chr 4, Chr 5, Chr 15, and Chr 17, indicated by the blue arrowheads.
A Replication Dataset
To examine the possibility that the innate immune network is activated in a naturally occurring model of glaucoma, we examined data from the DBA/2J mouse. We analyzed the data of Howell et al.28 to explore this model of glaucoma using the GeneNetwork suite of tools. Howell and coworkers28 had examined changes in gene expression in the DBA/2J mouse using Affymetrix microarrays (Santa Clara, CA). In their study, they related changes in the transcriptome of retinas from DBA/2J mice directly to the severity of the glaucoma (as defined by determining the degree of degeneration in the optic nerve). After processing their microarray data (which they had deposited in the public domain at GEO under accession number GSE26299) in GeneNetwork, we plotted C4b expression levels from all of the individual animals and severity groups (Fig. 5). After identifying the top 100 correlates of C4b within the DBA/2J dataset,28 we compared these correlates with the list of correlates of C4b in the HEI Retina Dataset. Approximately one-quarter of the probe sets (26%), were in common, and these genes form the core of the innate immune network. The ONC network we derived in our study (Fig. 2), reveals that a similar set of genes in the innate immune network is activated in the DBA/2J model of glaucoma and the degree of activation is proportional to the degree of optic nerve damage.28 Howell et al.28 also identified members of the complement cascade that are upregulated. We observed similar findings after ONC in our database (Table 2). Howell et al.28 found that the innate immune network was upregulated in early stages of glaucoma in their optic nerve head dataset. The data from their optic nerve head study28 was placed in GeneNetwork and the variation across individual animals was analyzed. It appears that the members of the innate immune system are upregulated relatively early in the optic nerve head sample; however, using our approach, it was difficult to separate the changes occurring in the nerve head from the changes occurring in an adjacent portion of retina included in the microarray sample.
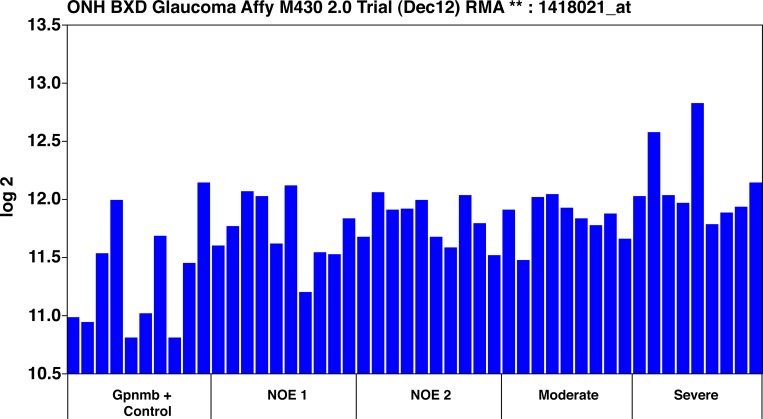
Figure 5. .
C4b RNA levels. The levels of C4b in a meta-analysis of mouse pigmentary glaucoma dataset from Howell et al.28 The ordinate represents the C4b mRNA level from microarrays, expressed in log base 2, scaled with the mean set to 8. The mice were classified as wild-type Gpnmb+/+ controls (Gpnmb + Controls), no detectable glaucoma 1 (NOE 1), no detectable glaucoma 2 (NOE 2), moderate glaucoma (Moderate), and severe glaucoma (Severe). Other than the wild-type group, all the groups were Gpnmb−/−. There were no average differences among the ages of the mouse groups, and the reasons that the Gpnmb−/− were different in severity are not obvious. But it is essential to note that the expression of C4b in the retina increases as the severity of glaucoma increases.
Discussion
The single most novel and exciting aspect of the present study is the identification of a genetic network in the retina that modulates innate immunity in the retina. The internal environment of the retina and optic nerve are protected from outside influences by the blood–brain and blood–retina barriers, creating an immunologically partially privileged environment where peripheral blood components do not enter the parenchyma of the retina or optic nerve. Within this immunologically privileged environment, we demonstrated that many components of the innate immune system are expressed intrinsically by retinal cells (Retinal Database, GeneNetwork.org32,40). Others find that components of the complement cascade are expressed in human RPE.5,23 The genes within this internal retinal network are activated by injury. This innate immune network is activated by ONC34 and in mouse models of glaucoma.28 In the ONC database, the top 100 correlates of C4b have correlation values greater than 0.66. When we examined the DBA/2J glaucoma database in a similar manner (meta-analysis of the Howell et al.28 data), we find the top 100 correlates for C4b to have correlation values above 0.78. Many innate immune system genes are in both of the ONC and glaucoma innate immune networks. Of the top 200 correlates from each dataset, 78 probes (representing 66 different genes) are in common among the three experimental groups (Fig. 3), and they include Serping1, C3, Tlr4, H2-Q2, H2-Tk23, CD74, C1q, and Cfi. Thus, different types of insults to the retina activate this same innate immune network.
Members of this innate immune network play significant roles in the development of the brain. One specific example of the functional role of the innate immune system is best illuminated by a recent developmental study of the lateral geniculate nucleus (LGN).3,41 In the developing brain, complement components are involved in synaptic elimination,3 marking the specific synaptic contacts that are disposed to elimination. During development, as the LGN receives input from both eyes, there is significant overlap in projections from the two eyes.42 As the visual system matures, the nucleus goes from an intermixing of the retinal inputs to a complete segregation of the inputs of one eye to the other eye. One layer of the LGN receives inputs exclusively from one eye, whereas the other layer receives input exclusively from the other.42 The synapses that are to be eliminated express C3 and in animals in which this gene is knocked out, segregation does not completely occur.3 Thus, members of the complement cascade form part of the system by which elements of the developing CNS are tagged for elimination.
In the adult retina, the C4b network, as defined by the expression pattern in the 80 BXD RI mouse strains, is an intrinsic network within retinal cells. The mRNA expression levels are upregulated by ONC (above the mean expression levels of mRNA in the retina as measured by our microarray platform). Since we have not found invading peripheral blood components in the retinas after ONC,34 these high levels of message must represent an increased expression in intrinsic retinal cells. In addition, several cellular markers are associated with the innate immune network, including astrocyte and Müller cell markers such as Gfap, Stat3, and Cp. Microglial signature genes are also present within the network, including Aif1, Cd68, and Cd74. This indicates that at least glial cells intrinsic to the retina are associated with this innate immune network. In the brain, others have found that glial cells are sources of many members of the complement cascade.1,43,44 All of these data indicate that, within the immunologically privileged environment of the retina and other CNS structures, the components of the complement cascade are produced and play a functional role. We are currently conducting experiments to define the specific cell types expressing the major members of the innate immune network in the retina.
An examination of the genes within the innate immune network points not only to genes associated with a response to injury, but also reveals many genes that are risk factors for retinal disease. There is a particularly strong association of the members of the complement system to human disease, specifically AMD. Many of the allelic variants of the components of the complement system are risk factors for AMD.1,14,15,23,45 Many of these complement proteins form deposits that are found within drusen, a pathological deposit that characterizes AMD.7,21,30,46
The associations of the complement system with neuronal diseases are not restricted to the retina. The role of the complement cascade in the brain is relatively well documented (for review see Refs. 11, 47). In the brain, the complement cascade is strongly associated with Alzheimer's disease.8,10 Members of this innate immune system are active after brain trauma.47,48 The innate immune system is associated with Parkinson's disease49 and multiple sclerosis29 in the CNS. Not only are individual members of the complement system associated with these diseases, it may be the case that the entire innate immune network is activated in each of these disease processes. One of the most instructive disease models is experimental autoimmune encephalomyelitis, a model for multiple sclerosis. In this disease model, the terminal pathway involving the membrane attack complex is not critical for the development of the disease.50 However, mice deficient in either C3 or Cfb displayed markedly reduced disease response and severity.29,50 These data suggest that the components of the complement system are not interacting with exactly the same components of the traditional complement pathways.
In the retina, these same components of the innate immune system are expressed and, as we have shown here, form a coregulated group of transcripts that form a novel genetic network. When members of this network carry mutations, the network can become dysregulated; it can drive the retina to accumulate drusen, and it can result in AMD (for review see Ref. 5). Clearly, the specific interactions of the innate immune network and its role in injury and disease are fertile grounds for future studies.
Supplementary Material
Acknowledgments
Supported by Grants R01 EY017841 (EEG), Department of Defense W81XWH-12-1-0255 (EEG), University of Tennessee Health Science Center Center for Integrative and Translational Genomics, U01 AA016662 (RWW), U01 AA013499 (RWW), R01 EY021200 (MMJ), unrestricted funds from Research to Prevent Blindness (University of Tennessee, Emory University, Vanderbilt University), R01 EY016470 (JMN); R01 EY021592 (JMN); R01 EY0211200 (MMJ); P30EY006360 (Emory University); R01EY022349 (TSR); and Department of Defense W81XWH-10-1-0528 (TSR).
Disclosure: J.P. Templeton, None; N.E. Freeman, None; J.M. Nickerson, None; M.M. Jablonski, None; T.S. Rex, None; R.W. Williams, None; E.E. Geisert, None
References
- 1. Veerhuis R, Nielsen HM, Tenner AJ. Complement in the brain. Mol Immunol. 2011; 48: 1592–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Prasad SS, Cynader MS. Identification of cDNA clones expressed selectively during the critical period for visual cortex development by subtractive hybridization. Brain Res. 1994; 639: 73–84 [DOI] [PubMed] [Google Scholar]
- 3. Stevens B, Allen NJ, Vazquez LE, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007; 131: 1164–1178 [DOI] [PubMed] [Google Scholar]
- 4. Anderson DH, Radeke MJ, Gallo NB, et al. The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog Retin Eye Res. 2010; 29: 95–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sparrow JR, Ueda K, Zhou J. Complement dysregulation in AMD: RPE-Bruch's membrane-choroid. Mol Aspects Med. 2012; 33: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonifati DM, Kishore U. Role of complement in neurodegeneration and neuroinflammation. Mol Immunol. 2007; 44: 999–1010 [DOI] [PubMed] [Google Scholar]
- 7. Alexander JJ, Anderson AJ, Barnum SR, Stevens B, Tenner AJ. The complement cascade: Yin-Yang in neuroinflammation—neuro-protection and -degeneration. J Neurochem. 2008; 107: 1169–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rogers J, Schultz J, Brachova L, et al. Complement activation and beta-amyloid-mediated neurotoxicity in Alzheimer's disease. Res Immunol. 1992; 143: 624–630 [DOI] [PubMed] [Google Scholar]
- 9. Jiang H, Burdick D, Glabe CG, Cotman CW, Tenner AJ. Beta-Amyloid activates complement by binding to a specific region of the collagen-like domain of the C1q A chain. J Immunol. 1994; 152: 5050–5059 [PubMed] [Google Scholar]
- 10. Bradt BM, Kolb WP, Cooper NR. Complement-dependent proinflammatory properties of the Alzheimer's disease beta-peptide. J Exp Med. 1998; 188: 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stoltzner SE, Grenfell TJ, Mori C, et al. Temporal accrual of complement proteins in amyloid plaques in Down's syndrome with Alzheimer's disease. Am J Pathol. 2000; 156: 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anderson AJ, Najbauer J, Huang W, Young W, Robert S. Upregulation of complement inhibitors in association with vulnerable cells following contusion-induced spinal cord injury. J Neurotrauma. 2005; 22: 382–397 [DOI] [PubMed] [Google Scholar]
- 13. Anderson AJ, Robert S, Huang W, Young W, Cotman CW. Activation of complement pathways after contusion-induced spinal cord injury. J Neurotrauma. 2004; 21: 1831–1846 [DOI] [PubMed] [Google Scholar]
- 14. Edwards AO, Ritter R III, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005; 308: 421–424 [DOI] [PubMed] [Google Scholar]
- 15. Hageman GS, Anderson DH, Johnson LV, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005; 102: 7227–7232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005; 308: 419–421 [DOI] [PubMed] [Google Scholar]
- 17. Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005; 308: 385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gold B, Merriam JE, Zernant J, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006; 38: 458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson LV, Leitner WP, Staples MK, Anderson DH. Complement activation and inflammatory processes in Drusen formation and age related macular degeneration. Exp Eye Res. 2001; 73: 887–896 [DOI] [PubMed] [Google Scholar]
- 20. Mullins RF, Johnson LV, Anderson DH, Hageman GS. Characterization of drusen-associated glycoconjugates. Ophthalmology. 1997; 104: 288–294 [DOI] [PubMed] [Google Scholar]
- 21. Crabb JW, Miyagi M, Gu X, et al. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci U S A. 2002; 99: 14682–14687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hageman GS, Mullins RF, Russell SR, Johnson LV, Anderson DH. Vitronectin is a constituent of ocular drusen and the vitronectin gene is expressed in human retinal pigmented epithelial cells. Faseb J. 1999; 13: 477–484 [DOI] [PubMed] [Google Scholar]
- 23. Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007; 39: 1200–1201 [DOI] [PubMed] [Google Scholar]
- 24. Ennis S, Jomary C, Mullins R, et al. Association between the SERPING1 gene and age-related macular degeneration: a two-stage case-control study. Lancet. 2008; 372: 1828–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ahmed F, Brown KM, Stephan DA, Morrison JC, Johnson EC, Tomarev SI. Microarray analysis of changes in mRNA levels in the rat retina after experimental elevation of intraocular pressure. Invest Ophthalmol Vis Sci. 2004; 45: 1247–1258 [DOI] [PubMed] [Google Scholar]
- 26. Steele MR, Inman DM, Calkins DJ, Horner PJ, Vetter ML. Microarray analysis of retinal gene expression in the DBA/2J model of glaucoma. Invest Ophthalmol Vis Sci. 2006; 47: 977–985 [DOI] [PubMed] [Google Scholar]
- 27. Fan W, Li X, Wang W, Mo JS, Kaplan H, Cooper NG. Early involvement of immune/inflammatory response genes in retinal degeneration in DBA/2J mice. Ophthalmol Eye Dis. 2010; 1: 23–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Howell GR, Macalinao DG, Sousa GL, et al. Molecular clustering identifies complement and endothelin induction as early events in a mouse model of glaucoma. J Clin Invest. 2011; 121: 1429–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bo L, Vedeler CA, Nyland HI, Trapp BD, Mork SJ. Subpial demyelination in the cerebral cortex of multiple sclerosis patients. J Neuropathol Exp Neurol. 2003; 62: 723–732 [DOI] [PubMed] [Google Scholar]
- 30. Anderson MG, Smith RS, Hawes NL, et al. Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice. Nat Genet. 2002; 30: 81–85 [DOI] [PubMed] [Google Scholar]
- 31. Howell GR, Libby RT, Marchant JK, et al. Absence of glaucoma in DBA/2J mice homozygous for wild-type versions of Gpnmb and Tyrp1. BMC Genet. 2007; 8: 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Freeman NE, Templeton JP, Orr WE, Lu L, Williams RW, Geisert EE. Genetic networks in the mouse retina: growth associated protein 43 and phosphatase tensin homolog network. Mol Vis. 2011; 17: 1355–1372 [PMC free article] [PubMed] [Google Scholar]
- 33. Templeton JP, Geisert EE. A practical approach to optic nerve crush in the mouse. Mol Vis. 2012; 18: 2147–2152 [PMC free article] [PubMed] [Google Scholar]
- 34. Templeton JP, Nassr M, Vazquez-Chona F, et al. Differential response of C57BL/6J mouse and DBA/2J mouse to optic nerve crush. BMC Neurosci. 2009; 10: 90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morse HC III, Chused TM, Hartley JW, Mathieson BJ, Sharrow SO, Taylor BA. Expression of xenotropic murine leukemia viruses as cell-surface gp70 in genetic crosses between strains DBA/2 and C57BL/6. J Exp Med. 1979; 149: 1183–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taylor BA, Wnek C, Kotlus BS, Roemer N, MacTaggart T, Phillips SJ. Genotyping new BXD recombinant inbred mouse strains and comparison of BXD and consensus maps. Mamm Genome. 1999; 10: 335–348 [DOI] [PubMed] [Google Scholar]
- 37. Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004; 5: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang B, Khanna H, Hawes N, et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet. 2006; 15: 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Geisert EE, Lu L, Freeman-Anderson NE, et al. Gene expression in the mouse eye: an online resource for genetics using 103 strains of mice. Mol Vis. 2009; 15: 1730–1763 [PMC free article] [PubMed] [Google Scholar]
- 40. Rosen GD, Chesler EJ, Manly KF, Williams RW. An informatics approach to systems neurogenetics. Methods Mol Biol. 2007; 401: 287–303 [DOI] [PubMed] [Google Scholar]
- 41. Boulanger LM, Shatz CJ. Immune signalling in neural development, synaptic plasticity and disease. Nat Rev Neurosci. 2004; 5: 521–531 [DOI] [PubMed] [Google Scholar]
- 42. Godement P, Salaun J, Imbert M. Prenatal and postnatal development of retinogeniculate and retinocollicular projections in the mouse. J Comp Neurol. 1984; 230: 552–575 [DOI] [PubMed] [Google Scholar]
- 43. Walker DG, Kim SU, McGeer PL. Complement and cytokine gene expression in cultured microglial derived from postmortem human brains. J Neurosci Res. 1995; 40: 478–493 [DOI] [PubMed] [Google Scholar]
- 44. Trouw LA, Blom AM, Gasque P. Role of complement and complement regulators in the removal of apoptotic cells. Mol Immunol. 2008; 45: 1199–1207 [DOI] [PubMed] [Google Scholar]
- 45. Yates JR, Sepp T, Matharu BK, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007; 357: 553–561 [DOI] [PubMed] [Google Scholar]
- 46. Johnson PT, Betts KE, Radeke MJ, Hageman GS, Anderson DH, Johnson LV. Individuals homozygous for the age-related macular degeneration risk-conferring variant of complement factor H have elevated levels of CRP in the choroid. Proc Natl Acad Sci U S A. 2006; 103: 17456–17461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alexander JJ, Anderson AJ, Barnum SR, Stevens B, Tenner AJ. The complement cascade: Yin-Yang in neuroinflammation—neuro-protection and -degeneration. J Neurochem. 2008; 107: 1169–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Charbel Issa P, Chong NV, Scholl HP The significance of the complement system for the pathogenesis of age-related macular degeneration—current evidence and translation into clinical application. Graefes Arch Clin Exp Ophthalmol. 2011; 249: 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yamada T, McGeer PL, McGeer EG. Lewy bodies in Parkinson's disease are recognized by antibodies to complement proteins. Acta Neuropathol. 1992; 84: 100–104 [DOI] [PubMed] [Google Scholar]
- 50. Weerth SH, Rus H, Shin ML, Raine CS. Complement C5 in experimental autoimmune encephalomyelitis (EAE) facilitates remyelination and prevents gliosis. Am J Pathol. 2003; 163: 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.