Abstract
Background
Coronary artery disease (CAD) is the major cause of death in patients with type 2 diabetes (DM). While demographic and clinical factors associated with extent of CAD in patients with DM have been described, genetic factors have not. We hypothesized that genetic variation in peroxisome proliferator-activated receptor (PPAR)-pathway genes, important in DM and atherosclerosis, would be associated with extent of CAD in patients with DM.
Methods and Results
1,043 patients (702 Caucasian; 175 African-Americans) from the Bypass Angioplasty Revascularization Investigation 2 Diabetes (BARI 2D) genetic cohort were genotyped for 3,351 variants in 223 PPAR-pathway genes using a custom targeted-genotyping array. Angiographic endpoints were determined by a core laboratory. In Caucasians, a single variant (rs1503298) in TLL1 was significantly (p = 5.5 × 10−6) associated with extent of CAD, defined as number of lesions with % diameter stenosis (DS) ≥ 20%, after stringent Bonferroni correction for all 3,351 SNPs. This association was validated in the diabetic subgroups of two independent cohorts, the TRIUMPH post-myocardial infarction registry and the prospective Family Heart Study of individuals at risk for CAD. rs1503298 was also significantly associated with extent of severe CAD (≥ 70% DS; p = 3.7 × 10−2) and myocardial jeopardy index (p = 8.7 × 10−4). In general linear regression modeling, rs1503298 explained more variance of extent of CAD than the previously determined clinical factors.
Conclusions
We identified a variant in one PPAR-pathway gene, TLL1, that is associated with the extent of CAD, independent of clinical predictors, specifically in patients with type 2 DM and CAD.
Keywords: coronary artery disease, coronary calcium, diabetes mellitus type 2, genetic variation, PPAR, TLL1
Diabetes mellitus (DM) currently affects more than 17 million people in the U.S. and approximately 1.6 million new cases are diagnosed each year.1 Among persons with DM, coronary atherosclerosis is highly prevalent and accounts for the majority of deaths.1 In patients with coronary artery disease (CAD), patients with DM have lower 10-year survival than those without diabetes,2 and the extent of CAD predicts mortality.3 Atherosclerosis in patients with DM has an accelerated phenotype, with more diffuse and extensive disease that shows more rapid progression, suggesting a distinctive pathogenesis.4 The distinct pathogenesis of the accelerated atherosclerosis observed among patients with DM is poorly understood,5 and the role of genetic factors is unknown.
Bypass Angioplasty Revascularization Investigation 2 Diabetes (BARI 2D) was a multicenter randomized clinical trial that investigated the effect of different approaches to the treatment of both CAD and DM on long term outcomes in patients with type 2 DM.6, 7 All patients in BARI 2D had DM and CAD suitable for, but not requiring, revascularization. Baseline extent of CAD in BARI 2D was defined angiographically and quantified by a core laboratory. Clinical factors contributing to extent of CAD among patients in BARI 2D, defined as the number of coronary lesions ≥ 20% diameter stenosis (DS), were recently reported,8 however, the total variance in extent of CAD explained by baseline clinical factors was less than 10%,8 providing an important rationale for examining the putative role of genetic factors.
Peroxisome-proliferator activated receptor (PPAR)-pathway genes are involved in cellular processes relevant to both CAD and DM. PPARs are master regulators of lipid and glucose homeostasis, cardiac energy metabolism, vascular inflammation and cell differentiation and have been implicated in the development and progression of both type 2 DM and atherosclerosis in animal studies.9-12 We therefore hypothesized that investigating genetic variation within the PPAR gene pathway would identify novel genes involved in diabetic atherosclerosis.
Using a custom PPAR-pathway gene SNP microarray containing 3,351 single nucleotide polymorphisms (SNPs) in 223 PPAR-pathway genes, we investigated genetic associations of PPAR-pathway genes with extent of CAD among patients with DM in the BARI 2D trial. We then sought to validate the discovered associations in two additional well-phenotyped cohorts of patients with CAD and DM.
Research Design and Methods
BARI 2D Cohort and Baseline Phenotypic Data
Eligibility criteria for BARI 2D have been previously described. 6, 7 Patients with type 2 DM having angiographically-documented CAD involving at least one major epicardial coronary artery (≥ 50% DS with a positive stress test or ≥ 70% DS with classic angina) suitable for treatment with either medical therapy or coronary revascularization, were eligible for inclusion.6, 7 Exclusion criteria included significant left main disease (≥ 50% DS), class III or IV congestive heart failure, and coronary revascularization within the preceding 12 months. All patients underwent coronary angiography and the angiograms were reviewed by a central core laboratory (Stanford University) with a standard protocol used in previous trials and masked to patient randomization or intended revascularization strategy.7, 8 This assessment included quantifying the number and % DS of each lesion, its location in the coronary tree using the BARI modification of the Coronary Artery Surgery Study (CASS) map, and the lesion morphology according to the American College of Cardiology/American Heart Association (ACC/AHA) classification scheme.8 Percent DS was the ratio of the minimum diameter divided by the reference diameter (the mean of the proximal and distal diameter of the normal appearing artery adjacent to the lesion). All lesions ≥ 20% DS were counted. In addition to number of lesions, total myocardial jeopardy index was calculated. The myocardial jeopardy index was calculated as the percentage of the myocardium that was jeopardized by lesions ≥ 50% DS in any of the three main coronary arteries or their branches.8, 13 In accordance with the published analysis of clinical predictors of extent of CAD in BARI 2D,8 number of coronary lesions ≥ 20% DS was selected to represent overall extent of atherosclerotic CAD; additional angiographic variables were included to represent extent of severe atherosclerotic CAD (number of lesions ≥ 70% DS and myocardial jeopardy index).
BARI 2D Genetic Cohort
A total of 2,368 patients were enrolled in BARI 2D from January 2001 through March 2005. 1,439 BARI 2D patients consented to genetic analysis. 1,072 genomic samples (702 Caucasian; 175 African American) were available for genotyping (353 samples from Brazil were not available due to international restrictions on DNA shipment). To determine if the genetic cohort was representative of the entire BARI 2D cohort, clinical covariates and association with number of lesions ≥ 20% DS were determined for the entire genetic cohort (not stratified by race). Using the same clinical covariates and the same analytic strategy (variable entry and retention criteria; entry p<0.15, retain p < 0.05; age, and sex included irrespective of statistical significance) as was used in the entire cohort,8 the magnitude of individual variables associated with extent of CAD (presented as semi-partial correlation coefficients) were determined.
Validation Cohorts
TRIUMPH
Translational Research Investigating Underlying disparities in acute Myocardial infarction Patients' Health status (TRIUMPH) is a large, prospective, observational cohort study of consecutive patients with acute myocardial infarction presenting to 24 US hospitals from 4/2005 – 12/2008.14 Myocardial infarction was diagnosed using contemporary definitions15 and all patients had an elevated troponin blood test. 31% of TRIUMPH patients had DM on enrollment. Extent of coronary artery disease was determined by angiography and defined as the number of vessels with greater than or equal to 20% diameter stenosis. 386 Caucasians were genotyped using the PPAR-pathway custom array constituting the genetic cohort for the present analysis. The study was approved by the Institutional Review Boards of all participating institutions and written informed consent was provided by each participant. A separate consent form for the acquisition of blood for genetic analysis was signed by each participant.
Family Heart Study
The Family Heart Study (FHS) has been described in detail.16, 17 Briefly, adults (ages 45-69) were recruited from one of three studies, the Framingham study, the Utah study and two ARIC sites. Subjects, and their family members, were either 1) randomly selected or 2) selected because they had a high relative risk of coronary heart disease.17 Of these, 14% had DM. The cardiac CT subset of FHS (Family Heart Study – SubClinical Atherosclerosis Network; FHS-SCAN) recruited patients and their family members previously enrolled in FHS who had been genotyped by GWAS to undergo cardiac CT to determine extent of coronary artery and aortic calcification16, 17 Coronary artery calcification was determined by a core laboratory (Wake Forest University Health Sciences, Winston Salem, N.C.) as previously described.16, 17 In brief, two sequential scans were performed 1 minute apart and the amount of calcium was quantified by the Agatston scoring method adjusted for slice thickness, and the total coronary artery calcification score was calculated as the sum of the Agatston scores from each scan and then averaged. 974 Caucasians were genotyped with the Illumina 550K SNP array.
Genotyping and Custom Array
DNA was isolated and extracted using the Puregene genomic DNA purification kit (Gentra, Minneapolis, MN). Four micrograms of DNA were genotyped using a custom designed PPAR-pathway targeted-genotyping 3K Affymetrix array containing 3,351 single nucleotide polymorphism (SNPs) in 223 genes within the PPAR-pathway as well as 100 SNPs allowing identification of ethnic origin by mapping by admixture linkage disequilibrium (MALD) (Supplemental Table S1).18
Method of Selection of Genes on Custom Array
The PPAR-pathway genes were selected if they met one of two criteria: (1) identified as a PPAR-pathway gene in the published literature, or (2) showing greater than 2-fold up- or down-regulation in in vitro gene expression profiling experiments performed with mouse hearts overexpressing PPARα as compared to non-transgene littermates (data graciously provided by Dr. Daniel P. Kelly, Scientific Director, Burnham Institute for Medical Research, Orlando, Florida).
Method of Selection of SNPs on Custom Array
SNPs were chosen from HapMap data. The projected populations selected for tagging were Africans and Caucasians with a bias towards Caucasian coverage. A tagging SNP approach (from 5kb upstream of the gene to 5kb downstream of the gene; minor allele frequency of 10% or greater in Caucasian and African American populations; method of Carlson 19 used to select tagging SNPs; r2 > 0.8 required) was used. If a tagging SNP was in a large “bin” (containing 5 or more SNPs with r2 > 0.8), we included 2 tagging SNPs on the array to ensure maximal data acquisition (while just genotyping one tagging SNP is highly efficient, if it fails to genotype, a large amount of data could potentially be lost). In addition to the tagging SNPs the array also includes all non-synonymous coding SNPs in each gene (regardless of minor allele frequency). The SNP coverage provided by the final panel was 8.5K/∼10K Caucasian and 7K/∼11K Yoruban.
Genotyping QC Data
Data completeness was 99.64% and repeatability was 100%. To ensure high quality data and to minimize false positive results, filtering criteria were set as follows: genotype call rate ≥ 0.80 per SNP, minor allele frequency ≥ 0.01, and P value for Hardy-Weinberg equilibrium test ≥ 0.0001. Using the PLINK analysis program (v1.06), five Caucasian subjects were identified whose reported sex did not match estimated sex based on X chromosome data. These five samples were excluded from further analysis. Cluster analysis implemented in PLINK was used for screening any evidence of subtle population stratifications based on pairwise identity-by-state (IBS) sharing distance among all possible pairs of the SNPs. Across the samples of the BARI2D genetic cohort, four well-separated subgroups correlated with self-reported ethnic groups, and the two major groups of African Americans and Caucasians were clearly distinguished. In African Americans, no further population stratification was detected according to multidimensional scaling plots (Supplement; Figure S1). In Caucasians, subtle clustering was noted; we therefore estimated 10 principal components using Eigenstrat.
Statistical Methods
Clinical Covariates
The primary endpoint for our analysis was number of lesions ≥ 20% DS documented on the coronary angiogram. Myocardial jeopardy index and number of lesions ≥ 70% DS were also determined for variant(s) achieving significance in replication cohorts. Number of lesions and myocardial jeopardy index were evaluated as continuous variables with kernel density estimators used to plot their overall distributions. Continuous variables were compared by student t tests or Wilcox tests (depending on distributional properties); categorical variables were compared by chi-square tests. Stepwise linear regression was used to identify baseline factors associated with number of lesions (log transformed) and myocardial jeopardy index.
SNP association
To prevent confounding from racial admixture, we performed our analyses separately in our two largest racial groups (Caucasians, n=702; African-Americans, n=175). Single SNP associations for the number of lesions ≥ 20% DS were tested in general linear regression analyses using an additive genetic model. Age, sex, BMI and prior revascularization were included in the model as covariates. Significance level was 1.5 × 10-5 using stringent Bonferroni correction for all 3,351 SNPs.
Validation
We validated putative statistically significant associations in two separate cohorts of patients systematically assessed for extent of CAD described above: 1) TRIUMPH and 2) FHS. Since the significant association was observed in Caucasian BARI 2D subjects, Caucasian TRIUMPH and FHS subjects were used for validation. None of the subjects were related in TRIUMPH. Since FHS had related subjects, we used mixed models to take family structure into account. P <0.05 was considered statistically significant for validation analyses. Statistical analyses were performed using SAS 9.1 (SAS Institute Inc, Cary, North Carolina) and plots were generated by R 2.3.1 (CRAN, the comprehensive R archive network).
Results
The BARI 2D Genetics Cohort
The baseline characteristics of the patients comprising the genetic cohort of BARI 2D were similar to the entire group (Table 1). The primary endpoint representing extent of CAD (number of coronary lesions ≥ 20% DS) was the same in the BARI 2D genetic cohort (mean number of coronary lesions ≥ 20% DS = 4.6 ± 2.2) as in the entire BARI 2D cohort (mean number of coronary lesions ≥ 20% DS = 4.6 ± 2.3). To further confirm that the extent of CAD in the genetics subgroup was representative of the entire cohort, the magnitude of association between clinical covariates and number of coronary lesions ≥ 20% DS was compared between the two groups. As shown in Figure 1, individual clinical covariates in the BARI 2D genetic cohort showed similar correlations with the extent of CAD as the entire BARI 2D cohort. Similar to the entire BARI 2D cohort, female sex, black race, higher body mass index and higher thirty second pulse were associated with less extensive CAD whereas older age, longer duration of DM, hypertension and higher tissue-type plasminogen activator antigen levels were associated with more extensive CAD.
Table 1. Baseline characteristics of the entire BARI 2D cohort, the BARI 2D genetic cohort, and the Caucasian subgroups of BARI 2D, TRIUMPH and FHS.
| All patients (N = 2368) | Genetics Cohort (N = 1072) | Genetics Cohort Caucasian (N = 702) | FHS DM Cauc. (N = 139) | Triumph DM Cauc. (N = 100) | |
|---|---|---|---|---|---|
| Age, mean, SD | 62.4, 8.9 | 63.1, 9.2 | 64.3, 8.8 | 63.6, 11.3 | 58.8, 11.5 |
| Male, % | 70.4 | 69.4 | 75.2 | 42.4 | 67.0 |
| Race, % | |||||
| White | 70.4 | 66.6 | 100.00 | 100.00 | 100.00 |
| Black | 17 | 17.1 | 0 | 0 | 0 |
| Asian | 4.2 | 5.5 | 0 | 0 | 0 |
| Indian/Native American | 4.3 | 0.1 | 0 | 0 | 0 |
| Other | 4.1 | 5.9 | 0 | 0 | 0 |
| Hispanic, ethnicity | 12.5 | 14.0 | 5.1 | ||
| Region of World, % | |||||
| USA | 63.3 | 76.1 | 75.5 | 100 | 100 |
| Canada | 14.9 | 15.3 | 17.2 | 0 | 0 |
| Mexico | 3.6 | 4.0 | 0 | 0 | 0 |
| Czech | 3.2 | 4.6 | 7.3 | 0 | 0 |
| BMI categories (kg/m2), % | |||||
| Normal or underweight, <25 | 9.7 | 7.9 | 5.1 | 23.0 | 11.1 |
| Overweight, 25 to <30 | 34 | 30.9 | 28.8 | 41.7 | 26.3 |
| Class 1 obese, 30 to <35 | 32.1 | 31.8 | 35.3 | 24.5 | 35.4 |
| Class 2 obese, 35 to <40 | 15.3 | 17.1 | 19.4 | 7.9 | 11.1 |
| Class 3 obese, ≥40 | 9 | 12.2 | 11.6 | 2.9 | 16.2 |
| Blood pressure >130/80 mmHg,% | 52.4 | 48.8 | 45.1 | 47.5 | 79.0 |
| Hypertension, % | 82.5 | 81.9 | 81.6 | 53.2 | 77.0 |
| Myocardial infarction, % | 32 | 29.5 | 30.0 | 12.6 | 24.0 |
| Congestive heart failure, % | 6.6 | 8.4 | 8.9 | 0.7 | 13.0 |
| Anginal status, % | |||||
| None | 17.9 | 21.1 | 22.7 | 87.8 | 27.0 |
| Anginal equivalents only | 21.4 | 25.2 | 26.4 | ||
| Stable CCS1 | 14.3 | 14.4 | 14.7 | ||
| Stable CCS2 | 28.8 | 24.5 | 23.5 | ||
| Stable CCS3 | 7.5 | 5.2 | 5.3 | ||
| Stable CCS4 | 1.2 | 0.7 | 0.7 | ||
| Unstable angina | 9.5 | 8.9 | 6.8 | ||
| Duration of DM categories, % | |||||
| <5 yrs | 33.3 | 32.1 | 32.18 | ||
| 5 - <10 yrs | 23.5 | 23.1 | 22.99 | ||
| 10 - <20 yrs | 29.2 | 30.4 | 30.17 | ||
| ≥ 20 yrs | 14.1 | 14.3 | 14.66 | ||
| History of insulin use, % | 29.3 | 29.0 | 27.4 | ||
| Glycemia measurements, % | |||||
| HbA1c ≤ 7.0% | 41.7 | 45.5 | 48.0 | 47.4 | |
| 7.0% < HbA1c ≤ 8.0% | 25.3 | 27.9 | 23.6 | 12.6 | |
| HbA1c >8.0% | 33 | 26.6 | 28.5 | 40.0 | |
| Total cholesterol ≥200 mg/dL, % | 19.0 | 18.5 | 16.6 | 29.6 | 34.0 |
| Triglycerides ≥200 mg/dL, % | 31.0 | 32.3 | 34.8 | 27.4 | 41.0 |
| HDL <40 male <50 female mg/dL, % | 72.4 | 74.4 | 78.8 | 44.4 | 68.0 |
| LDL ≥100 mg/dL, % | 40.5 | 40.4 | 36.0 | 55.0 | 33.0 |
Figure 1.
Comparison of clinical covariates associated with number of lesions ≥ 20% in the BARI 2D genetic cohort (right) compared with the entire BARI 2D cohort (left).
PPAR-pathway SNP Associations with Extent of CAD
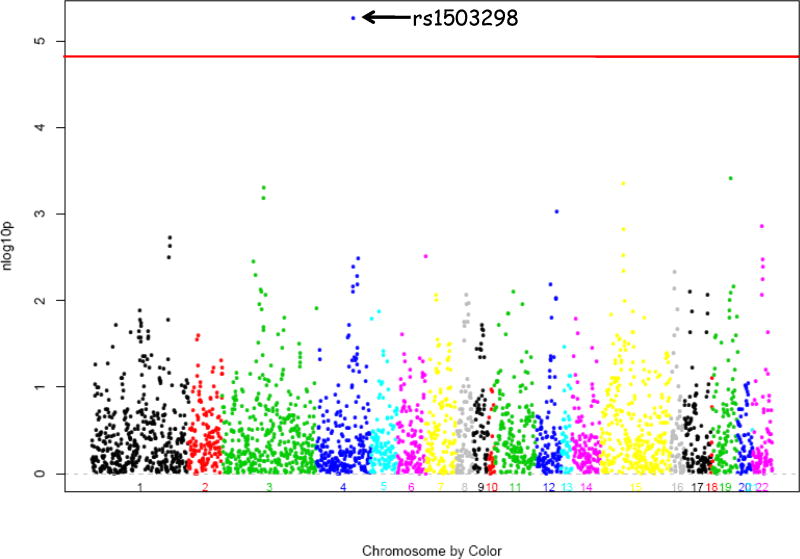
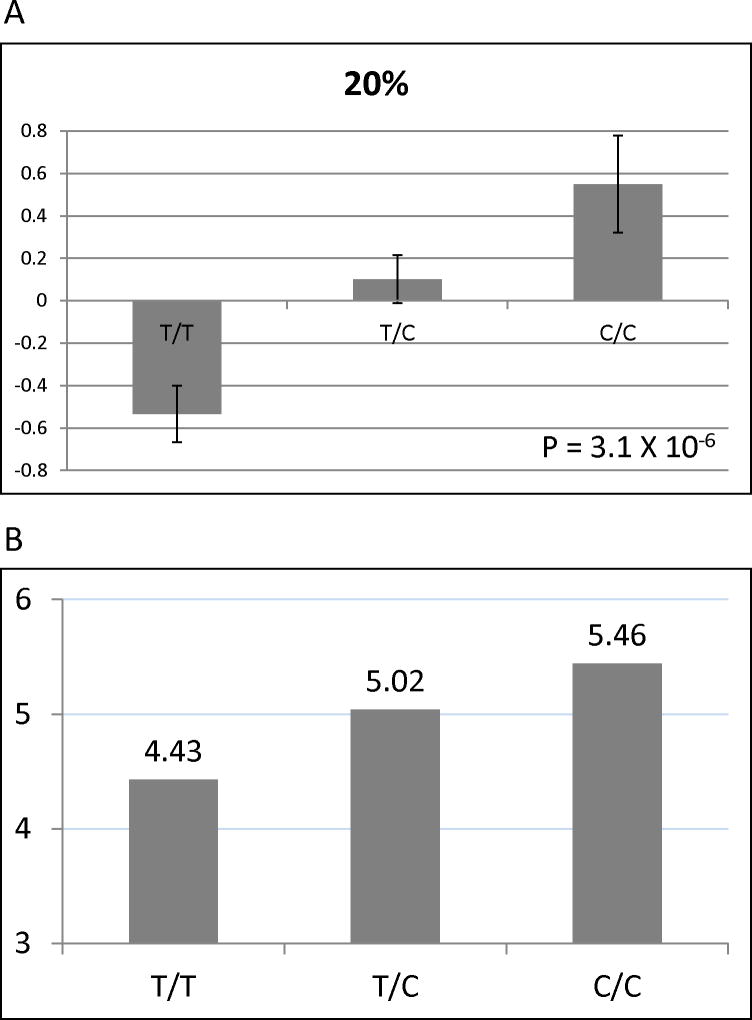
To prevent confounding from racial admixture, we performed our analyses separately in our two largest racial groups (Caucasians, n=702; African Americans, n=175). In Caucasian BARI 2D subjects, single SNP analysis identified one SNP (rs1503298) that was highly significantly associated (P = 5.5 × 10 −6) with number of coronary lesions ≥ 20% DS (Figure 2), even after stringent Bonferroni correction for all 3,351 SNPs. This SNP is located in intron 12 of the gene that encodes Tolloid-like 1 (TLL1), a metalloproteinase that regulates bone morphogenetic protein-2 (BMP-2) and transforming growth factor β. This association remained significant after including all baseline clinical covariates and principal components in the model. The mean (±SE) residual of number of lesions ≥ 20% by TLL1 rs1503298 genotype after adjustment for age, sex and BMI in Caucasian BARI 2D subjects is shown in Figure 3A. To put this data into a clinically relevant context, an average BARI 2D subject (a 63 year old male with a BMI of 30) with the TT genotype would have 4.43 coronary lesions ≥ 20% DS, with the CT genotype would have 5.02 coronary lesions ≥ 20% DS, and with the CC genotype would have 5.46 coronary lesions ≥ 20% DS (Figure 3B). Of note, in general linear regression modeling, this SNP explained more variance of the phenotype (number of coronary lesions ≥ 20% DS) than the previously determined clinical factors. TLL1 rs1503298 explained 2.75% of the variance as compared to sex (1.61%), age (0.12%), and BMI (0.01%). Subjects with TLL1 rs1503298 CT genotype had 22% more coronary lesions ≥ 20% DS as compared with those with TT genotype and those with the CC genotype had 37% more coronary lesions ≥ 20% DS.
Figure 2.
Results of tests for a trend in the association between number of lesions ≥ 20% and each SNP on custom chip in Caucasians in BARI 2D. Red line depicts threshold for statistical significance after Bonferroni correction for multiple comparisons.
Figure 3.
(A) Mean (±SE) residual of number of lesions ≥ 20% by TLL1 rs1503298 genotype after adjustment for age, sex and BMI in Caucasian BARI 2D subjects. (B) Number of lesions ≥ 20% by genotype for the average BARI 2D subject (a 63 year old male with BMI of 30).
In the smaller subgroup of African American BARI 2D subjects (n=175), no SNP was found to be significantly associated with number of coronary lesions ≥ 20% DS after stringent Bonferroni correction for all 3,351 SNPs. This included TLL1 rs1503298 (p=0.25).
Validation
For validation, the genetic association observed in BARI 2D was tested in both the overall group and the diabetic subgroups of two independent cohorts of patients with CAD, TRIUMPH and FHS. These validation cohorts were chosen because both TRIUMPH and FHS were independent populations with detailed information on extent of coronary artery disease by quantitative imaging studies, both had detailed information on subjects with and without carefully defined DM, both had extensive phenotypic data to allow consistent adjustment for covariates, and both were genotyped for relevant polymorphisms. Validation analyses were performed on (Caucasian) subjects with and without DM within these same cohorts in order to obtain insight as to whether the association was specific to individuals with DM.
Applying the same model in TRIUMPH with the identical SNPs used in BARI 2D, a significant association with extent of CAD was selectively demonstrated for TLL1 rs1503298, among patients with DM (n=98; P = 3.3 × 10−2), but not in the overall cohort (n=386; P = 1.8 × 10−1). A congruent trend was found in FHS using a less sensitive measure of CAD extent. The association between rs1503298 and extent of CAD defined by coronary calcium score was demonstrated selectively in FHS individuals with DM (n=139; P = 8.5 × 10−2) but did not replicate in the larger FHS cohort (n=974; P = 5.7 × 10−1).
Associations with Severe CAD
We also investigated whether the rs1503298 variant was associated with the extent of severe CAD and myocardial jeopardy index in BARI 2D, variables previously associated with mortality in other studies.3, 13 TLL1 rs1503298 was significantly associated with both number of coronary lesions ≥ 70% DS (P = 3.7 × 10−2) and myocardial jeopardy index (P = 8.7 × 10−4; Figure 4).
Discussion
Using a gene-pathway microarray approach, we have identified a novel PPAR-pathway genetic polymorphism that is significantly associated with extent of CAD in patients with DM. The TLL1 rs1503298 variant, located in intron 12 of TLL1, showed associations with rigorous and objectively derived measures of atherosclerotic coronary disease in BARI 2D and explained more variance of the phenotype than the previously determined clinical factors. The association of extent of CAD observed with TLL1 rs1503298 in BARI 2D was validated by replication in the diabetic patients in two independent populations, TRIUMPH and FHS.
The association of TLL1 rs1503298 with extent of CAD was observed among Caucasian patients with type 2 DM in the BARI 2D trial, where clinically stable CAD was defined and quantified by core laboratory interpretation of coronary angiograms. This association was validated by replication in the Caucasian diabetic subgroups of two independent populations with CAD, the TRIUMPH cohort, where CAD in patients with acute myocardial infarction was defined angiographically, and the FHS cohort, where individuals at high risk of CAD had their extent of coronary artery calcification determined by CT scanning. The observation that this variant was reproducibly associated with extent of disease quantified by disparate methods among individuals with disparate clinical manifestations supports a potential role for this genetic polymorphism in the pathogenesis of diabetic atherosclerosis. Further reinforcing the clinical relevance of the association between this SNP and CAD in patients with DM, the TLL1 rs1503298 polymorphism showed strong associations not only with the primary endpoint of number of lesions ≥ 20% DS, but also with number of angiographically severe (≥70% DS) lesions, and with myocardial jeopardy index, a semi-quantitative method used to estimate the amount of potential myocardial ischemia attributable to the location and severity of coronary lesions in an individual; these measures of extent of severe CAD have previously been shown to correlate with prognosis in patients with CAD.3, 13 To our knowledge, this is the first demonstration of a significant association of genetic variation specifically with extent of the atherosclerotic disease that develops among patients with DM, where atherosclerosis has been known to display a distinctively aggressive phenotype.20-22
The observation of an association between extent of CAD in patients with DM and this particular gene in the PPAR-pathway regulated processes is noteworthy. TLL1 encodes a protein, Tolloid-like1, that has been identified within the cascade of cellular processes related to vascular inflammation and calcification. Since vascular calcification is a prominent feature of the phenotype of diabetic atherosclerosis,23 it is intriguing to speculate that genetic variability in TLL1 is may have strong biologic plausibility as a contributor to its pathogenesis.23 Tolloid-like1 (TLL1) is a metalloproteinase that cleaves the BMP-2 antagonists Chordin and gremlin, thereby upregulating BMP-2/4 pathway genes that promote inflammation and extracellular matrix neogenesis.24, 25 BMP-2 itself is an important mediator of tissue calcification, and endothelial BMP-2 expression can be up-regulated by vascular oxidative stress, intravascular pressure, and elevated glucose.26-29 BMP-2 appears to play a critical role in programming the phenotype of the calcifying vascular cell, the mural cell type that directs osteogenic mineralization in the vessel wall.24, 25, 27, 30-32 Moreover, TLL1 is also a member of the BMP-1-related activators of the procollagen C-proteinase enhancer.33, 34 Because TLL1 is at the intersection of the BMP-2/4 and frizzled-related protein-2 signals that drive collagen deposition, TLL1 is also likely to be a critical regulator of collagenous matrix deposition – a hallmark of robust calcification processes in cardiovascular tissues as well as in bone.33, 34 BMP-2 is upregulated in human atherosclerotic plaques,26, 35 and has been identified as a key multifunctional regulator of vascular calcium accrual in atherosclerosis and diabetic arteriosclerosis.36, 37 Thus, TLL1 has a direct link to fundamental cellular processes involved in vascular calcification, a widely recognized feature of atherosclerosis that is characteristically increased in patients with DM compared to those without DM.4, 20-22
Previous genetic association studies have observed associations between polymorphisms of PPAR-pathway genes and the risk of developing type 2 DM, obesity, insulin resistance, abnormal lipid profiles, and cardiovascular disease in European and American populations (Reviewed in 9, 38). In separate candidate gene studies, a SNP in intron 7 of the PPARα gene (PPARA IVS7 2498)39 and two SNPs in PPARG (the gene encoding PPARγ) were associated with increased risk of myocardial infarction in patients with type 2 DM.40 Also using a candidate gene approach, Regieli JJ et al.41 reported an association between the Pro12Ala variant (rs1801282) of PPARG and both angiographic extent of CAD and 10-year risk of ischemic events including death among 679 participants in the Regression Growth Evaluation Statin Study (REGRESS) without differentiation of result by diabetic status. This association was not confirmed in other studies.42 Of note, in the current analysis, the IVS7 2498 SNP in PPARA and thePro12Ala polymorphism in PPARG (both present on the PPAR-pathway array utilized) were not identified as variants with a significant association with extent of CAD in the BARI 2D cohort.
Our findings should be considered in the context of several potential limitations. First, to prevent confounding from racial admixture, we performed analyses in the largest racial groups (Caucasians and African American) separately. The association between TLL1 and extent of CAD was observed among Caucasian patients with DM. Similar analyses did not confirm an association between TLL1 and extent of CAD among African-American participants in BARI 2D, although the smaller sample size may have limited the power to detect a significant relationship if present, or there may be different patterns of linkage disequilibrium between TLL1 rs1503298 and the causal variant in different ethnic groups. Our findings can therefore not be extrapolated to other racial groups and should be examined independently in adequately powered, racially characterized cohorts, particularly given the burden of DM and CAD in African-American and Hispanic patients. In addition, this study used the genetic cohort of BARI 2D and, although the genetic cohort had similar baseline characteristics as the overall BARI 2D cohort, selection bias is possible. However, since patients (and their treating physicians) were unaware of their genetic code, it is unlikely that there was a direct association between genotype and participation in the genetic cohort of BARI 2D to make selection bias significantly problematic. Finally, it should be noted that diabetic patients with the most extensive, or unstable, CAD for whom immediate revascularization was considered clinically indicated, by design, were excluded from BARI 2D. The observed association, therefore, has not been tested in this potentially important subgroup of patients with DM and CAD. Furthermore, it should be noted that the mechanisms of action of TLL1 could potentially be via a pathway other than PPAR. Without further confirmation, caution is warranted in attributing the observed association to a direct or indirect effect of PPAR activation.
Conclusions
Using a gene pathway microarray approach, we identified, and validated, a novel PPAR pathway gene variant, TLL1 rs1503298, that is significantly associated with extent of CAD specifically in Caucasian patients with DM. The reproducibility of the observed association across populations, despite varied clinical manifestations of CAD and varied methods of quantifying the extent of atherosclerosis, underscore its potential importance and relevance to the fundamental pathophysiology of diabetic atherosclerosis. Moreover, the physiologic pathways affected by this genetic variant may provide novel targets for further investigation and therapeutic intervention to address the accelerated rate of progression and the high risk of adverse events associated with CAD in patients with DM.
Supplementary Material
Figure 4.
Mean (±SE) residual of number of lesions ≥ 70% (A) and myocardial jeopardy index (B) by TLL1 rs1503298 genotype after adjustment for age, sex and BMI in Caucasian BARI 2D subjects.
In patients with diabetes, atherosclerosis is more extensive and rapidly progressive. The reasons for this are not known, but may relate in part to genetic variation that influences vascular disease severity. Using microarray technology to interrogate over 3,000 tagged single nucleotide polymorphism markers of PPAR-pathway genes, we have discovered a polymorphism in the TLL1 genexs that is significantly associated with angiographic extent of coronary artery disease among patients with type 2 diabetes. This genetic polymorphism explains more variance of the phenotype than previously determined clinical factors. This association was initially observed in Caucasian patients with type 2 diabetes in the BARI 2D trial and then validated specifically in the diabetic subgroups of two other independent Caucasian patient cohorts with quantitatively coronary imaging. TLL1 encodes a metalloproteinase that regulates procollagen processing, and the metabolism of inhibitors of bone morphogenetic proteins (BMPs) involved with atherosclerotic calcification. These observations identify a novel putative genetic contributor to the biology of diabetic coronary atherosclerosis, and suggest that targeting the TLL1/BMP pathway may afford specific therapeutic intervention for diabetic vascular disease.
Acknowledgments
The authors would like to thank Dr. Dwight Towler for critical review of this manuscript.
Funding Sources: “The Bypass Angioplasty Revascularization Investigation 2 Diabetes (BARI 2D) is funded by the National Heart, Lung and Blood Institute and the National Institute of Diabetes and Digestive and Kidney Diseases, Nos. U01 HL061744, U01 HL061746, U01 HL061748, and U01 HL063804. Significant supplemental funding is provided by GlaxoSmithKline, Collegeville, PA, Bristol-Myers Squibb Medical Imaging, Inc., North Billerica, MA, Astellas Pharma US, Inc., Deerfield, IL, Merck & Co., Inc., Whitehouse Station, NJ, Abbott Laboratories, Inc., Abbott Park, IL, and Pfizer, Inc, New York, NY. Generous support is given by Abbott Laboratories Ltd., MediSense Products, Mississauga, Canada, Bayer Diagnostics, Tarrytown, NY, Becton, Dickinson and Company, Franklin Lakes, NJ, J. R. Carlson Labs, Arlington Hts., IL, Centocor, Inc., Malvern, PA, Eli Lilly and Company, Indianapolis, IN, LipoScience, Inc., Raleigh, NC, Merck Sante, Lyon, France, Novartis Pharmaceuticals Corporation, East Hanover, NJ, and Novo Nordisk, Inc. Princeton, NJ.
“Please note that as an NIH funded trial, we are required to abide by the NIH PubMed Central Policy that we retain the right to provide a copy of the final manuscript to the NIH upon acceptance for publication by your journal, for public archiving in PubMed Central as soon as possible, but no later than 12 months after publication.”
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, And Blood Institute, the National Institute of Diabetes And Digestive And Kidney Diseases, or the National Institutes of Health.”
SC is funded by NIH R21HL089681, NIH Specialized Center for Clinically-Oriented Research (SCCOR) in Cardiac Dysfunction and Disease P50 HL077113, P60 DK20579, and The Longer Life Foundation.
Footnotes
Disclosures: No conflicts to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, Dai S, De SG, Ferguson TB, Ford E, Furie K, Gillespie C, Go A, Greenlund K, Haase N, Hailpern S, Ho PM, Howard V, Kissela B, Kittner S, Lackland D, Lisabeth L, Marelli A, McDermott MM, Meigs J, Mozaffarian D, Mussolino M, Nichol G, Roger VL, Rosamond W, Sacco R, Sorlie P, Roger VL, Thom T, Wasserthiel-Smoller S, Wong ND, Wylie-Rosett J. Heart disease and stroke statistics--2010 update: a report from the American Heart Association. Circulation. 2010;121:e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- 2.The final 10-year follow-up results from the BARI randomized trial. J Am Coll Cardiol. 2007;49:1600–1606. doi: 10.1016/j.jacc.2006.11.048. [DOI] [PubMed] [Google Scholar]
- 3.Natali A, Vichi S, Landi P, Severi S, L'Abbate A, Ferrannini E. Coronary atherosclerosis in Type II diabetes: angiographic findings and clinical outcome. Diabetologia. 2000;43:632–641. doi: 10.1007/s001250051352. [DOI] [PubMed] [Google Scholar]
- 4.Hayden MR, Tyagi SC, Kolb L, Sowers JR, Khanna R. Vascular ossification-calcification in metabolic syndrome, type 2 diabetes mellitus, chronic kidney disease, and calciphylaxis-calcific uremic arteriolopathy: the emerging role of sodium thiosulfate. Cardiovasc Diabetol. 2005;4:4. doi: 10.1186/1475-2840-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacoby RM, Nesto RW. Acute myocardial infarction in the diabetic patient: pathophysiology, clinical course and prognosis. J Am Coll Cardiol. 1992;20:736–744. doi: 10.1016/0735-1097(92)90033-j. [DOI] [PubMed] [Google Scholar]
- 6.Brooks MM, Frye RL, Genuth S, Detre KM, Nesto R, Sobel BE, Kelsey SF, Orchard TJ. Hypotheses, design, and methods for the Bypass Angioplasty Revascularization Investigation 2 Diabetes (BARI 2D) Trial. Am J Cardiol. 2006;97:9G–19G. doi: 10.1016/j.amjcard.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 7.Frye RL, August P, Brooks MM, Hardison RM, Kelsey SF, MacGregor JM, Orchard TJ, Chaitman BR, Genuth SM, Goldberg SH, Hlatky MA, Jones TL, Molitch ME, Nesto RW, Sako EY, Sobel BE. A randomized trial of therapies for type 2 diabetes and coronary artery disease. N Engl J Med. 2009;360:2503–2515. doi: 10.1056/NEJMoa0805796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz L, Kip KE, Alderman E, Lu J, Bates ER, Srinivas V, Bach RG, Mighton LD, Feit F, King S, III, Frye RL. Baseline coronary angiographic findings in the Bypass Angioplasty Revascularization Investigation 2 Diabetes trial (BARI 2D) Am J Cardiol. 2009;103:632–638. doi: 10.1016/j.amjcard.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 9.Cresci S. The PPAR genes, cardiovascular disease and the emergence of PPAR pharmacogenetics. Expert Opin Pharmacother. 2005;6:2577–2591. doi: 10.1517/14656566.6.15.2577. [DOI] [PubMed] [Google Scholar]
- 10.Libby P, Plutzky J. Inflammation in diabetes mellitus: role of peroxisome proliferator-activated receptor-alpha and peroxisome proliferator-activated receptor-gamma agonists. Am J Cardiol. 2007;99:27B–40B. doi: 10.1016/j.amjcard.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Madrazo JA, Kelly DP. The PPAR trio: regulators of myocardial energy metabolism in health and disease. J Mol Cell Cardiol. 2008;44:968–975. doi: 10.1016/j.yjmcc.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 12.Bouhlel MA, Staels B, Chinetti-Gbaguidi G. Peroxisome proliferator-activated receptors--from active regulators of macrophage biology to pharmacological targets in the treatment of cardiovascular disease. J Intern Med. 2008;263:28–42. doi: 10.1111/j.1365-2796.2007.01892.x. [DOI] [PubMed] [Google Scholar]
- 13.Graham MM, Faris PD, Ghali WA, Galbraith PD, Norris CM, Badry JT, Mitchell LB, Curtis MJ, Knudtson ML. Validation of three myocardial jeopardy scores in a population-based cardiac catheterization cohort. Am Heart J. 2001;142:254–261. doi: 10.1067/mhj.2001.116481. [DOI] [PubMed] [Google Scholar]
- 14.Smolderen KG, Spertus JA, Reid KJ, Buchanan DM, Krumholz HM, Denollet J, Vaccarino V, Chan PS. The association of cognitive and somatic depressive symptoms with depression recognition and outcomes after myocardial infarction. Circ Cardiovasc Qual Outcomes. 2009;2:328–337. doi: 10.1161/CIRCOUTCOMES.109.868588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alpert JS, Thygesen K, Antman E, Bassand JP. Myocardial infarction redefined--a consensus document of The Joint European Society of Cardiology/American College of Cardiology Committee for the redefinition of myocardial infarction. J Am Coll Cardiol. 2000;36:959–969. doi: 10.1016/s0735-1097(00)00804-4. [DOI] [PubMed] [Google Scholar]
- 16.Djousse L, Arnett DK, Carr JJ, Eckfeldt JH, Hopkins PN, Province MA, Ellison RC. Dietary linolenic acid is inversely associated with calcified atherosclerotic plaque in the coronary arteries: the National Heart, Lung, and Blood Institute Family Heart Study. Circulation. 2005;111:2921–2926. doi: 10.1161/CIRCULATIONAHA.104.489534. [DOI] [PubMed] [Google Scholar]
- 17.Higgins M, Province M, Heiss G, Eckfeldt J, Ellison RC, Folsom AR, Rao DC, Sprafka JM, Williams R. NHLBI Family Heart Study: objectives and design. Am J Epidemiol. 1996;143:1219–1228. doi: 10.1093/oxfordjournals.aje.a008709. [DOI] [PubMed] [Google Scholar]
- 18.Collins-Schramm HE, Phillips CM, Operario DJ, Lee JS, Weber JL, Hanson RL, Knowler WC, Cooper R, Li H, Seldin MF. Ethnic-difference markers for use in mapping by admixture linkage disequilibrium. Am J Hum Genet. 2002;70:737–750. doi: 10.1086/339368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter A, Murphy MO, Turner NJ, Halka AT, Ghosh J, Serracino-Inglott F, Walker MG, Syed F. Intimal neovascularisation is a prominent feature of atherosclerotic plaques in diabetic patients with critical limb ischaemia. Eur J Vasc Endovasc Surg. 2007;33:319–324. doi: 10.1016/j.ejvs.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 21.Hayden MR, Tyagi SC. Vasa vasorum in plaque angiogenesis, metabolic syndrome, type 2 diabetes mellitus, and atheroscleropathy: a malignant transformation. Cardiovasc Diabetol. 2004;3:1. doi: 10.1186/1475-2840-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayden MR, Tyagi SC. Intimal redox stress: accelerated atherosclerosis in metabolic syndrome and type 2 diabetes mellitus. Atheroscleropathy. Cardiovasc Diabetol. 2002;1:3. doi: 10.1186/1475-2840-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Virmani R, Burke AP, Kolodgie F. Morphological characteristics of coronary atherosclerosis in diabetes mellitus. Can J Cardiol. 2006;22(B):81B–84B. doi: 10.1016/s0828-282x(06)70991-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demer LL, Tintut Y. Vascular calcification: pathobiology of a multifaceted disease. Circulation. 2008;117:2938–2948. doi: 10.1161/CIRCULATIONAHA.107.743161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao JS, Cheng SL, Sadhu J, Towler DA. Inflammation and the osteogenic regulation of vascular calcification: a review and perspective. Hypertension. 2010;55:579–592. doi: 10.1161/HYPERTENSIONAHA.109.134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bostrom K, Watson KE, Horn S, Wortham C, Herman IM, Demer LL. Bone morphogenetic protein expression in human atherosclerotic lesions. J Clin Invest. 1993;91:1800–1809. doi: 10.1172/JCI116391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Csiszar A, Smith KE, Koller A, Kaley G, Edwards JG, Ungvari Z. Regulation of bone morphogenetic protein-2 expression in endothelial cells: role of nuclear factor-kappaB activation by tumor necrosis factor-alpha, H2O2, and high intravascular pressure. Circulation. 2005;111:2364–2372. doi: 10.1161/01.CIR.0000164201.40634.1D. [DOI] [PubMed] [Google Scholar]
- 28.Csiszar A, Lehoux S, Ungvari Z. Hemodynamic forces, vascular oxidative stress, and regulation of BMP-2/4 expression. Antioxid Redox Signal. 2009;11:1683–1697. doi: 10.1089/ars.2008.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M, Zhou SH, Zhao S, Li XP, Liu LP, Shen XQ. Pioglitazone can downregulate bone morphogenetic protein-2 expression induced by high glucose in human umbilical vein endothelial cells. Pharmacology. 2008;81:312–316. doi: 10.1159/000119118. [DOI] [PubMed] [Google Scholar]
- 30.Csiszar A, Ahmad M, Smith KE, Labinskyy N, Gao Q, Kaley G, Edwards JG, Wolin MS, Ungvari Z. Bone morphogenetic protein-2 induces proinflammatory endothelial phenotype. Am J Pathol. 2006;168:629–638. doi: 10.2353/ajpath.2006.050284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zebboudj AF, Shin V, Bostrom K. Matrix GLA protein and BMP-2 regulate osteoinduction in calcifying vascular cells. J Cell Biochem. 2003;90:756–765. doi: 10.1002/jcb.10669. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Yang HY, Giachelli CM. BMP-2 promotes phosphate uptake, phenotypic modulation, and calcification of human vascular smooth muscle cells. Atherosclerosis. 2008;199:271–277. doi: 10.1016/j.atherosclerosis.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge G, Zhang Y, Steiglitz BM, Greenspan DS. Mammalian tolloid-like 1 binds procollagen C-proteinase enhancer protein 1 and differs from bone morphogenetic protein 1 in the functional roles of homologous protein domains. J Biol Chem. 2006;281:10786–10798. doi: 10.1074/jbc.M511111200. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi K, Luo M, Zhang Y, Wilkes DC, Ge G, Grieskamp T, Yamada C, Liu TC, Huang G, Basson CT, Kispert A, Greenspan DS, Sato TN. Secreted Frizzled-related protein 2 is a procollagen C proteinase enhancer with a role in fibrosis associated with myocardial infarction. Nat Cell Biol. 2009;11:46–55. doi: 10.1038/ncb1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon HM, Hong BK, Kang TS, Kwon K, Kim HK, Jang Y, Choi D, Park HY, Kang SM, Cho SY, Kim HS. Expression of osteopontin in calcified coronary atherosclerotic plaques. J Korean Med Sci. 2000;15:485–493. doi: 10.3346/jkms.2000.15.5.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scatena M, Liaw L, Giachelli CM. Osteopontin: a multifunctional molecule regulating chronic inflammation and vascular disease. Arterioscler Thromb Vasc Biol. 2007;27:2302–2309. doi: 10.1161/ATVBAHA.107.144824. [DOI] [PubMed] [Google Scholar]
- 37.Dhore CR, Cleutjens JP, Lutgens E, Cleutjens KB, Geusens PP, Kitslaar PJ, Tordoir JH, Spronk HM, Vermeer C, Daemen MJ. Differential expression of bone matrix regulatory proteins in human atherosclerotic plaques. Arterioscler Thromb Vasc Biol. 2001;21:1998–2003. doi: 10.1161/hq1201.100229. [DOI] [PubMed] [Google Scholar]
- 38.Cresci S. Pharmacogenetics of the PPAR genes and cardiovascular disease. Pharmacogenomics. 2007;8:1581–1595. doi: 10.2217/14622416.8.11.1581. [DOI] [PubMed] [Google Scholar]
- 39.Doney AS, Fischer B, Lee SP, Morris AD, Leese G, Palmer CN. Association of common variation in the PPARA gene with incident myocardial infarction in individuals with type 2 diabetes: A Go-DARTS study. Nucl Recept. 2005;3:4. doi: 10.1186/1478-1336-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doney AS, Fischer B, Leese G, Morris AD, Palmer CN. Cardiovascular risk in type 2 diabetes is associated with variation at the PPARG locus: a Go-DARTS study. Arterioscler Thromb Vasc Biol. 2004;24:2403–2407. doi: 10.1161/01.ATV.0000147897.57527.e4. [DOI] [PubMed] [Google Scholar]
- 41.Regieli JJ, Jukema JW, Doevendans PA, Zwinderman AH, van der GY, Kastelein JJ, Grobbee DE. PPAR gamma variant influences angiographic outcome and 10-year cardiovascular risk in male symptomatic coronary artery disease patients. Diabetes Care. 2009;32:839–844. doi: 10.2337/dc08-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pischon T, Pai JK, Manson JE, Hu FB, Rexrode KM, Hunter D, Rimm EB. Peroxisome proliferator-activated receptor-gamma2 P12A polymorphism and risk of coronary heart disease in US men and women. Arterioscler Thromb Vasc Biol. 2005;25:1654–1658. doi: 10.1161/01.ATV.0000171993.78135.7e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.