Near-infrared fluorescence (NIRF) imaging is emerging as an important tool in preclinical studies.[1–5] As the technology is being established and implemented worldwide, a wave of effort has been spurred to develop novel imaging probes that can accurately recognize and report diseases, such as cancer.[3,6] One common drawback of NIRF imaging, however, is its high background and as a consequence of that, low specificity.[7,8] To address this issue, we and others have developed activatable probes with various approaches. Such probes, constructed by bringing into proximity an energy donor and receptor, are designed to be fluorescently quenched at a quiescent stage but activatable when encountering a specific trigger, which leads to a structural or conformational change and restores fluorescence activities.[7–9] This technique allows the signals to be only amplified at diseased areas upon a designated environment change, a feature which can greatly improve the signal-to-background ratio.
One such trigger that has been intensively investigated is proteases–enzymes that recognize and hydrolyze peptide substrates.[8,10,11] The overexpression of proteases was found to be constantly associated with pathological events like cancer. Matrix metalloproteinases (MMPs), for instance, are a family of enzymes critical to extracellular matrix remodeling. Their upregulation was found to play an important role in tumor invasiveness, metastasis, and angiogenesis.[12–15] Such an implication has made MMP a good tumor marker and a common target in the design of activatable probes.[16–19] We and others have previously developed activatable probes, based on either peptide,[16,19,20] polymer,[17,21,22] or inorganic nanoparticles,[23,24] of MMP targeting specificity. On the other hand, the use of protein as a building block to construct MMP-activatable probes has, to the best of our knowledge, never been reported. Compared with other macromolecules such as polymers, there are a limited number of chemical groups on the protein surface that can be used for conjugation. It is therefore challenging to integrate an MMP substrate, a dye molecule, and its corresponding quencher into such a platform with accurate control, while not disturbing the functions of the parent proteins.
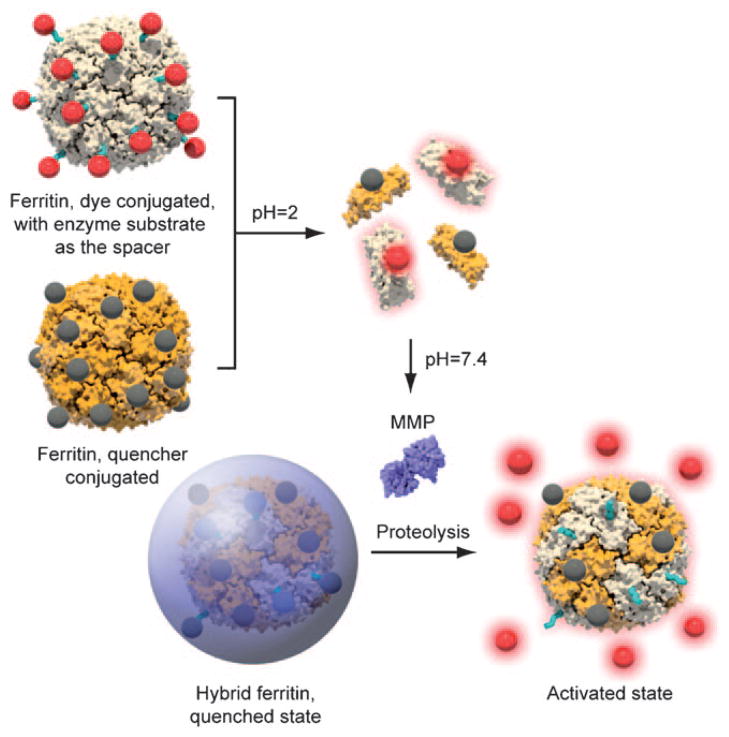
Herein, we report a successful case of using a heavy-chain ferritin cage as the scaffold to build MMP-specific activatable probes. Ferritins are a family of proteins that are found in many types of animals, plants, and prokaryotes, with the primary function being iron storage. Each ferritin molecule is made of 24 subunits, which are self-assembled, in a non-covalent fashion, into a cagelike nanostructure.[25] Such an intriguing architecture of ferritins has attracted previous research interest in their use as nanoreactors to synthesize nanomaterials.[26–28] Their potential in the field of optical imaging, however, has seldom been studied. Recently, it was found that despite the rigid architecture, the association among the ferritin subunits is pH-dependent, which means that the nanostructure can be broken down in an acidic environment and can be restored at neutral pH.[29] By mixing two or more types of ferritin cages and subjecting the mixture to such a transition, one can arrive at ferritins with hybrid features.[30] The use of such a strategy to load multiple motifs onto a single ferritin nanostructure, however, has never been exploited.
In the current study, we harnessed such a disassemble/ reassemble nature as the driving force to pack an energy pair into a ferritin nanostructure to arrive at activatable probes (Scheme 1). Specifically, we coupled Cy5.5-tagged peptide (Cy5.5-Gly-Pro-Leu-Gly-Val-Arg-Gly-Cys) and BHQ-3 (BHQ =black hole quencher), a widely utilized quencher of Cy5.5, onto ferritins to create two sets of protein cages. The details of conjugation can be found in the Supporting Information. For peptide conjugation, a bifunctional compound, N-succinimidyl-4-maleimidobutyrate (TCI America), was first coupled onto the ferritin surface to bridge the particle with Cy5.5-Gly-Pro-Leu-Gly-Val-Arg-Gly-Cys. We then mixed and broke down both types of proteins to subunits at pH 2, and subsequently tuned the pH back to neutral, with the anticipation of restoring the protein nanostructure, this time, in a hybrid fashion. The core peptide sequence, Pro-Leu-Gly-Val-Arg (PLGVR), has proven selectivity for multiple types of MMPs (such as MMP-2, -9, -13), which we validated in our previous investigations.[23] We assumed that the conglomeration of both dyes and quenchers on the protein surface will induce a quenched state to the overall nanostructure. Subsequently, when the probes are exposed to an MMP-rich environment, the PLGVR substrate will be cut off, thus leading to the release of Cy5.5 and the restoration of fluorescence activity (Scheme 1). The detailed ligand coupling and purification procedures can be found in the Supporting Information. By tuning the reaction ratio, we were able to obtain ferritins with a ratio of one dye/quencher to one ferritin subunit, which was validated by mass analysis (Figure S1 in the Supporting Information).
Scheme 1.
Formation of ferritin cage-based activatable probes.
To assess the feasibility of such an approach, we subjected the ferritin particles to different conditions and monitored the fluorescence change by fluorospectrometry. We first mixed the two types of ferritins, coupled with Cy5.5 (C-Fn) and BHQ-3 (B-Fn), respectively, in Tris buffer (500 μL; 50 mM Tris, 10 mM CaCl2, 150 mM NaCl, pH 2) at a C/B ratio of 1:1 (final concentration 0.43 μM for both C-Fn and B-Fn). In such an acidic environment, the ferritin cages were decomposed into two sets of ferritin subunits. This caused a spatially disassociated state for the dyes and the quenchers, which explained the overall maximized fluorescence activities (excitation at 675 nm, emission at 690 nm, Figure 1). Then 1M NaOH was added to adjust the pH back to neutral. The amount of NaOH was predetermined to make sure the final pH was 7.4. It can be seen from Figure 1 that such an addition of NaOH led to a dramatic decrease of fluorescence activity. This was attributed to the re-formation of nanocages, which brought together the quenching pairs and enforced a state of energy transfer among them. Subsequently, MMP-13 (final concentration 100 nM) was added to the solution. We anticipated that the enzyme would cleave the PLGVR substrate that bridged Cy5.5 and the ferritin scaffold, and as a consequence release Cy5.5 and restore the fluorescence. Indeed, we observed an increase in fluorescence activity upon the addition of MMP-13. After incubation, an almost full restoration of fluorescence was observed, thus confirming the capacity of the hybrid ferritins as activatable probes. A similar test was also performed with other MMPs (MMP-2, -9, -13, -14, -16) and similar activation patterns were observed (data not shown). The activation, however, was not as dramatic as that for MMP-13 (data not shown), which is in accordance with our previous observation with other activatable formulas.[17] Therefore, MMP-13 was used in the following in vitro studies.
Figure 1.
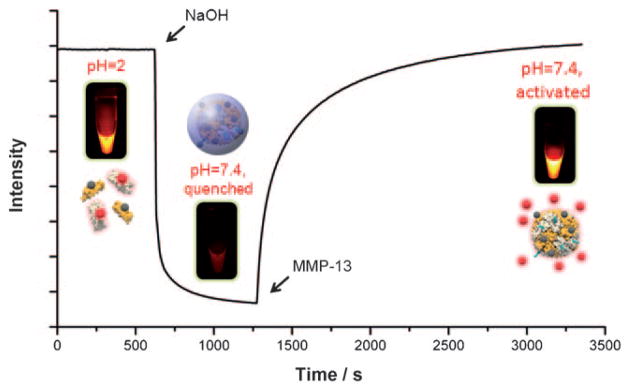
Fluorescence intensity change of hybrid ferritin (C/B ratio =1:1) during the course of disassembly, reassembly, and activation.
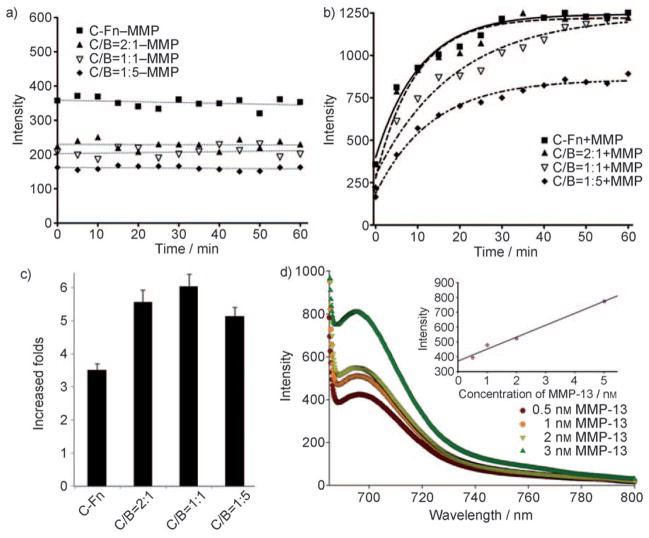
The composition of the hybrid ferritins, that is, the C/B ratio in the final product, may have an impact on the probes3 activation capacity. To optimize the formula, we started with a fixed concentration of C-Fn (0.43 μM) but varied the amount of B-Fn to arrive at a group of chimeric ferritins (C/B ratio = 2:1, 1:1, and 1:5). As a comparison, C-Fn alone (0.43 μM) was also investigated. In the absence of MMP-13, the hybrid ferritins were very stable under the incubation conditions (50 mM Tris, 150 mM NaCl, 10 mM CaCl2, pH 7.4, 37 °C), and showed no change of activities over the course of observation (excitation at 675 nm, emission at 690 nm, Figure 2a). This is not surprising since all the dye molecules were covalently conjugated to the nanostructure. Notably, although all the hybrid formulas were in a fluorescently quenched state, their intensities varied, with those of higher BHQ-3 composition showing lower fluorescence activities (intensities of 224.1, 207, and 165.3 a.u. for hybrids with a C/B ratio of 2:1, 1:1, and 1:5, respectively, at t =0). C-Fn alone also showed a certain degree of quenching effect (357 a.u. at t =0), which was attributed to self-quenching among the Cy5.5 fluorophores.[7,8]
Figure 2.
Kinetics of fluorescence intensity change of hybrid ferritins with different C/B ratios a) in the absence and b) in the presence of MMP-13. c) Activation capacities of the different ferritin formulas derived from (a) and (b). d) Activation of the C/B =1:1 formula when incubated with different concentrations of MMP-13 at 37°C. The activation time was 30 min.
The nanocages were then subjected to MMP-13 treatment (100 nM, 37°C) and their intensity change was monitored at 690 nm. All the ferritin formulas, including C-Fn alone, showed enzyme-dependent activation, and the efficiency was again composition-dependent (Figure 2b,c). In particular, while hybrids with C/B ratios of 2:1 and 1:1 showed a close-to-full restoration after 1 h of incubation, the formula with a C/B ratio of 1:5 showed only 60% activation capacity. Although the detailed mechanism is unknown at this stage, we believe it was because the excess amount of bulky BHQ-3 molecules on the ferritin surface blocked the access of MMP enzyme to its substrate. Overall, the hybrid ferritin with a C/B ratio of 1:1 showed the highest-fold activation (about sixfold under the described conditions, Figure 2d) and was selected for further tests. We subsequently incubated such a hybrid ferritin (C/B =1:1) with MMP-13 at different concentrations (0.5, 1, 2, and 5 nM) for 1 h, and studied its fluorescence properties (Figure 2d). We found that the activation depended on MMP concentration, and in the tested range the activation fold was almost linearly correlated with the MMP concentration.
The in vivo evaluation was performed on a UM-SCC-22B (head and neck squamous cell carcinoma) xenograft tumor model. We chose such a model because of its known high MMP expression.[31] For animal model preparation, about 5 million cells were inoculated subcutaneously into the right flanks of mice. The imaging was performed about one week later when the tumor reached a size of 0.5 cm3. A total of 0.02 mg hybrid ferritin (C/B =1:1) in phosphate-buffered saline (100 μL) was injected intratumorally and the imaging was performed at 30, 60, 90, 120, and 180 min post injection on a Maestro imaging system (Cambridge Research & Instrumentation, Inc.). For the control group, MMP inhibitor III (EMD Biosciences), a broad-spectrum inhibitor of various MMPs, was injected into the tumor 30 min prior to the probe injection.[23] As shown in Figure 3, without MMP inhibitor the probes demonstrated an instant response upon injection, and the activity developed steadily in the first 2 hours. On the contrary, the control group showed dramatically decreased intensities at the tumor area at all the time points, thus confirming that the activation was specifically governed by MMPs. We also performed a similar study on an SCC7 xenograft model and observed a similar activation feature (Figure S2 in the Supporting Information).
Figure 3.
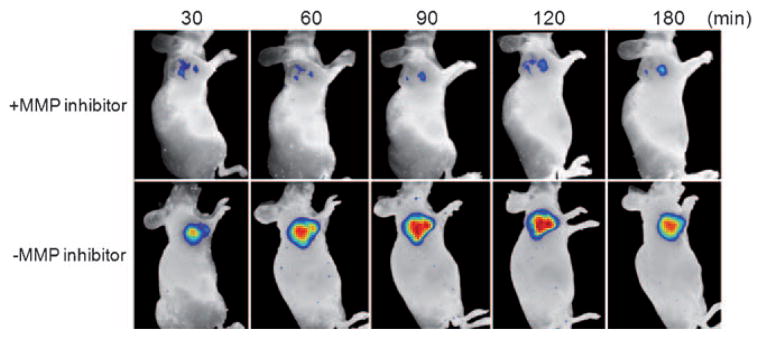
In vivo NIRF imaging of 22B tumor xenografted mice after intratumoral injection of ferritin-cage-based activatable probes (C/B =1:1) with and without MMP inhibitor.
In summary, we have reported the first case of using protein cages as a scaffold to build protease-activatable probes. Unlike previous approaches, we harnessed the self-assembly nature of the protein cages as a driving force to bring energy donors and receptors close to each other. By the use of such a strategy, we were able to generate a group of hybrid ferritins with different dye/quencher ratios. We then identified the formula with the highest-fold activation against MMPs, and confirmed the feasibility of such a probe in a xenograft mouse model. Such a technique can be readily applied to introduce other kinds of functionalities onto ferritin nanoparticles with precise control. For example, we prepared RGD-4C-labeled ferritins by following a previously published protocol.[32] We then mixed them with Cy5.5-labeled ferritins and, by using the same disassemble/reassemble technique, were able to arrive at hybrid ferritin nanocages with both Cy5.5 and RGD on the surface. The cell binding assay and the in vitro fluorescence staining confirmed the integrin αvβ3 targeting feature of the hybrid nanocages (Figures S3 and S4 in the Supporting Information). This finding indicates the broad role such a nanoplatform can play in the field of nanomedicine. Studies of the related applications are currently under way.
Supplementary Material
Footnotes
This work was supported by the Intramural Research Program of the National Institute of Biomedical Imaging and Bioengineering, NIH. J.X. is partially supported by the NIH pathway to independence (K99/R00) award; S.L. is partially supported by a NIBIB/NIST fellowship; G.N. is an Imaging Sciences Training Program (ISTP) Fellow. We thank Dr. Henry S. Eden for proofreading the manuscript.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201006757.
Contributor Information
Dr. Xin Lin, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH) (USA), Fax: (+1)301-480-1613.
Dr. Jin Xie, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH) (USA), Fax: (+1)301-480-1613.
Dr. Lei Zhu, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH) (USA), Fax: (+1)301-480-1613
Dr. Seulki Lee, Email: seulki.lee@nih.gov, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH) (USA), Fax: (+1)301-480-1613
Dr. Gang Niu, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH) (USA), Fax: (+1)301-480-1613
Dr. Ying Ma, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH) (USA), Fax: (+1)301-480-1613
Dr. Kwangmeyung Kim, Biomedical Research Center, Korea Institute of Science and Technology, Seoul (Korea)
Dr. Xiaoyuan Chen, Email: shawn.chen@nih.gov, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH) (USA), Fax: (+1)301-480-1613
References
- 1.Hilderbrand SA, Weissleder R. Curr Opin Chem Biol. 2010;14:71. doi: 10.1016/j.cbpa.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 2.Rao J, Dragulescu-Andrasi A, Yao H. Curr Opin Biotechnol. 2007;18:17. doi: 10.1016/j.copbio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Ballou B, Ernst LA, Waggoner AS. Curr Med Chem. 2005;12:795. doi: 10.2174/0929867053507324. [DOI] [PubMed] [Google Scholar]
- 4.Ntziachristos V, Bremer C, Weissleder R. Eur Radiol. 2003;13:195. doi: 10.1007/s00330-002-1524-x. [DOI] [PubMed] [Google Scholar]
- 5.Frangioni JV. Curr Opin Chem Biol. 2003;7:626. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S, Xie J, Chen X. Curr Top Med Chem. 2010;10:1135. doi: 10.2174/156802610791384270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, Park K, Kim K, Choi K, Kwon IC. Chem Commun. 2008:4250. doi: 10.1039/b806854m. [DOI] [PubMed] [Google Scholar]
- 9.Elias DR, Thorek DL, Chen AK, Czupryna J, Tsourkas A. Cancer Biomarkers. 2008;4:287. doi: 10.3233/cbm-2008-4602. [DOI] [PubMed] [Google Scholar]
- 10.Mahmood U, Weissleder R. Mol Cancer Ther. 2003;2:489. [PubMed] [Google Scholar]
- 11.Funovics M, Weissleder R, Tung CH. Anal Bioanal Chem. 2003;377:956. doi: 10.1007/s00216-003-2199-0. [DOI] [PubMed] [Google Scholar]
- 12.Kessenbrock K, Plaks V, Werb Z. Cell. 2010;141:52. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vihinen P, Ala-aho R, Kahari VM. Curr Cancer Drug Targets. 2005;5:203. doi: 10.2174/1568009053765799. [DOI] [PubMed] [Google Scholar]
- 14.Aureli L, Gioia M, Cerbara I, Monaco S, Fasciglione GF, Marini S, Ascenzi P, Topai A, Coletta M. Curr Med Chem. 2008;15:2192. doi: 10.2174/092986708785747490. [DOI] [PubMed] [Google Scholar]
- 15.Friedl P, Wolf K. Cancer Res. 2008;68:7247. doi: 10.1158/0008-5472.CAN-08-0784. [DOI] [PubMed] [Google Scholar]
- 16.Lee S, Park K, Lee SY, Ryu JH, Park JW, Ahn HJ, Kwon IC, Youn IC, Kim K, Choi K. Bioconjugate Chem. 2008;19:1743. doi: 10.1021/bc800264z. [DOI] [PubMed] [Google Scholar]
- 17.Lee S, Ryu JH, Park K, Lee A, Lee SY, Youn IC, Ahn CH, Yoon SM, Myung SJ, Moon DH, Chen X, Choi K, Kwon IC, Kim K. Nano Lett. 2009;9:4412. doi: 10.1021/nl902709m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryu JH, Lee A, Lee S, Ahn CH, Park JW, Leary JF, Park S, Kim K, Kwon IC, Youn IC, Choi K. Bioconjugate Chem. 2010;21:1378. doi: 10.1021/bc100008b. [DOI] [PubMed] [Google Scholar]
- 19.Jiang T, Olson ES, Nguyen QT, Roy M, Jennings PA, Tsien RY. Proc Natl Acad Sci USA. 2004;101:17867. doi: 10.1073/pnas.0408191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pham W, Choi Y, Weissleder R, Tung CH. Bioconjugate Chem. 2004;15:1403. doi: 10.1021/bc049924s. [DOI] [PubMed] [Google Scholar]
- 21.Bremer C, Tung CH, Weissleder R. Nat Med. 2001;7:743. doi: 10.1038/89126. [DOI] [PubMed] [Google Scholar]
- 22.Scherer RL, VanSaun MN, McIntyre JO, Matrisian LM. Mol Imaging. 2008;7:118. [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S, Cha EJ, Park K, Lee SY, Hong JK, Sun IC, Kim SY, Choi K, Kwon IC, Kim K, Ahn CH. Angew Chem. 2008;120:2846. doi: 10.1002/anie.200705240. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2008;47:2804. [Google Scholar]
- 24.Shi L, De Paoli V, Rosenzweig N, Rosenzweig Z. J Am Chem Soc. 2006;128:10378. doi: 10.1021/ja063509o. [DOI] [PubMed] [Google Scholar]
- 25.Uchida M, Kang S, Reichhardt C, Harlen K, Douglas T. Biochim Biophys Acta Gen Subj. 2010;1800:834. doi: 10.1016/j.bbagen.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosein HA, Strongin DR, Allen M, Douglas T. Langmuir. 2004;20:10283. doi: 10.1021/la0491100. [DOI] [PubMed] [Google Scholar]
- 27.Klem MT, Resnick DA, Gilmore K, Young M, Idzerda YU, Douglas T. J Am Chem Soc. 2007;129:197. doi: 10.1021/ja0667561. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Laug L, Munchgesang W, Pippel E, Gosele U, Brandsch M, Knez M. Nano Lett. 2010;10:219. doi: 10.1021/nl903313r. [DOI] [PubMed] [Google Scholar]
- 29.Santambrogio P, Levi S, Arosio P, Palagi L, Vecchio G, Lawson DM, Yewdall SJ, Artymiuk PJ, Harrison PM, Jappelli R, et al. J Biol Chem. 1992;267:14077. [PubMed] [Google Scholar]
- 30.Kang S, Oltrogge LM, Broomell CC, Liepold LO, Prevelige PE, Young M, Douglas T. J Am Chem Soc. 2008;130:16527. doi: 10.1021/ja807655t. [DOI] [PubMed] [Google Scholar]
- 31.Werner JA, Rathcke IO, Mandic R. Clin Exp Metastasis. 2002;19:275. doi: 10.1023/a:1015531319087. [DOI] [PubMed] [Google Scholar]
- 32.Uchida M, Flenniken ML, Allen M, Willits DA, Crowley BE, Brumfield S, Willis AF, Jackiw L, Jutila M, Young MJ, Douglas T. J Am Chem Soc. 2006;128:16626. doi: 10.1021/ja0655690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.