Abstract
BACKGROUND:
Bacterial pathogens belonging to the genus Acinetobacter cause serious infections in immunocompromised individuals that are very difficult to treat due to their extremely high resistance to many antibiotics.
OBJECTIVE:
To investigate the role of resistance-nodulation-division (RND) pumps and porins in the antibiotic resistance of Acinetobacter species collected from Canadian hospitals.
METHODS:
Clinical isolates of Acinetobacter species collected from Canadian hospitals were analyzed for the expression of genes encoding RND pumps (adeB, adeG, adeJ, AciBau_2746 and AciBau_2436) and outer membrane porins (carO, 33 kDa porin and oprD) using quantitative reverse transcription (qRT) polymerase chain reaction. Species identification of the isolates was performed using a multiplex polymerase chain reaction method for gyrB.
RESULTS:
The expression of RND pump-encoding genes was widespread in the clinical isolates of Acinetobacter species, with each of the isolates expressing at least one RND pump. adeG was found to be overexpressed in all of the isolates, while adeB was found to be overexpressed in only two isolates. Among the porin-encoding genes, the expression of carO was considerably downregulated among the majority of isolates.
CONCLUSION:
The present study was the first to analyze the expression of RND pump- and porin-encoding genes in the clinical isolates of Acinetobacter species from Canadian hospitals. The overexpression of genes encoding RND pumps and the downregulation of genes encoding porins was common in clinical isolates of Acinetobacter species from Canadian hospitals, with the AdeFGH pump being the most commonly expressed RND pump.
Keywords: Acinetobacter, Antibiotic resistance, Gene expression
Abstract
HISTORIQUE :
Les pathogènes bactériens qui appartiennent au gène Acinetobacter provoquent de graves infections chez les personnes immunocompromises. Ces infections sont très difficiles à traiter en raison de leur résistance extrêmement élevée à de nombreux antibiotiques.
OBJECTIF :
Examiner le rôle des pompes RND (resistance-nodulation-division) et des porines dans l’antibiorésistance des espèces d’Acinetobacter prélevées dans des hôpitaux canadiens.
MÉTHODOLOGIE :
Les chercheurs ont analysé des isolats cliniques d’espèces d’Acinetobacter prélevés dans des hôpitaux canadiens afin de déceler l’expression des gènes codants des pompes RND (adeB, adeG, adeJ, AciBau_2746 et AciBau_2436) et des porines de la membrane externe (carO, porine 33 kDa et oprD) à l’aide de la réaction en chaîne de la polymérase après transcription inverse quantitative (qRT). Ils ont identifié les espèces d’isolats à l’aide d’une méthode de réaction en chaîne de la polymérase multiplexe pour le gyrB.
RÉSULTATS :
L’expression de gènes codants de la pompe RND était généralisée dans les isolats cliniques des espèces d’Acinetobacter, chaque isolat exprimant au moins une pompe RND. Les chercheurs ont observé une surexpression de l’adeG dans tous les isolats, mais la surexpression de l’adeB dans seulement deux isolats. Parmi les gènes codants des porines, l’expression du carO présentait une régulation négative marquée dans la majorité des isolats.
CONCLUSION :
La présente étude était la première à analyser l’expression des gènes codants de la pompe d’efflux RND et des porines dans les isolats cliniques d’espèces d’Acinetobacter provenant d’hôpitaux canadiens. La surexpression des gènes codants des pompes RND et la régulation négative des gènes codants des porines était courante dans les isolats cliniques d’espèces d’Acinetobacter provenant d’hôpitaux canadiens, la pompe AdeFGH étant la pompe d’efflux RND la plus exprimée.
The Acinetobacter calcoaceticus–Acinetobacter baumannii complex consists of A calcoaceticus, A baumannii, Acinetobacter pittii (formerly Acinetobacter GS3) and Acinetobacter nosocomialis (formerly Acinetobacter 13TU) species (1). This complex is notorious for causing serious infections in immunocompromised individuals. Although recent studies have shown that infections caused by A pittii and A nosocomialis may not be as uncommon as previously believed (2), A baumannii, responsible for a variety of infections, is still considered to be the most prominent species in this genera. These infections include ventilator-associated pneumonia, endocarditis, surgical-site infections, septicemia and urinary tract infections (3). The mortality rates associated with infections caused by A baumannii can be very high, with the mortality rate of pneumonia reported to be as high as 73% (4). Outbreaks in hospitals have been reported from various geographical areas (5), and the incidence of A baumannii-mediated infection has been increasing steadily over the past two decades (6).
The multidrug-resistant (MDR) nature of A baumanniii infections has proven to be very challenging for clinicians. The organism frequently exhibits resistance to many classes of antimicrobial drugs, including β-lactams (penicillins and cephalosporins), carbapenems, aminoglycosides, fluoroquinolones and tetracyclines (7). Therefore, the emergence of MDR strains of A baumannii is becoming a major concern in hospital settings. Mechanisms of resistance include production of β-lactamase (8), the presence of aminoglycoside-modifying enzymes (9), target-site mutations (10), loss of outer membrane permeability (11) and multidrug efflux systems (12). The latter two, outer membrane permeability and multidrug efflux systems (particularly those belonging to the resistance-nodulation-division [RND] family), constitute the most important mechanisms of intrinsic resistance in Acinetobacter species.
Little is known about the mechanisms of antibiotic resistance in A baumannii from Canadian hospitals. The research objectives of this study were to analyze the expression of RND efflux pump- and outer membrane porin-encoding genes in clinical isolates of Acinetobacter species and to determine whether there is a correlation between the expression of these genes and the intrinsic antimicrobial resistance of this organism.
METHODS
Bacterial strains and growth conditions
Clinical isolates of Acinetobacter species used in the present study are listed in Table 1. Isolates, displaying varying antibiotic susceptibility profiles, were obtained from Canadian hospitals, specifically, from intensive care units from 2006 to 2009, inclusively, as part of the Canadian Hospital Ward Antibiotic Resistance Surveillance (CANWARD) study (www.can-r.ca). A baumannii ATCC 19606 was used as the control strain. Lysogeny broth (LB) medium (Biobasic, Canada) (37°C) was used for the culturing of bacterial strains.
TABLE 1.
Antibiotic susceptibilities of Acinetobacter species isolates
| Strain | Source | AMK | CFZ | FEP | CRO | CIP | CLI | IPM | GEN | LVX | MEM | MXF | TZP | TGC | SXT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AB004 | Winnipeg, MB | 32 | >128 | 128 | 32 | 2 | 2 | 0.25 | 32 | 1 | 2 | 0.5 | 64 | 2 | 0.12 |
| AB005 | Winnipeg, MB | <1 | 128 | ≤1 | ≤1 | ≤0.06 | 1 | 0.25 | ≤0.5 | 0.12 | 0.25 | ≤0.06 | ≤1 | 0.5 | ≤0.12 |
| AB006 | Vancouver, BC | 2 | >128 | 4 | 16 | 0.5 | 2 | 0.25 | 2 | 0.25 | 1 | 0.25 | 8 | 1 | ≤0.12 |
| AB007 | Vancouver, BC | 2 | >128 | 8 | 8 | 1 | 1 | 0.25 | 1 | 0.25 | 0.5 | 0.12 | 16 | 0.5 | 0.5 |
| AB008 | Hamilton, ON | 2 | >128 | 2 | 4 | 0.12 | 1 | 0.25 | 0.5 | ≤0.06 | 0.5 | ≤0.06 | ≤1 | 0.5 | 0.25 |
| AB009 | Hamilton, ON | 2 | >128 | 2 | 8 | 0.5 | 0.5 | 0.25 | 0.5 | 0.12 | 0.5 | 0.12 | ≤1 | 0.5 | 0.25 |
| AB010 | Vancouver, BC | 2 | >128 | 4 | 16 | 0.5 | 1 | 0.25 | 1 | 0.25 | 1 | 0.12 | 8 | 0.5 | >8 |
| AB011 | Winnipeg, MB | 32 | >128 | 64 | 16 | 2 | 2 | 0.5 | 32 | 0.5 | 2 | 0.5 | 32 | 2 | ≤0.12 |
| AB012 | Victoria, BC | 2 | >128 | 2 | 16 | 0.12 | 1 | 0.12 | 0.5 | 0.12 | 0.5 | ≤0.06 | 4 | 0.25 | ≤0.12 |
| AB013 | Vancouver, BC | 16 | 128 | 16 | 16 | 1 | 1 | 0.25 | 32 | 0.5 | 1 | 0.5 | ≤1 | 2 | ≤0.12 |
| AB014 | Montreal, QC | 2 | >128 | 8 | 16 | 0.5 | 0.25 | 0.25 | ≤0.25 | 0.25 | 1 | 0.12 | 16 | 0.5 | 0.25 |
| AB027 | Montreal, QC | >64 | >128 | >128 | >256 | >16 | 1 | 32 | 32 | 16 | 16 | 8 | >512 | 4 | >8 |
| AB028 | Winnipeg, MB | 2 | >28 | 32 | 64 | >16 | 0.5 | 0.25 | >32 | 16 | 2 | 8 | 128 | 0.5 | 8 |
| AB029 | Winnipeg, MB | ≤1 | 128 | 16 | 2 | 2 | 0.5 | 0.25 | ≤0.5 | 1 | 0.5 | 0.5 | ≤1 | 2 | ≤0.12 |
| AB030 | Winnipeg, MB | 64 | >128 | >64 | >64 | >16 | 0.5 | >32 | >32 | 16 | >32 | >16 | >512 | >16 | >8 |
| AB031 | Toronto, ON | 2 | >128 | 4 | 16 | 0.25 | 0.5 | 0.25 | <0.5 | 0.25 | 1 | 0.12 | 4 | 8 | 4 |
| ATCC19606 | ATCC | 8 | 128 | 16 | 32 | 1 | 1 | 0.25 | 8 | 0.5 | 1 | 0.5 | 4 | 1 | 8 |
Data expressed as μg/mL. AMK Amikacin; ATCC American Type Culture Collection; BC British Columbia; CFZ Cefazolin; CIP Ciprofloxacin; CLI Colistin; CRO Ceftriaxone; FEP Cefepime; GEN Gentamicin; IPM Imipenem; LVX Levofloxacin; MB Manitoba; MEM Meropenem; MXF Moxifloxacin; ON Ontario; QC Quebec; SXT trimethoprim/sulfamethoxazole; TGC Tigecycline; TZP Piperacillin-tazobactam
Genomic DNA extraction
Genomic DNA extraction was performed, following the manufacturer’s instructions, using a commercially available kit (DNeasy, Qiagen, Canada). DNA was stored at −20°C until used.
Genotyping of Acinetobacter species isolates
Genotyping of Acinetobacter species isolates was performed using up to 5 ng of genomic DNA according to the multiplex polymerase chain reaction (PCR) method for gyrB described in the study by Higgins et al (13).
Antibiotic susceptibility assays
Antibiotic susceptibility of Acinetobacter species isolates to antibiotics commonly used for the treatment (Table 1) of Acinetobacter species infections was determined using the microbroth dilution method, which has been previously described (14).
RNA extraction and complementary DNA (cDNA) synthesis
Overnight cultures grown in LB medium at 37°C with shaking (200 rpm) were subcultured (1:100) in fresh LB medium (without antibiotics) and incubated. Cells were harvested at an A600 of approximately 0.8 and were frozen as pellets (−80°C) to facilitate lysis. RNA extractions were performed using a commercially available kit (RNeasy, Qiagen) according to manufacturer’s instructions. RNA samples were treated with DNAse I (Qiagen), according to manufacturer’s instructions, to remove any genomic DNA carryover from RNA extraction. Synthesis of complementary DNA (cDNA) was performed using the GoScript Reverse Transcriptase kit (Promega, USA) using 1.2 μg of total RNA in a 20 μL reaction volume according to the manufacturer’s instructions. To confirm the absence of genomic DNA contamination of RNA samples in the quantitative reverse transcription (qRT)-PCR described below, the minus-RT control reaction was performed by excluding the reverse transcriptase enzyme.
qRT-PCR
qRT-PCR reactions were performed using SsoFast Evagreen Supermix (BioRad, Canada) to study the expression of RND pump- and outer membrane porin-encoding genes. The RND pump-encoding genes analyzed included adeB, adeG, adeJ, AciBau_2436 and AciBau_2746; the outer membrane porin-encoding genes analyzed included carO, oprD and the 33 kDa porin. All primers used for analysis (designed using Oligoperfect software, Invitrogen, Canada, www.invitrogen.com), except those used for adeE and the 33 kDa porin, have been described elsewhere (15). The primers used for adeE were adeE_RT_F (5′–gaaacagagcgggttggtaa–3′) and adeE_RT_R (5′–tgcctgcgttatttc-taccc–3′), and those used for the 33 kDa porin gene were 33KD_RT_F (5′–atccaaaacgaccaagatgc–3′) and 33KD_RT_R (5′–caaaaccgattgccat-gtta–3′). 16S ribosomal RNA (rRNA) was used as the housekeeping gene, while A baumannii ATCC 19606 was used as the reference strain. The efficiencies of primers were tested by pooling the cDNA from all isolates, serially diluting the pool 10-fold and then generating a standard curve. Reactions were performed in a total volume of 15 μL using 300 nM of each primer and 5 μL of the cDNA template (diluted 1:20 from the cDNA synthesis reaction described above). At least two independent samples were analyzed for each target gene and all reactions were performed in triplicate. A no-template control was used in all reactions. The 16S rRNA reaction was used to rule out genomic DNA contamination from the minus-RT control reaction.
RESULTS
Genotyping of Acinetobacter species isolates
Results of the multiplex PCR for gyrB used for the genotyping of the Acinetobacter species isolates are shown in Figure 1. This method identified two of 16 (13%) isolates as A pittii (Acinetobacter species AB006 and Acinetobacter species AB007), four of 16 (25%) as A nosocomialis (Acinetobacter species AB004, Acinetobacter species AB005, Acinetobacter species AB011 and Acinetobacter species AB013) and nine of 16 (56%) isolates were identified as A baumannii. One isolate, Acinetobacter AB012, could not be identified conclusively because the PCR products observed could have originated from either A baumannii or A nosocomialis. Multiple individual colonies of Acinetobacter AB012 were tested to rule out contamination, and the results consistently indicated (data not shown) that it was, in fact, a pure culture.
Figure 1).
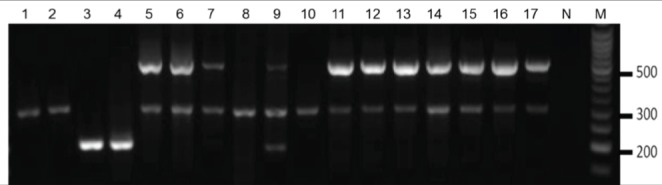
Genotyping of Acinetobacter species isolates. Multiplex PCR for the gyrB gene identified clinical isolates as Acinetobacter baumannii, Acinetobacter pittii or Acinetobacter nosocomialis. Presence of a 294 bp band indicates A nosocomialis, and a 193 bp band indicates A pittii. Presence of two bands at 294 bp and 490 bp indicates A baumannii. Lane 1 Acinetobacter species AB004, Lane 2 Acinetobacter species AB005, Lane 3 Acinetobacter species AB006, Lane 4 Acinetobacter species AB007, Lane 5 Acinetobacter species AB008, Lane 6 Acinetobacter species AB009, Lane 7 Acinetobacter species AB010, Lane 8 Acinetobacter species AB011, Lane 9 Acinetobacter species AB012, Lane 10 Acinetobacter species AB013, Lane 11 Acinetobacter species AB014, Lane 12 Acinetobacter species AB027, Lane 13 Acinetobacter species AB028, Lane 14 Acinetobacter species AB029, Lane 15 Acinetobacter species AB030, Lane 16 Acinetobacter species AB031, Lane 17 A baumannii ATCC 19606. M Molecular weight marker; N No-template control
Antibiotic susceptibility assays
Results of the antibiotic susceptibility assays are summarized in Table 1. The susceptibility profiles of the Acinetobacter species isolates varied considerably from extensively susceptible to MDR (concomitantly resistant to ≥3 different drug classes). Some of the most resistant isolates in the present study were A baumannii (A baumannii AB027, A baumannii AB028 and A baumannii AB030), particularly with respect to resistance to aminoglycosides, cephalosporins, fluoroquinolones, carbapenems, piperacillin/tazobactam, tigecycline and trimethoprim/sulfamethoxazole. Among the non-A baumannii isolates, AB004 and AB011 (both identified as A nosocomialis) displayed resistance to aminoglycosides, cephalosporins and piperacillin/tazobactam.
Expression analysis of RND pump- and outer membrane porin-encoding genes
The expression analysis of RND pump- and outer membrane porin-encoding genes is summarized in Table 2. The expression analysis of outer membrane porin-encoding genes carO, the 33 kDa porin and oprD revealed that the expression of carO was either considerably lower than or similar to the A baumannii ATCC 19606 control, with the exception of A baumannii AB009 (16-fold higher expression), A baumannii AB028 and A baumannii AB030 (four-fold higher expression for both). Expression of the 33 kDa porin gene was found to be higher in seven isolates, with >10-fold higher expression seen in four isolates (A baumannii AB009, A baumannii AB027, A baumannii AB028 and A baumannii AB030). Six isolates (A nosocomialis AB004, A pittii AB006, A pittii AB007, A baumannii AB010, A pittii AB011, and A nosocomialis AB013) exhibited lower expression of oprD compared with the control strain A baumannii ATCC 19606. All other strains showed at least a two-fold higher expression of oprD compared with the A baumannii ATCC 19606 control. One exception was A baumannii AB030, which showed a similar level of expression of oprD as the control strain.
TABLE 2.
Expression* of resistance-nodulation-division pump†- and outer membrane porin-encoding genes in clinical isolates of Acinetobacter species
| Isolate strain |
Gene
|
|||||||
|---|---|---|---|---|---|---|---|---|
| adeB | adeG | adeJ | AciBau_2746 | AciBau_2436 | carO | oprD | 33kDa porin | |
| AB004 | 0.589±0.109 | 4.452±0.81 | 0.005±0.0005 | 0.004±0.001 | 0.006±0.003 | 0.003±0.0004 | 0.007±0.002 | 0.183±0.028 |
| AB005 | 0.005±0.001 | 6.271±0.81 | 0.0002±0.000 | 0.155±0.017 | 0.051±0.007 | 0.003±0.0002 | 1.790±0.139 | 1.932±0.168 |
| AB006 | 0.035±0.005 | 3.339±0.337 | 0.128±0.027 | 0.004±0.0020 | 0.012±0.002 | 0.002±0.0005 | 0.008±0.001 | 5.149±0.405 |
| AB007 | 0.004±0.001 | 3.594±0.357 | 0.001±0.0004 | 0.247±0.026 | 0.003±0.0005 | 1.043±0.126 | 0.004±0.001 | 1.253±0.205 |
| AB008 | 0.030±0.004 | 8.658±0.662 | 0.104±0.335 | 4.167±0.304 | 2.401±0.157 | 0.005±0.0004 | 5.636±0.451 | 0.492±0.024 |
| AB009 | 0.004±0.001 | 4.264±0.502 | 5.477±0.763 | 0.009±0.005 | 0.020±0.003 | 16.328±2.171 | 3.513±0.436 | 12.855±2.431 |
| AB010 | 0.013±0.004 | 3.749±0.331 | 0.232±0.299 | 0.007±0.001 | 0.007±0.002 | 0.008±0.001 | 0.008±0.001 | 7.439±0.606 |
| AB011 | 1.256±0.179 | 6.754±0.688 | 0.255±0.017 | 0.007±0.002 | 0.019±0.009 | 0.005±0.001 | 0.008±0.001 | 0.324±0.023 |
| AB012 | 0.027±0.021 | 5.396±0.915 | 2.350±0.052 | 0.002±0.0003 | 0.011±0.003 | 0.004±0.0004 | 2.016±0.083 | 8.480±1.926 |
| AB013 | 0.804±0.243 | 5.048±0.763 | 0.038±0.043 | 0.008±0.005 | 0.008±0.001 | 0.004±0.0007 | 0.012±0.003 | 2.178±0.26 |
| AB014 | 0.105±0.014 | 8.803±1.356 | 3.916±0.269 | 3.684±0.178 | 0.020±0.001 | 2.254±0.218 | 3.710±0.302 | 32.295±11.429 |
| AB027 | 0.471±0.018 | 2.122±0.206 | 3.721±0.155 | 1.474±0.084 | 1.686±0.045 | 0.001±0.0002 | 2.971±0.114 | 23.381±3.238 |
| AB028 | 0.108±0.006 | 7.498±0.564 | 7.592±1.078 | 2.400±0.239 | 0.020±0.003 | 4.051±0.502 | 4.160±0.275 | 0.387±0.045 |
| AB029 | 0.037±0.007 | 4.800±0.514 | 0.691±0.047 | 2.431±0.31 | 0.013±0.004 | 0.225±0.022 | 3.426±0.301 | 0.223±0.017 |
| AB030 | 5.674±0.334 | 6.771±0.414 | 8.097±0.605 | 1.904±0.121 | 0.086±0.011 | 4.485±0.326 | 1.397±0.095 | 21.052±1.256 |
| AB031 | 1.876±0.17 | 5.872±0.638 | 7.641±1.118 | 0.852±0.601 | 0.019±0.002 | 0.151±0.023 | 4.822±0.464 | 1.897±0.349 |
Data expressed as relative quantity ± SE.
Relative to the control strain ATCC 19606;
A >2-fold overexpression of resistance-nodulation-division efflux pump-encoding genes compared with the control strain ATCC 19606 is indicated in bold
As for the RND efflux pump-encoding genes, adeB was over-expressed only by A baumannii AB030 (5.5-fold) and A baumannii AB031 (approximately two-fold), while adeG was overexpressed by all isolates. Seven isolates (A baumannii AB009, Acinetobacter species AB012, A baumannii AB014, A baumannii AB027, A baumannii AB028, A baumannii AB030 and A baumannii AB031) were found to overexpress adeJ. Among the two as yet uncharacterized RND pump encoding genes, AciBau_2746 was overexpressed in six isolates while AciBau_2436 was overexpressed in two.
DISCUSSION
In the past few decades, A baumannii has emerged as a major nosocomial pathogen capable of causing serious drug-resistant infections. The drug resistance of this organism poses a considerable challenge in the treatment of infections. In spite of the ever-increasing clinical importance of Acinetobacter species worldwide, little is known about the antibiotic resistance mechanisms used by these organisms. In the present study, we analyzed isolates of Acinetobacter species, collected from various Canadian hospital intensive care units, for expression of RND pump- and outer membrane porin-encoding genes.
RND pumps are believed to be the major determinants of intrinsic antibiotic resistance in Gram-negative pathogens and their over-expression is often linked to the MDR phenotype of these organisms. At present, three different RND pumps have been described in Gram-negative pathogens, namely, AdeABC (16), AdeIJK (17) and AdeFGH (18). AdeDE is another RND pump that has been reported in A pittii(19). In addition, a survey of the A baumannii ATCC 19606 genome revealed the presence of two more RND complex-encoding operons, namely AciBau_2434-5-6 and AciBau_2747-6.
In all 16 isolates, we analyzed the expression of three characterized and two as yet uncharacterized RND pump-encoding genes and compared their expression with that found in A baumannii ATCC 19606. We observed that all of the isolates expressed at least one RND pump-encoding gene. Previous studies have shown the AdeABC pump to be the major MDR pump in clinical isolates of Gram-negative pathogens (12,20), which is responsible for the efflux of amikacin, chloramphenicol, cefotaxime, erythromycin, gentamicin, kanamycin, norfloxacin, netilmicin, ofloxacin, pefloxacin, sparfloxacin, tetracycline, tobramycin and trimethoprim (16). In the present study, A baumannii AB030, an isolate found to be one of the most resistant, was among the two isolates exhibiting expression of adeB (Table 2). Interestingly, the levels of adeB in A baumannii AB027, another MDR isolate, were not found to be any different from any of the susceptible isolates (Table 1) suggesting that the expression of adeB is not solely responsible for the MDR phenotype of A baumannii AB030. However, we did observe a correlation between adeB expression and resistance to tigecycline. Both isolates, AB030 and AB031, which exhibited higher expression of adeB, were also found to be resistant to tigecycline. A similar correlation has also been reported previously (21).
AdeFGH and AdeIJK are two other pumps characterized from A baumannii. The AdeFGH pump is known to efflux chloramphenicol, trimethoprim, ciprofloxacin and clindamicin (18), while the AdeIJK pump effluxes β-lactams, chloramphenicol, tetracycline, erythromycin, lincosamides, fluoroquinolones, fusidic acid, novobiocin, rifampin, trimethoprim, acridine, pyronine, safranin and sodium dodecyl sulfate (17). We found adeG to be the most commonly expressed RND efflux pump gene among the isolates, followed by adeJ. A baumannii AB030, one of the most resistant isolates, was the only isolate that exhibited overexpression of adeG and adeJ in addition to adeB. We have previously shown the overexpression of adeG in some of these isolates using end-point RT-PCR (22), and the persent work further confirms that data using qRT-PCR. Three other isolates found to express three different RND pump-encoding genes, A baumannii AB008 (adeG, AciBau_2436 and AciBau_2746), A baumannii AB014 (adeG, adeJ and AciBau_2746) and A baumannii AB028 (adeG, adeJ and AciBau_2746) did not exhibit susceptibilities different from isolates expressing fewer pumps. The lack of any obvious correlation between the expression of RND pump-encoding genes and resistance to their respective substrates tested in the present study was most likely due to the presence of other resistance mechanisms present in these isolates.
We also analyzed two as yet uncharacterized RND pump-encoding genes, namely, AciBau_2436 and AciBau_2746. Sequence alignment indicated that these two genes likely encode metal efflux proteins and are not likely to play a role in the antibiotic resistance of Acinetobacter species isolates.
In addition, we analyzed the expression of adeE in two isolates identified as A pittii and found its expression to be nine-fold higher in A pittii AB007 compared with A pittii AB006 (data not shown). A pittii AB007 was also found to be moderately more resistant to ciprofloxacin, tazobactam, and trimethoprim/sulfamethoxazole (Table 1) than A pittii AB006, which could be due to the activity of the AdeDE pump.
Downregulation of outer membrane porins, sometimes with concurrent overexpression of RND pumps, is linked with antibiotic resistance in Gram-negative bacterial pathogens (23,24). All three porins analyzed in the present study have been implicated in carbapenem resistance in Acinetobacter species (25). Although we observed a considerable down-regulation of carO in all but three isolates, namely, A baumannii AB009, A baumannii AB028 and A baumannii AB030 (Table 2), we did not notice any correlation with carbapenem resistance. Similarly, we did not observe any obvious correlations between oprD or 33 kDA porin gene expression and carbapenem resistance in the isolates (Tables 1 and 2). It is possible that the carbapenem resistance in the isolates was due to mutational alterations in the porin structures instead of changes in the expression levels (26). In addition, carbapenem resistance could also be a result of carbapenemase activity in the resistant isolates, the production of which was not analyzed in the present study.
Although the approach used in the present study only reveals RND pump-encoding genes that are transcribed constitutively due to regulatory mutations or the experimental conditions used, to our knowledge, it is the first to analyze the simultaneous expression of all known RND pump- and carbapenem resistance-associated outer membrane porin-encoding genes in clinical isolates of Acinetobacter species. We analyzed expression of all characterized and as yet uncharacterized RND pump-encoding genes in Acinetobacter species, and also analyzed the expression of all three porin-encoding genes known to be involved in the antibiotic resistance of Acinetobacter species. Although we did not observe any correlation between the expression of RND pump- and porin-encoding genes, most likely due to the presence of other resistance mechanisms, we conclude that the overexpression of RND pump-encoding genes and downregulation of porin-encoding genes appeared to be quite common in clinical isolates of Acinetobacter species from Canadian hospitals.
Footnotes
FUNDING: The present study was supported by grants from the Natural Science and Engineering Research Council, the Canada Foundation for Innovation and the Ontario Research Foundation.
REFERENCES
- 1.Gerner-Smidt P, Tjernberg I, Ursing J. Reliability of phenotypic tests for identification of Acinetobacter species. J Clin Microbiol. 1991;29:277–82. doi: 10.1128/jcm.29.2.277-282.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee YC, Huang YT, Tan CK, et al. Acinetobacter baumannii and Acinetobacter genospecies 13TU and 3 bacteraemia: Comparison of clinical features, prognostic factors and outcomes. J Antimicrob Chemother. 2011;66:1839–46. doi: 10.1093/jac/dkr200. [DOI] [PubMed] [Google Scholar]
- 3.Bergogne-Berezin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–65. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jain R, Danziger LH. Multidrug-resistant Acinetobacter infections: An emerging challenge to clinicians. Ann Pharmacother. 2004;38:1449–59. doi: 10.1345/aph.1D592. [DOI] [PubMed] [Google Scholar]
- 5.Bergogne-Berezin E. Acinetobacterspecies. In: Yu VL, Weber R, Raoult D, editors. Antimicrobial therapy and vaccines. 2nd edn. New York: Apple Trees Production, LLC; 2002. pp. 11–8. [Google Scholar]
- 6.Gaynes R, Edwards JR. Overview of nosocomial infections caused by Gram-negative bacilli. Clin Infect Dis. 2005;41:848–54. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 7.Abbo A, Navon-Venezia S, Hammer-Muntz O, et al. Multidrug-resistant Acinetobacter baumannii. Emerg Infect Dis. 2005;11:22–9. doi: 10.3201/eid1101.040001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hujer KM, Hujer AM, Hulten EA, et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother. 2006;50:4114–23. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akers KS, Chaney C, Barsoumian A, et al. Aminoglycoside resistance and susceptibility testing errors in Acinetobacter baumannii-calcoaceticus complex. J Clin Microbiol. 2010;48:1132–8. doi: 10.1128/JCM.02006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamouda A, Amyes SGB. Novel gyrA and parC point mutations in two strains of Acinetobacter baumannii resistant to ciprofloxacin. J Antimicrob Chemother. 2004;54:695–6. doi: 10.1093/jac/dkh368. [DOI] [PubMed] [Google Scholar]
- 11.Mussi MA, Relling VM, Limansky AS, Viale AM. CarO, an Acinetobacter baumannii outer membrane protein involved in carbapenem resistance, is essential for L-ornithine uptake. FEBS Letts. 2007;581:5573–8. doi: 10.1016/j.febslet.2007.10.063. [DOI] [PubMed] [Google Scholar]
- 12.Lin L, Ling B-D, Li X-Z. Distribution of the multidrug efflux pump genes, adeABC, adeDE and adeIJK, and class 1 integron genes in multiple-antimicrobial-resistant clinical isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus complex. Int J Antimicrob Agents. 2009;33:27–32. doi: 10.1016/j.ijantimicag.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 13.Higgins PG, Lehmann M, Wisplinghoff H, Seifert H. gyrB multiplex PCR To differentiate between Acinetobacter calcoaceticus and Acinetobacter genomic species 3. J Clin Microbiol. 2010;48:4592–4. doi: 10.1128/JCM.01765-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. M7-A7, Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, approved standard. 7th edn. 2006. [Google Scholar]
- 15.Fernando D, Kumar A. Growth phase-dependent expression of RND efflux pump- and outer membrane porin-encoding genes in Acinetobacter baumannii ATCC 19606. J Antimicrob Chemother. 2012;67:569–72. doi: 10.1093/jac/dkr519. [DOI] [PubMed] [Google Scholar]
- 16.Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001;45:3375–80. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damier-Piolle L, Magnet S, Bremont S, Lambert T, Courvalin P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob Agents Chemother. 2008;52:557–62. doi: 10.1128/AAC.00732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coyne S, Rosenfeld N, Lambert T, Courvalin P, Perichon B. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54:4389–93. doi: 10.1128/AAC.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chau SL, Chu YW, Houang ET. Novel resistance-nodulation-cell division efflux system AdeDE in Acinetobacter genomic DNA group 3. Antimicrob Agents Chemother. 2004;48:4054–5. doi: 10.1128/AAC.48.10.4054-4055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coyne S, Guigon G, Courvalin P, Perichon B. Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrob Agents Chemother. 2009;54:333–40. doi: 10.1128/AAC.01037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bratu S, Landman D, Martin DA, Georgescu C, Quale J. Correlation of antimicrobial resistance with beta-lactamases, the OmpA-like porin, and efflux pumps in clinical isolates of Acinetobacter baumannii endemic to New York City. Antimicrob Agents Chemother. 2008;52:2999–3005. doi: 10.1128/AAC.01684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortez-Cordova J, Kumar A. Activity of the efflux pump inhibitor phenylalanine-arginine beta-naphthylamide against the AdeFGH pump of Acinetobacter baumannii. Int J Antimicrob Agents. 2011;37:420–4. doi: 10.1016/j.ijantimicag.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Bornet C, Davin-Regli A, Bosi C, Pages JM, Bollet C. Imipenem resistance of Enterobacter aerogenes mediated by outer membrane permeability. J Clin Microbiol. 2000;38:1048–52. doi: 10.1128/jcm.38.3.1048-1052.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez-Alles S, Alberti S, Alvarez D, et al. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiol. 1999;145:673–9. doi: 10.1099/13500872-145-3-673. [DOI] [PubMed] [Google Scholar]
- 25.Mussi MA, Limansky AS, Viale AM. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: Natural insertional inactivation of a gene encoding a member of a novel family of beta-barrel outer membrane proteins. Antimicrob Agents Chemother. 2005;49:1432–40. doi: 10.1128/AAC.49.4.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siroy A, Cosette P, Seyer D, et al. Global comparison of the membrane subproteomes between a multidrug-resistant Acinetobacter baumannii strain and a reference strain. J Prot Res. 2006;5:3385–98. doi: 10.1021/pr060372s. [DOI] [PubMed] [Google Scholar]


