Abstract
The Neotropics harbors a high diversity of species and several hypotheses have been proposed to account for this pattern. However, while species of forested domains are frequently studied, less is known of species from open vegetation formations occupying, altogether, a larger area than the Amazon Forest. Here we evaluate the role of historical barriers and the riverine hypothesis in the speciation patterns of small mammals by analyzing an ancient rodent lineage (Thrichomys, Hystricomorpha). Phylogenetic and biogeographic analyses were carried out with mitochondrial and nuclear DNA markers to analyze the evolutionary relationships between Thrichomys lineages occurring in dry domains along both banks of the Rio São Francisco. This river is one of the longest of South America whose course and water flow have been modified by inland tectonic activities and climate changes. Molecular data showed a higher number of lineages than previously described. The T. inermis species complex with 2n = 26, FN = 48 was observed in both banks of the river showing a paraphyletic arrangement, suggesting that river crossing had occurred, from east to west. A similar pattern was also observed for the T. apereoides complex. Thrichomys speciation occurred in Late Miocene when the river followed a different course. The current geographic distribution of Thrichomys species and their phylogenetic relationships suggested the existence of frequent past connections between both banks in the middle section of the Rio São Francisco. The extensive palaeodune region found in this area has been identified as a centre of endemism of several vertebrate species and is likely to be a center of Thrichomys diversification.
Introduction
The Neotropics harbors a high diversity of species [1] across different biomes, from forest to open vegetation formations. Several hypotheses for explaining its biodiversity, like the refugia and the riverine barrier hypotheses have been tested resulting in contradictory results [2]–[6]. The riverine hypothesis was postulated based on the distribution of primate species with respect to the major Amazonian rivers [7]. This hypothesis predicted that sister taxa would be separated by rivers and that gene flow was more likely to occur in narrow headwater regions rather than downriver sites [8], [9].
Studies of the mammalian fauna across extensive regions of Brazil, a country with both forested and open biomes, will contributed to a better understanding of mammalian speciation timing, in view of its controversial dating to the Tertiary or Quaternary [1], [10]. Furthermore, South American open vegetation domains occupy, altogether, a larger area [11] and may harbor a larger number of mammal species and of endemic species than Amazonia [12], a reason why its biodiversity deserves special attention. Our study focuses on two less frequently studied biomes, the Cerrado and Caatinga.
The Cerrado is the largest open vegetation biome in South America, encompassing an area of approximately 20% of the Brazilian territory and small enclaves in Bolivia and Paraguay [13], [14]. It is the second largest South American biome and one of the most threatened tropical savannas in the world [14], [15]. The Caatinga is one of the largest areas of Seasonally Dry Tropical Forests (SDTFs). It is a poorly studied dry domain encompassing an area of approximately 800,000 Km2 and entirely located in Brazil.
Species distribution, biogeography and patterns of historic diversification of open vegetation domains have been recently reviewed by Werneck [11]. This author suggested that the origin and patterns of biodiversity could not be attributed to one or few events during key time intervals. It most likely resulted from complex ecologic and evolutionary trends triggered by Neogene tectonic events and palaeogeographic reorganizations maintained by Quaternary climatic changes and vegetation fluctuations. These areas, infrequently included in phylogeographic studies [16], have become a matter of recent studies [17]–[21] which resulted in earlier estimates of divergence and cryptic diversity.
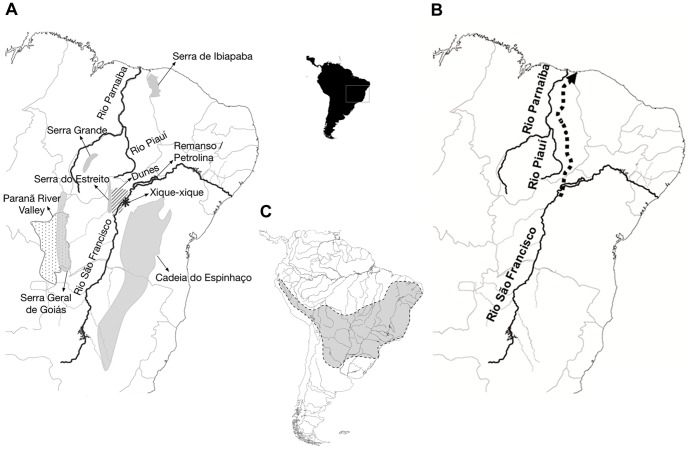
The Rio São Francisco flows through portions of the Cerrado, Caatinga and part of the Atlantic Forest. This river is one of the longest of South America, with the third largest river basin in Brazil, covering an area of approximately 645,000 Km2 (nearly 7.6% of the Brazilian territory) [22], [23] within the limits of the São Francisco craton [24]. The maximum width and depth of this river accounts for 850 m and 80 m, respectively, and its annual average flow has been estimated as 2,850 m3/s [22]. These characteristics support the proposition that the Rio São Francisco is a barrier to gene flow for several animal taxa. Due to inland tectonic activities, this river is likely to have changed its course [25]–[28] although, presently, it flows towards the north, curving abruptly towards the southeast and to the Atlantic Ocean (Figure 1A). Mabesoone [27] postulated that this river previously flowed in a different direction, probably connecting with the current Rio Piauí and Rio Parnaíba to the equatorial Atlantic Ocean (Figure 1B). This has been supported by the finding of the same gravel deposits of the middle section of the Rio São Francisco and the dry gap between this river and Rio Piauí. Mabesoone [27] also suggested that the course of the Rio São Francisco was interrupted by the uplift of Serra Grande and Ibiapaba cuestas (Figure 1A), subsequently becoming endorheic (stagnated, forming lakes due to lack of outflow) in the Remanso-Petrolina area (Figure 1A), and later changing to its present water course during the Mindel glaciation (ca. 450.000 years ago).
Figure 1. Maps with geological and floristic characteristics showing (A) main rivers and mountain chains; (B) hypothetical course (dotted line) of the Rio São Francisco before switching to its present course; and (C) extent of the "Pleistocenic Arc" following Prado and Gibbs [97].
(A) Map showing the main rivers and mountain chains mentioned in the text; (B) Map showing a hypothetical course (dotted line) for the Rio São Francisco before changing to its present course; (C) Map showing the extent of the “Pleistocenic Arc” following Prado and Gibbs [97].
During the dry periods of the Late Pleistocene, sand islands emerged due to the lower water level of the Rio São Francisco, favoring the formation of wind-activated dunes [24]. In the Early Holocene, the water volume of the Rio São Francisco was augmented due the increase of humidity and, in the Middle Holocene, the decrease of humidity due to a dry and hot climate favored once again wind activities and expansion of the caatinga vegetation throughout the dunes [24]. At this time, the water volume of the Rio São Francisco probably decreased, once again forming islands similar to the present islands near Xique-Xique municipality [24]. Cyclic climate changes during the Quaternary suggest that the middle course of the Rio São Francisco, close to the sand dunes, is still more prone to forming islands and decreasing its water volume. Furthermore, Barreto et al. [29] proposed that the large sand thickness of the dunes area may be attributed to Early Quaternary or even to Late Tertiary events.
To evaluate the riverine hypothesis and how historical barriers have influenced mammalian speciation patterns, we analyzed the evolution of the genus Thrichomys based on mitochondrial and nuclear DNA. This rodent genus occurs only in open vegetation biomes and is mainly found in sandy soils and granitic formations including lowland lajeiros and elevated serrotes and serras [30], [31]. Thrichomys comprises at least five allopatric species, T. fosteri, T. pachyurus, T. inermis, T. laurentius and T. apereoides; the last three representing a species complex [32]–[34]. Thrichomys laurentius, T. apereoides and T. inermis are distributed in the Caatinga and Cerrado domains, in both banks of the Rio São Francisco, providing valuable insights on the process of speciation along this river.
Methods
Ethics Statement
Small mammals were collected and handled according to recommended safety procedures [35]. Permits for field collection were provided by the Instituto Chico Mendes de Conservação da Biodiversidade (Permit Numbers: 02001.003618/03-06, 02001.006721/2004 and 11375-1). Animal handling procedures were approved by the Animal Research Ethics Committee of the Oswaldo Cruz Foundation, Rio de Janeiro, RJ (CEUA P-0336-07).
Each capture station was sampled with Sherman (7.62×9.53×30.48 cm) or Tomahawk (40.64×12.70×12.70 cm) live traps, placed 10 m apart, in linear transects on the ground. The bait was a mixture of bacon, peanut butter, banana, and oatmeal. Traps were set in the late afternoon and inspected in the early morning.
Each animal was anesthetized and euthanized by intramuscular injection of ketamine hydrochloride (Ketalar; Laboratorio ELEA S.A.C.I.F. y A.) based on an anesthetic protocol according to allometric scaling [36].
Sample collection and karyotyping
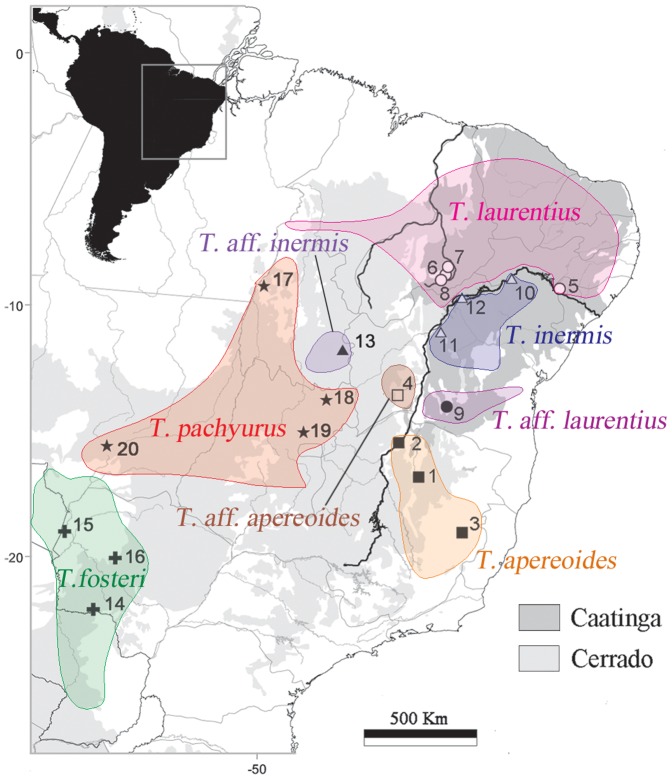
Forty-eight Thrichomys were collected in 12 Brazilian municipalities in the Cerrado and Caatinga biomes (Table S1 and Figure 2). We followed the species nomenclature of Braggio and Bonvicino [33], Borodin et al. [37] and reassessed the present nomenclature in view of our findings: T. apereoides (2n = 28, FNa = 50 located at the right bank of the Rio São Francisco), T. aff. apereoides (2n = 28, FNa = 52 at the left bank), T. laurentius (2n = 30, FNa = 54 at the left bank), T. aff. laurentius (2n = 30, FNa = 54 at the right bank), T. inermis (2n = 26, FNa = 48 at the right bank), T. aff. inermis (2n = 26, FNa = 48 at the left bank), T. pachyurus (2n = 30, FNa = 56) and T. fosteri (2n = 34, FNa = 64). Thrichomys laurentius was not recognized as a valid species in the latest mammal compilation [38] although a more recent report [37] clearly showed that hybrids between T. apereoides and T. laurentius were not fertile, supporting species validation. Recently, T. laurentius has been recognized as a valid species in the latest Brazilian mammal checklist [39]. In addition, the karyomorphotype 2n = 34, FNa = 64 was considered T. pachyurus in the latest mammal compilation. However, specimens collected in Paraguay at Piribebuy, 35 km North of the type locality of T. fosteri, showed 2n = 34 and specimens collected by us in the type locality of T. pachyurus (Cuiabá, Mato Grosso state) showed 2n = 30, FNa = 56. An outline of Thrichomys distribution is shown in Figure 2.
Figure 2. Location of sites of collection of Thrichomys (see Table S1 for a list of sites) and outline of Thrichomys distribution (modified from [110]).
Nineteen specimens were karyotyped in this study (Table S1). Chromosome preparations were obtained from bone marrow cultures in RPMI 1640 medium with 20% fetal calf serum, ethidium bromide [5 μg/mL] and colchicine [10−6 M] for two hours. Estimates of fundamental number were restricted to autosome pairs. Voucher specimens are in the mammal collections of the Museu Nacional (MN; Universidade Federal do Rio de Janeiro), and the Laboratório de Biologia e Parasitologia de Mamíferos Reservatórios Silvestres (LBCE; IOC, Fiocruz). Other acronyms refer to field number of C. R. Bonvicino (CRB).
Gene choice, DNA extraction, PCR assays and sequencing
The haploid mitochondrial genome is uniparently inherited in most animals, is not subjected to recombination and evolves at a faster rate than nuclear DNA. It accounts for a lower effective population size (Ne) when compared to nuclear DNA (nuDNA). These characteristics make mitochondrial DNA (mtDNA) a useful material for evolutionary studies, especially for recently diverged taxa [40]–[42]. On the other hand, noncoding nuDNA harbors a higher variation than coding regions, and is also valuable for analyzing rapidly evolving taxa with a good phylogenetic resolution. Furthermore, the majority of nucleotide positions in introns are free to vary unlike protein coding sequences [43], [44]. Although poorly resolved phylogenies can result by nuDNA analyses due to few informative sites, some studies have shown its utility in phylogenetic reconstructions (for a review see [43]). In this study, we analyzed the mitochondrial cytochrome b gene (cytb) and the nuclear β-fibrinogen intron 7 (FGB) of Thrichomys.
DNA was isolated from 48 liver samples preserved in 100% ethanol with GenomicPrep™ Cells and Tissue DNA Isolation Kit (GE Healthcare, Brazil) or with the standard phenol-chloroform protocol [45].
The cytb (1,140 bp) was PCR-amplified using primers L14724 [46] and MVZ14 [47] following a pre-denaturation step at 94°C for 2 min and three series of cycles, of 10, 10 and 15 cycles at 94°C for 30 sec, annealing at 55°C for 30 sec, 53°C for 30 sec, and 51°C for 30 sec respectively, extension at 72°C for 90 sec, and final extension of 72°C for 2 min.
The FGB (ca. 800 bp) was amplified in a sample subset (Table S1) with primers β17-mammL and βfib-mammU [48] following a pre-denaturation step at 94°C for 3 min, 5 cycles of denaturation at 94°C for 45 sec, annealing at 62°C (decreasing 0.4°C per cycle) for 45 sec and extension at 72°C for 45 sec; 31 cycles of denaturation at 94°C for 45 sec, annealing at 60°C for 45 sec and extension at 72°C for 45 sec with a final extension of 72°C for 5 min. Due to lack of FGB sequences in GenBank, FGB was amplified in single specimens of Myocastor coypus (GenBank JX459849), Makalata macrura (JX459850) and Makalata sp. (JX459851) to use as outgroups in phylogenetic reconstructions.
Amplicons were purified with GFX™ PCR DNA and Gel Band Purification kit (GE Healthcare). Cytochrome b amplicons were labeled with primer L14724 and internal primers TricR (5′-TGTGATGACTGTCGCTCCTC-3′), cytbTrich (5′-CCCTACATCGGACCTTCACT-3′) and MVZ16 [47], while FGB amplicons were labeled with primers β17-mammL and βfib-mammU. Sequencing was carried out with ABI PrismTM 377 and MegaBACE 1000 (GE Healthcare) platforms. Electropherograms were manually checked using Bioedit (version 7.0.8.0) [49].
Alignment and genetic distance estimation
A total of 70 Thrichomys cytb were analyzed. These included the complete (1,140 bp) cytb sequence of three T. laurentius, two from Coronel José Dias municipality (AY083333, AY083334) and another from João Costa municipality (AY083332) previously analyzed by Braggio and Bonvicino [33] based on 817, 1,109 and 801 base pairs, respectively. Twenty-two of these 70 sequences have been previously published [33], [50], [51] (Table S1). Furthermore, one sequence of Ctenomys boliviensis (AF155869), Ctenomys maulinus (AF370703), Euryzygomatomys spinosus (U34858), Dasyprocta leporina (AF437789), Hydrochoeris hydrochaeris (GU136721), Erethizon dorsatum (FJ357428), Proechimys cuvieri (AY206631), Makalata macrura (EU302702) and Myocastor coypus (EU544663) were used as outgroups. A total of 18 FGB sequences (ca. 800 bp), representative of each Thrichomys lineages, were analyzed. One sequence of Myocastor coypus (JX459849) and Makalata macrura (JX459850) were also included in phylogenetic analyses. All sequences were manually aligned with Bioedit.
Pairwise genetic distances were estimated using the modified Log-Det for closely related taxa [52] using Mega 5 [53].
Gene tree estimation
A DNA substitution model was selected prior to phylogenetic reconstructions using Modelgenerator version 0.85 [54] and the Bayesian information criterion (BIC).
Gene tree estimation was based on cytb and FGB carried out with maximum likelihood (ML) using PhyML version 3.0 [55] and Bayesian inference (BI) using MrBayes 3.2 [56].
The ML tree topology was searched with the best of Nearest Neighbor Interchange and Subtree Pruning and Regrafting algorithms from five random, starting trees generated by the BioNJ algorithm [55], [57]. Branch support was estimated using the approximate likelihood ratio test with Shimodaira-Hasegawa-like interpretation (SH-aLRT), a procedure as conservative and accurate as bootstrapping but less computationally intensive [57]–[59]. For BI, analyses were run for 4,000,000 generations with sampling every 100. Acceptable mixing and convergence to the stationary distribution were checked with Tracer 1.5 [60] and the first 10% were discarded as burn-in. A consensus tree was then generated. We used five complete cytb sequences of T. laurentius (one representative of each location).
Species tree and divergence date estimation
To estimate the Thrichomys species tree we used the *Beast (StarBeast) method [61] implemented in Beast 1.7.0 [62] using both cytb and FGB genes and unlinked substitution models, clock models and trees. An uncorrelated lognormal relaxed clock [63] and a Yule tree prior [64] were used and estimates of posterior distribution were obtained by Markov chain Monte Carlo (MCMC) sampling every 5,000 MCMC steps over a total of 100,000,000 steps. As a higher number of cytb sequences were generated, and to avoiding a bias resulting from missing data, the species tree was reconstructed based on specimens for which cytb and FGB data were available.
We calibrated Thrichomys divergence based on its sister fossil taxon Pampamys emmonsae, Echimyidae from Cerro Azul Formation, La Pampa Province, Argentina, of Chasicoan-Huayquerian age [65]–[67]. This accounted for a minimum age constraint of 6.0 Ma as suggested by Olivares et al. [65] and a maximum age constraint of 10 Ma based on the boundary between the Chasicoan and Huayquerian ages. Estimates of divergence times were calibrated using a lognormally distributed prior for the divergence of Thrichomys. Lognormal prior was preferred in view of its most appropriate distribution to summarize paleontologic information [68].
We included a user generated start tree based on a nonparametric rate smoothing (NPRS) transformed consensus tree estimated with MrBayes. Acceptable mixing and convergence to the stationary distribution were checked using Tracer and the first 10% were discarded as burn-in. The maximum clade credibility (MCC) tree was computed with TreeAnnotator and using mean heights for divergence date estimates.
Phylogeographic analysis
In order to reconstruct the phylogeographic history of Thrichomys species through time and in continuous space (by using location coordinate data for specimens), we used the relaxed random walk (RRW) approach proposed by Lemey et al. [69]. We used the Cauchy RRW model implemented in Beast and all other parameters were set as previously described in the species tree and divergence date estimation section. Because some specimen coordinates were duplicated we used a jitter option of 0.50. Estimates of the posterior distribution were obtained by MCMC sampling every 5,000 MCMC steps over a total of 50,000,000 steps. This analysis was performed for cytb excluding identical haplotypes and outgroups.
We also included a user generated start tree based on a nonparametric rate smoothing (NPRS) transformed consensus tree estimated with MrBayes. Acceptable mixing and convergence to the stationary distribution were also checked using Tracer and the first 10% were discarded as burn-in. The MCC tree was computed with TreeAnnotator and using mean heights for divergence date estimates.
To summarize the posterior distribution of ancestral locations using the Cauchy RRW model we annotated nodes in a MCC tree using the program TreeAnnotator. This MCC tree was used as input for the program Spread 1.0.4 [70] to generate a keyhole markup language (KML) file containing the reconstructed dispersal route paths to visualize in Google Earth. Although this reconstructed route of dispersal has been superimposed on a contemporary map rather than on an unavailable Tertiary map, it provides useful information on locations of ancestral Thrichomys populations and their distribution along SDTFs.
Biogeographic reconstructions
In order to reconstruct the biogeographic history of Thrichomys we employed a parsimony-based reconstruction method using a modified version of the dispersal-vicariance analysis (DIVA) [71]–the S-DIVA (Statistical DIVA). Analyses were carried out with cytb data with Rasp [72] and implementing the “Bayes-DIVA” approach [73]. We used the complete tree distribution obtained with the phylogeographic Beast analysis without the first 10% as burn-in to account for phylogenetic uncertainty and the MCC tree. The maximum area at each node was set to 2 [74]. Biogeographic regions were defined a priori (Table S1 and Figure S3).
Migration rates
To further test whether the Rio São Francisco might be a potential barrier to gene flow we estimated migration rates (M) between populations using both IMa2 [75], [76] and Migrate [77], [78] with the cytb dataset. Because we analyzed populations of different Thrichomys species, we compared population pairs: (1) between river banks and sharing the same karyotype (T. apereoides versus T. aff. apereoides, and T. inermis versus T. aff. inermis), (2) from the same river bank and geographically close (T. pachyurus versus T. fosteri, T. pachyurus versus T. aff. inermis, and T. inermis versus T. apereoides), (3) from the same river bank but geographically distant (T. laurentius versus T. fosteri). If the river were a barrier to gene flow, then lower migration rates would be expected between population pairs from opposite banks than between population pairs from the same bank.
After preliminary runs to check convergence of parameters, IMa2 was run in the L-mode with different start seeds and an initial burn-in of 100,000 steps followed by saving 500,000 genealogies every 100th MCMC steps. Migrate was run in the Bayesian inference mode with an initial burn-in of 50,000 steps for 500,000 (or 2,000,000) MCMC steps. We used the static heating with four chains sampled at every tenth interval using the temperature scheme suggested with the character # as described in the Migrate tutorial [79].
Results
Karyologic analyses
Thrichomys aff. apereoides from the left bank of the Rio São Francisco showed 2n = 28 and FNa = 52, while T. apereoides from the right bank of this river showed 2n = 28 and FNa = 50. Thrichomys inermis from the right bank of the Rio São Francisco and T. aff. inermis from the left bank of this river showed the same 2n = 26 and FNa = 48 karyotype. Thrichomys pachyurus from type locality showed 2n = 30 and FNa = 56. Thrichomys laurentius and T. aff. laurentius also showed the same 2n = 30 and FNa = 54 karyotype. Finally, T. fosteri showed 2n = 34 and FNa = 64 (Table S1).
Descriptive analyses of cytb and FGB intron
Table S1 lists sequence data from all Thrichomys specimens belonging to eight lineages herein analyzed. Analysis of cytb and FGB showed 59 and 10 haplotypes, respectively, none of them being shared between different taxa. We also observed a higher proportion of variable and parsimony-informative sites for cytb (24.3% and 18.7%, respectively) than FGB (4.1% and 2.9%, respectively).
Pairwise genetic distances for cytb within T. apereoides (from right bank of the Rio São Francisco) were higher (0.0–1.9%) than within T. aff. apereoides (from left bank) (0.0–0.3%), while genetic distance estimates between these lineages ranged from 2.8–3.9%. Pairwise genetic distances within T. laurentius (from left bank) ranged from 0.0–3.1% unlike T. aff. laurentius (from right bank) which showed a single haplotype. Genetic distance between these lineages ranged from 3.7–4.6%. Pairwise genetic distances within T. inermis (from right bank) were higher (0.0–9.6%) than within T. aff. inermis (from left bank) (0.0–1.3%) while estimates between these lineages equaled 7.5–10.4%. Genetic distance within T. fosteri and T. pachyurus equaled 0.0–1.5% and 0.0–2.9% respectively.
The FGB in T. apereoides specimens from the right bank showed a single haplotype that differed from the one found in the single specimen of T. aff. apereoides from the left bank. Genetic distance between these two lineages was 0.6%. FGB in T. laurentius from the left bank showed a single haplotype that differed from the single haplotype of T. aff. laurentius from the right bank. Genetic distances between these two lineages were 1.1%. Pairwise genetic distances in T. inermis from right bank were higher (0.0–1.6%) than in T. aff. inermis from left bank (0.0–0.1%), while estimates between these lineages ranged from 1.1–1.3%. Single haplotypes were observed in T. fosteri and T. pachyurus.
Gene trees
Phylogenetic trees based on cytb were constructed with HKY [80] and gamma distribution as DNA substitution model. Both ML and Bayesian trees (Figure S1) showed very similar topologies; all branches were well supported except for T. aff. inermis (branch support≈0.60; Figure S1). All analyses showed a paraphyletic arrangement for T. inermis between opposite banks of the Rio São Francisco despite sharing the same karyotype with conventional staining. In view of these findings, T. inermis from the left bank was renamed T. aff. inermis. A close relationship was also observed between T. aff. apereoides and T. aff. laurentius, currently separated by the Rio São Francisco. All analyses were also consistent in showing T. inermis, currently located in the right bank, as the most basal lineage.
Phylogenetic trees based on FGB (Figure S2), constructed with K80 [81] and gamma distribution as DNA substitution model yielded less robust resolution of the evolutionary relationships of Thrichomys. Branch support for the node between T. fosteri and the remaining lineages of the clade was low (0.67; Figure S2) as was the case of the node between T. pachyurus and the remaining lineages of the clade (0.52; Figure S2). These low estimates might have resulted from a lower number of variable and parsimony-informative sites in FGB than in cytb. Despite these limitations, however, FGB topologies were consistent in showing T. inermis as the most basal lineage.
Species tree, phylogeographic analysis and divergence dating
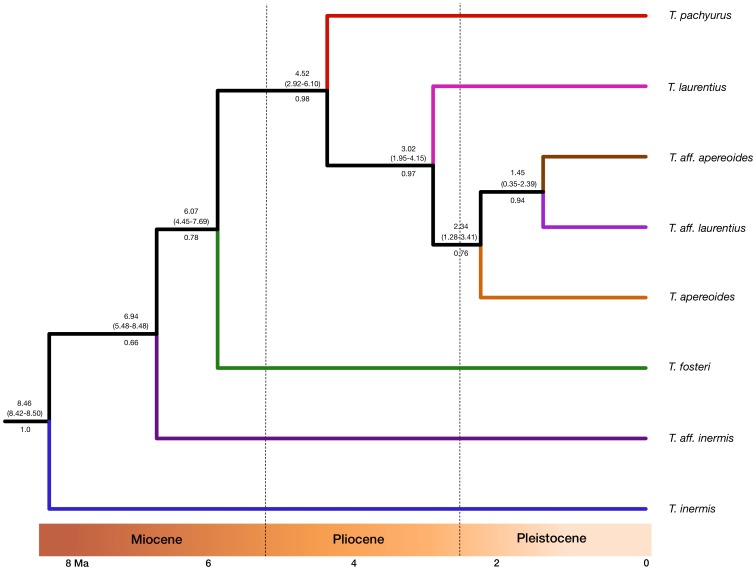
The Thrichomys species tree (Figure 3) obtained with *Beast showed an identical topology to the cytb gene tree (Figure S1). All branches were well supported except for those leading to T. aff. inermis, T. fosteri and T. apereoides.
Figure 3. MCC phylogeny representing Thrichomys species tree generated with the *Beast method.
Numbers above branches are mean height and 95% HPD interval (in brackets) of date estimates and numbers below branches are posterior probability values. This analysis was generated using Beast. Branch colors follow same colors of Figure 2.
Divergence time estimates showed that the ancestral Thrichomys population had been present in the Late Miocene [8.46 Ma (highest posterior density (HPD) = 8.42 to 8.50 Ma)] and that most lineages emerged in the Plio-Pleistocene (Figure 3). Interestingly, the ancestors of each Thrichomys lineage apparently originated at similar times, as indicated by overlapping between 95% HPD estimates (Figure 3). Thrichomys apereoides, T. aff. apereoides, T. laurentius and T. aff. laurentius comprise a clade that diverged in the Pliocene [3.02 Ma (1.95 to 4.15)] (Figure 3).
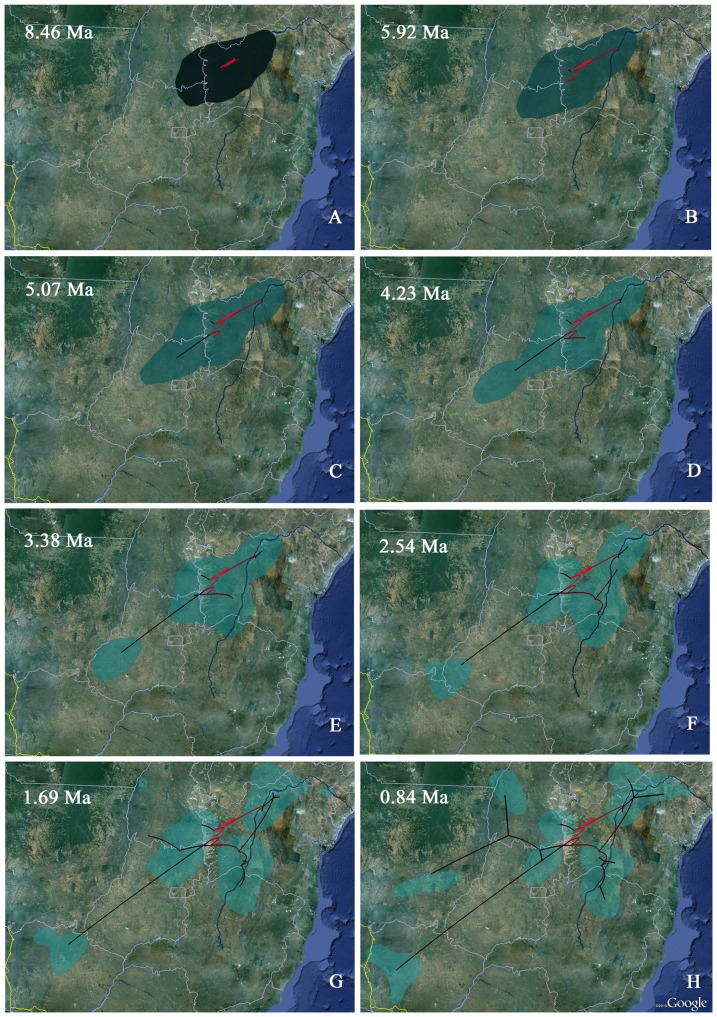
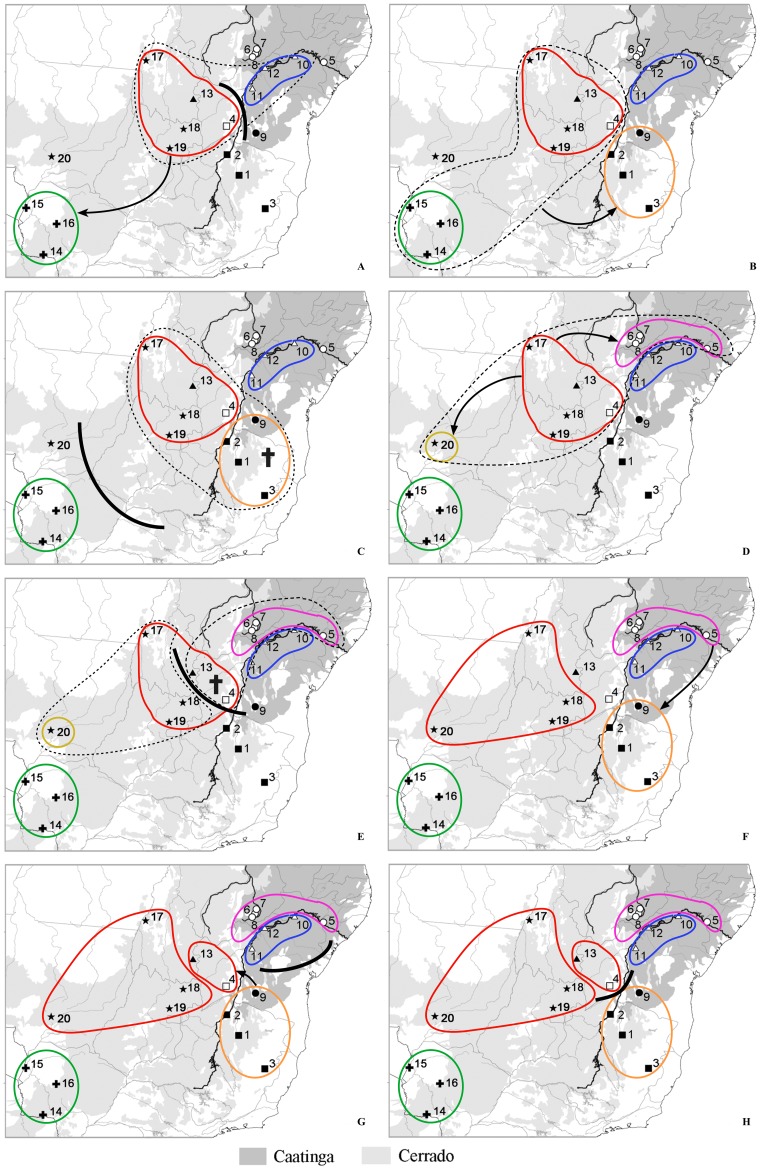
Phylogeographic analysis suggested that Thrichomys originated in central Caatinga and northern Cerrado, in both banks of the Rio São Francisco (Figure 4A). A similar area to the one presently existing remained unchanged for a long period, probably until 5.92 Ma (Figure 4B), while, approximately 5.07 to 4.23 Ma ago, a range expansion probably occurred towards the southwest (Figure 4C and 4D). The ancestral Thrichomys population split in two demes at approximately 3.38 Ma (Figure 4E). Approximately 2.54 Ma, the ancestral population expanded (Figure 4F) and, approximately 1.69 Ma, six ancestral populations emerged, one of them accounting for the current distribution of T. fosteri (Figure 4G).
Figure 4. Spatiotemporal dynamics of Thrichomys suggesting they originated in both banks of the Rio São Francisco from 8.46 to 5.92 Ma (A–B), a posterior range expansion occurred at ca.
5.07 to 4.23 Ma ago (C–D), a population split in two demes at ca. 3.38 Ma (E), another expansion of an ancestral population at ca. 2.54 Ma (F), and the emergence of six ancestral populations at ca. 1.69 Ma (G) that coexisted for approximately 0.84 Ma (H). Lines represent the MCC tree branches projected on the surface. Maps are based on satellite pictures made available with Google Earth. The current course of the Rio São Francisco is highlighted.
Six ancestral populations coexisted approximately 0.84 Ma, one of them still accounting for the current distribution of T. fosteri, three other to the current distribution of T. pachyurus and T. aff. inermis, a fifth one to the current distribution of T. laurentius and T. inermis, and a sixth one to the current distribution of T. aff. laurentius, T. apereoides and T. aff. apereoides (Figure 4H).
Biogeographic reconstructions
S-DIVA analyses indicated that the ancestral Thrichomys population originated with a probability of 65% in the central Caatinga and/or the northern Cerrado, probably in both banks of the Rio São Francisco. It also indicated that a vicariant event separated this ancestral population but soon after the population of the northern Cerrado dispersed to the southern Cerrado (Figure 5A). These two populations dispersed to the southern Caatinga across the river (Figure 5B), followed by a vicariant event separating the population of the southern Cerrado from the northern Cerrado and southern Caatinga, accounting for the current distribution of T. fosteri (Figure 5C).
Figure 5. Biogeographic hypotheses of colonization history of Thichomys based on S-DIVA analysis using RASP (A–H).
Solid line polygons represent the different ancestral populations; dotted line polygons represent past connected populations; solid lines represent vicariant events; crosses represent extinctions; arrows represent dispersions. Sites of collection are the same as Figure 2 and Table S1 and they are represented here for clarity.
Populations of the southern Caatinga and the one from northern Cerrado became extinct in the southern Caatinga (Figure 5C) and, subsequently, the remaining ancestral population of the northern Cerrado dispersed to the central Cerrado and northern Caatinga (Figure 5D). Soon after, a vicariant event split the ancestral populations of the northern Cerrado forming two separate populations: one with the population of the central Cerrado and another with the population of the northern Caatinga (Figure 5E).
Part of the ancestral population of the northern Cerrado which dispersed to the northern Caatinga became extinct (Figure 5E). The population of the northern Caatinga probably originated the current T. laurentius and dispersed to the southern Caatinga (Figure 5F), accounting for a second colonization event in this area. Soon after, a vicariant event separated populations of the northern from the southern Caatinga (Figure 5G), an event likely related to the initial alterations of the river course.
The ancestral population of the southern Caatinga dispersed to the northern Cerrado (Figure 5G) followed by a vicariant event separating both populations (Figure 5H).
Migration rates
Migrate detected low migration between population pairs (Table 1) while IMa2 did not detect migrations, suggesting that migration did not occur between any population pairs.
Table 1. List of populations tested with estimates of migration rates from Migrate.
| Pairs of populations tested | aM2→1 | aM1→2 |
| T. apereoides and T. aff. apereoides | 95.0 (0.0–830.7) | 145.0 (0.0–870.0) |
| T. inermis and T. aff. inermis | 52.3 (0.0–848.0) | 467.0 (3.3–900.7) |
| T. fosteri and T. pachyurus | 37.0 (0.0–771.3) | 15.7 (0.0–843.3) |
| T. pachyurus and T. aff. inermis | 15.0 (0.0–777.3) | 31.0 (0.0–847.3) |
| T. inermis and T. apereoides | 189.0 (8.7–863.3) | 461.0 (136.0–984.7) |
| T. laurentius and T. fosteri | 21.7 (0.0–866.7) | 29.0 (0.0–777.3) |
Modes are reported, as curves were positively skewed. Numbers in brackets represent 95% confidence intervals. Subscripts 1 and 2 represent the order in which species were listed in the first column.
Discussion
Mitochondrial and nuclear gene tree discordances and the gene tree versus species tree
Our results showed a disagreement between cytb and FGB gene trees, a common result in phylogenetic and phylogeographic studies (for a review see [82]). One reason for this pattern was attributed to long-term isolation followed by current secondary population contact or population range contact at some point in the past [82]. Our analyses showed Thrichomys as a very ancient lineage that possibly exchanged migrants in the past because of fluctuation in the river water volume allowing crossings between river banks (see below).
Furthermore, discordances between mitochondrial versus nuclear gene trees can be a consequence of the effective population size (Ne), which in most circumstances is smaller for mtDNA [83]. In general, smaller Ne increases the probability of congruence between gene and species trees and, in most cases, mtDNA will be more likely congruent to the species tree [84]. However, for some extreme scenarios a nuDNA would be favored over mtDNA for obtaining the species tree [84], [85]. Our results showed a species tree (Figure 3) identical to that using the cytb gene (Figure S1) suggesting that for Thrichomys, mtDNA would be a reliable choice for recovering the species tree. However, other nuclear genes should be analyzed for a better resolution of this assumption.
Climate change, diversification processes and the Rio São Francisco
Our results suggest that the ancestral Thrichomys population initially appeared in the Late Miocene [8.46 Ma (8.42 to 8.50 Ma)]. This very ancient origin presumably explained why Migrate and IMa2 were incapable of detecting migration between population pairs. Lack of gene flow also indicated that all eight Thrichomys evolutionary lineages (including the affinis species) are well defined and reproductively isolated species.
Our estimates of Thrichomys divergence differed from previous reports postulating an earlier origin [86]–[88]. Leite and Patton [86] used the same fossil record (Pampamys emmonsae) as the one used in this report albeit dated to the Huayquerian age (6.8 to 9.0 Ma) following Verzi et al. [67]. Galewski's et al. [87] single constraint for Caviomorph radiation was based on the fossil record of the Dasyproctidae and Chinchilidae from the Tinguiririca fauna of Chile dated between 31.5 to 37.5 Ma [89], [90]. Although their dating of Thrichomys/Proechimys divergence was estimated as 13.3 Ma, their 95% credibility interval was very high (7.4 to 20.7 Ma), probably due to relying on a single and ancient calibration point at the root of their phylogenetic tree. Upham and Patterson [88], based on five calibration points, dated the splitting of Thrichomys from Hoplomys/Proechimys to approximately 12 to 16 Ma. However, this node was not calibrated, a reason why the origin of Thrichomys could not be precisely estimated. Conversely, in our report, the node of the Thrichomys clade was calibrated based on the fossil record of its sister taxon P. emmonsae (see Candela and Rasia [91]) and the whole suite of congeneric species was analyzed.
Our results also suggest that the described scenario of water volume oscillations of the Rio São Francisco also occurred in other geological periods, like the Miocene. These oscillations lead to a decrease of the water volume and island formations around 10°–12° S, close to the sand dunes of the middle section of this river, suggesting ancient connections between river banks in the region close to the dunes and influencing the distribution and diversification of Thrichomys. This diversification occurred after the geologic origin of the Rio São Francisco, dated to the Early-to-Mid Cretaceous [92]. Furthermore, the species tree (Figure 3) suggests that the riverine hypothesis does not hold for Thrichomys species, with the exception of T. aff. apereoides and T. aff. laurentius. These two lineages are more related to each other and are currently separated by the river, following the riverine hypothesis.
The paraphyletic arrangement of T. inermis complex observed in phylogenetic analyses suggested two different evolutionary lineages. This taxon crossed the middle section of the Rio São Francisco, probably from east to west in view that T. inermis from Bahia (in the right bank of the river) appeared as the most basal and oldest lineage. This could also explain why a similar karyotype was found in populations of both banks of the river and why these populations are more closely related, implying a past continuous contact between them as indicated by phylogeographic analysis, at least before 0.84 Ma (Figure 4). Thrichomys aff. inermis, currently located in left bank of the Rio São Francisco, was collected in Novo Jardim, in areas of SDTF enclaves of the Cerrado, located in the Paranã River Valley (Figure 1A). This valley encompasses an area of 5,940,382 ha in southeast Tocantins and northeast Goiás, in a transition area of the Cerrado, Caatinga and Amazon Forest [93], [94], and delimited in the east by the Serra Geral de Goiás (Figure 1A), a mountain chain between the Cerrado and Caatinga [95]. This region shows a high diversity of phytophysiognomies and is one of the most heterogeneous SDTF enclaves of Brazil [93]. These SDTFs currently show a disjunct distribution, a likely relict of a more extensive and continuous region hypothesized as the “Pleistocenic Arc” [11], [96], [97] (Figure 1C). This arc, evident by the distribution of tree species [97], might have ranged from northeastern to southeastern Brazil, northwestern Argentina, southwestern Bolivia and northwards to the dry Andean valleys of Peru, indicated that populations showed a more continuous and wide distribution in the past. Recently, it has been shown that, during the Late Pleistocene, dry regions were more disjunct than present ones, suggesting their previous expansion in the Early Pleistocene or end of the Tertiary, subsequently fragmented in the last glacial period and later undergoing a further expansion in the Holocene [11]. In this scenario, Thrichomys populations from the Caatinga might have colonized regions of the Cerrado (Figure 5), occupying present SDTF enclaves, a pattern suggested by our biogeographic analysis in view that most Thrichomys lineages emerged in the Plio-Pleistocene. Subsequently, these populations became isolated, followed different evolutionary histories and suffered differentiation, establishing endemic centers [11], [15], [98]. Plant, lizard, bird and mammal species endemic to STDF enclaves have been reported [13], [15], [98]–[100]. Furthermore, lizard species endemic to Caatinga were found in these enclaves, being completely isolated from other populations of Caatinga [98], suggesting a dynamic historic connection between the Cerrado and Caatinga, a pattern similar to the one observed for T. inermis and T. aff. inermis populations, both with 2n = 26, FNa = 48. These findings suggested that a vicariant event separated the ancestor of these Thrichomys lineages (Figure 5). This ancestor probably occurred along the “Pleistocenic Arc” and later split when STDFs became discontinuous, originating two isolated populations.
Further evidence on the coincidence between climate change and dispersal across the Rio São Francisco was provided by the close relationship of T. aff. apereoides and T. aff. laurentius, both from Bahia state and currently separated by this river, as well as the close relation of T. laurentius from the left bank with other species from the right bank (T. aff. laurentius and T. apereoides). This pattern corroborated Mabesoone's [27] hypothesis that the course of the Rio São Francisco differed from the present one due to its opening to the equatorial Atlantic Ocean rather than to the northeast. A ancient river course similar to the present one would result in a closer relationship of T. laurentius with species of the left bank (T. aff. inermis, T. pachyurus and T. fosteri) rather than to species of the right bank.
The Rio São Francisco is also a natural geographic barrier influencing the genetic diversification and speciation of different taxa, like lizard species [101]–[103], rodents [104] and sand flies [105]. The middle course of the Rio São Francisco and its paleodunes appear to be a dry refuge for open vegetation inhabitants. We postulate that this dry refuge is a center of endemism in this region, well documented by one mammal, one bird species, 20 new species and four new genera of squamates [106]–[108]. This dry refuge apparently contributed to speciation patterns of Thrichomys, with the oldest species, T. inermis, occurring near the paleodune area.
The dry morphoclimatic domains and Thrichomys diversity
The open vegetation formations of the Caatinga and Cerrado domains were probably reduced to isolated patches associated to speciation of some faunal taxa during the Quaternary-Tertiary, affecting the distribution of T. apereoides, T. inermis and T. laurentius species complexes. Most localities of T. apereoides (2n = 28, FNa = 50), including its type locality, are found in the right bank of the Rio São Francisco [32], [33], [109], mainly in the Caatinga domain, but also in the Cerrado, while T. aff. apereoides (2n = 28, FNa = 52) occurs in the left bank of this river in the Cerrado [32]. Similarly, all T. inermis localities, including its type locality, are in the right bank of the Rio São Francisco, restricted to the Caatinga, while the left bank T. aff. inermis, is restricted to the Cerrado. However, T. laurentius complex, although separated in two evolutionary lineages, does not follow this clear pattern. All localities of T. laurentius, including its type locality, are found in the left bank of this river contrary to all localities of T. aff. laurentius in the right bank [33], both in the Caatinga domain. The relief around the middle course of the Rio São Francisco is complex with several mountain chains, like the Cadeia do Espinhaço, Serra Geral de Goiás and Serra do Estreito (Figure 1A) which are apparent barriers to Thrichomys.
Conclusion
The diversification in the studied area cannot be explained by the riverine hypothesis. Due to the older age of the Rio São Francisco compared to Thrichomys origin, it is more likely that the pattern observed was a consequence of climate changes and arid periods in the middle section of the river. The cyclic increase and decrease of water volume appear to have allowed for interruptions of gene flow or, alternatively, crossings across riverbanks, influencing speciation and biological diversity. Our results also suggest a role of the ancient course of the river in the diversification of Thrichomys. Contrary to forested domains where a high biodiversity is frequently found in a small geographic area, diversity in open vegetation domains appear to be reduced to isolated patches, like a mosaic with different species compositions showing a variety of diversity levels, associated to speciation of some animal taxa during the Quaternary-Tertiary and affecting the distribution of T. apereoides, T. inermis and T. laurentius species complexes. This finding has implications for conservation, suggesting that a higher number of conservation units should be created in the Cerrado to account for the preservation of extant biodiversity.
Supporting Information
Maximum likelihood phylogeny for cytb of Thrichomys. Similar topology was observed for Bayesian analysis. Numbers close to branches are SH-aLRT followed by posterior probability (pp) values. When identical values are observed, only one value is shown.
(PDF)
Maximum likelihood phylogeny for FGB of Thrichomys. Similar topology was observed for Bayesian analysis. Numbers close to branches are SH-aLRT followed by pp values. When identical values are observed, only one value is shown.
(PDF)
Summary of Bayes-DIVA analysis of Thrichomys. The tree is a MCC phylogeny generated with Beast for cytb. Circles at nodes show probabilities of ancestral ranges. Only the higher probabilities are shown. When two biogeographic regions are underlined in a node, it represents that ancestral range was at both regions. Biogeographic regions: A: southern Caatinga; B: northern Caatinga; C: central Caatinga; D: southern Cerrado; E: northern Cerrado; F: central Cerrado (see map in top left and Table S1).
(PDF)
List of Thrichomys specimens included in this study, their haplotype number (H), GenBank accession number, field or museum identification number (ID), localities, karyotypes and Biogeographic regions (Bio Regions).
(PDF)
Acknowledgments
We are grateful to H. N. Seuánez (INCA, Brazil) for reviewing a previous version of the manuscript. We are thankful to Dr. S. Y. W. Ho (University of Sydney, Australia) for discussion on Bayesian analysis and divergence time estimates, to Dr. P. Beerli (Florida State University, USA) for discussion on Migrate and IMa analyses, and to Dr. Philippe Lemey (Rega Institute, Belgium) for discussion on phylogeographic analyses. We appreciated the facilities provided by staffs of Serra da Capivara national park, the owners of Fazenda Jatobá and Fazenda São Francisco da Trijunção, and the state secretary of Health of Ceará and Mato Grosso states. The collaboration in fieldwork by the field team of the Laboratório de Biologia e Parasitologia de Mamíferos Silvestres Reservatórios, IOC/FIOCRUZ and our collaborators, was most useful. Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) granted license to collect the specimens.
Funding Statement
Work supported by CNPq (http://www.cnpq.br/) fellowships to FFN, CRB and PSD; FAPERJ (http://www.faperj.br/) to CRB, ALGS, ANM and JCM; and CAPES (http://www.capes.gov.br/) to ALGS and JCM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rull V (2008) Speciation timing and neotropical biodiversity: the Tertiary-Quaternary debate in the light of molecular phylogenetic evidence. Mol Ecol 17: 2722–2729. [DOI] [PubMed] [Google Scholar]
- 2. Lessa EP, Cook JA, Patton JL (2003) Genetic footprints of demographic expansion in North America, but not Amazonia, during the Late Quaternary. Proc Natl Acad Sci USA 100: 10331–10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anthony NM, Johnson-Bawe M, Jeffery K, Clifford SL, Abernethy KA, et al. (2007) The role of Pleistocene refugia and rivers in shaping gorilla genetic diversity in central Africa. Proc Natl Acad Sci USA 104: 20432–20436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haffer J (1969) Speciation in amazonian forest birds. Science 165: 131–137. [DOI] [PubMed] [Google Scholar]
- 5. Gascon C, Malcolm JR, Patton JL, da Silva MNF, Bogart JP, et al. (2000) Riverine barriers and the geographic distribution of Amazonian species. Proc Natl Acad Sci USA 97: 13672–13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jalil MF, Cable J, Sinyor J, Lackman-Ancrenaz I, Ancrenaz M, et al. (2008) Riverine effects on mitochondrial structure of Bornean orangutans (Pongo pygmaeus) at two spatial scales. Mol Ecol 17: 2898–2909. [DOI] [PubMed] [Google Scholar]
- 7. Wallace AR (1852) On the monkeys of the Amazon. Proc Zool Soc London 20: 107–110. [Google Scholar]
- 8.Patton JL, da Silva MNF (1998) Rivers, refuges and ridges. The geography of speciation of Amazonian mammals. In: Howard DJ, Berlocher SH, editors. Endless forms: Species and speciation. New York: Oxford University Press. pp. 202–213. [Google Scholar]
- 9. Moritz C, Patton JL, Schneider CJ, Smith TB (2000) Diversification of rainforest faunas: an integrated molecular approach. Annu Rev Ecol Syst 31: 533–563. [Google Scholar]
- 10. Hoorn C, Wesselingh FP, ter Steege H, Bermudez MA, Mora A, et al. (2010) Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science 330: 927–931. [DOI] [PubMed] [Google Scholar]
- 11. Werneck FP (2011) The diversification of eastern South American open vegetation biomes: Historical biogeography and perspectives. Quat Sci Rev 30: 1630–1648. [Google Scholar]
- 12. Mares MA (1992) Neotropical mammals and the myth of amazonian biodiversity. Science 255: 976–979. [DOI] [PubMed] [Google Scholar]
- 13.Oliveira-Filho AT, Ratter JA (2002) Vegetation physiognomies and woody flora of the Cerrado biome. In: Oliveira PS, Marquis RJ, editors. The Cerrados of Brazil: Ecology and natural history of a neotropical savanna. New York: Columbia University Press. pp. 91–120. [Google Scholar]
- 14. Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- 15. Silva JMC, Bates JM (2002) Biogeographic patterns and conservation in the South American Cerrado: A tropical savanna hotspot. BioScience 52: 225–233. [Google Scholar]
- 16. Beheregaray LB (2008) Twenty years of phylogeography: the state of the field and the challenges for the Southern Hemisphere. Mol Ecol 17: 3754–3774. [DOI] [PubMed] [Google Scholar]
- 17. Almeida FC, Bonvicino CR, Cordeiro-Estrela P (2007) Phylogeny and temporal diversification of Calomys (Rodentia, Sigmodontinae): implications for the biogeography of an endemic genus of the open/dry biomes of South America. Mol Phylogenet Evol 42: 449–466. [DOI] [PubMed] [Google Scholar]
- 18. D'Elía G, Pardiñas UFJ, Jayat JP, Salazar-Bravo J (2008) Systematics of Necromys (Rodentia, Cricetidae, Sigmodontinae): species limits and groups, with comments on historical biogeography. J Mammal 89: 778–790. [Google Scholar]
- 19. Duputié A, Salick J, McKey D (2011) Evolutionary biogeography of Manihot (Euphorbiaceae), a rapidly radiating Neotropical genus restricted to dry environments. J Biogeogr 38: 1033–1043. [Google Scholar]
- 20. Prado CP, Haddad CF, Zamudio KR (2012) Cryptic lineages and Pleistocene population expansion in a Brazilian Cerrado frog. Mol Ecol 21: 921–941. [DOI] [PubMed] [Google Scholar]
- 21. Gamble T, Colli GR, Rodrigues MT, Werneck FP, Simons AM (2012) Phylogeny and cryptic diversity in geckos (Phyllopezus; Phyllodactylidae; Gekkota) from South America's open biomes. Mol Phylogenet Evol 62: 943–953. [DOI] [PubMed] [Google Scholar]
- 22.Godinho AL, Godinho HP (2003) Breve visão do São Francisco. In: Godinho AL, Godinho HP, editors. Águas, peixes e pescadores do São Francisco das Minas Gerais. Belo Horizonte, MG: Pontifícia Universidade Católica de Minas Gerais. pp. 15–24. [Google Scholar]
- 23.Kohler HC (2003) Aspectos geoecológicos da bacia hidrográfica do São Francisco (primeira aproximação na escala 1:1 000 000). In: Godinho HP, Godinho AL, editors. Águas, peixes e pescadores do São Francisco das Minas Gerais. Belo Horizonte, MG: Pontifícia Universidade Católica de Minas Gerais. pp. 25–36. [Google Scholar]
- 24. Barreto AMF (1996) Interpretação paleoambiental do sistema de dunas fixadas do médio Rio São Francisco, Bahia [PhD]. São Paulo: Universidade de São Paulo [Google Scholar]
- 25. Domingues AJP (1948) Contribuição à geologia do sudoeste da Bahia. Rev Bras Geogr 10: 255–287. [Google Scholar]
- 26. Grabert H (1968) Postmesozoische entwässerung und oszillation am ostrande des Brasilianischen schildes. Geol Rundsch 58: 166–190. [Google Scholar]
- 27.Mabesoone JM (1994) Sedimentary basins of Northeast Brazil. Recife: Federal University of Pernambuco. 310 p.
- 28. King LC (1956) A geomorfologia do Brasil Oriental. Rev Bras Geogr 2: 147–265. [Google Scholar]
- 29.Barreto AMF, Suguio K, Oliveira PE, Tatumi SH (2002) Campo de Dunas Inativas do Médio Rio São Francisco, BA-Marcante registro de ambiente desértico do Quaternário brasileiro. In: Schobbenhaus C, Campos DA, Queiroz ET, Winge M, Berbert-Born MLC, editors. Sítios Geológicos e Paleontológicos do Brasil Brasilia: DNPM/CPRM-Comissão Brasileira de Sítios Geológicos e Paleobiológicos (SIGEP). pp. 223–231. [Google Scholar]
- 30. Lacher Jr TE, Alho CJR (1989) Microhabitat use among small mammals in the Brazilian Pantanal. J Mammal 70: 396–401. [Google Scholar]
- 31. Streilein KE (1982) Ecology of small mammals in the semiarid Brazilian Caatinga. I. Climate and faunal composition. Ann Carnegie Mus 51: 79–107. [Google Scholar]
- 32. Bonvicino CR, Otazú IB, D'Andrea PS (2002) Karyologic evidence of diversification of the genus Thrichomys (Rodentia, Echimyidae). Cytogenet Genome Res 97: 200–204. [DOI] [PubMed] [Google Scholar]
- 33. Braggio E, Bonvicino CR (2004) Molecular divergence in the genus Thrichomys (Rodentia, Echimyidae). J Mammal 85: 316–320. [Google Scholar]
- 34.Vilela RV (2005) Estudos em roedores da família Echimyidae, com abordagens em sistemática molecular, citogenética e biogeografia [MSc]. São Paulo: Universidade de São Paulo. 123 p.
- 35. Mills JN, Ksiazek TG, Peters CJ, Childs JE (1999) Long-term studies of hantavirus reservoir populations in the southwestern United States: a synthesis. Emerg Infect Dis 5: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pachaly JR, Brito HFV (2001) Interspecific allometric scaling. In: Fowler ME, Cubas ZS, editors. Biology, medicine, and surgery of South American wild animals. Ames, IA: Iowa State University Press. pp. 475–481. [Google Scholar]
- 37. Borodin PM, Barreiros-Gomez SC, Zhelezova AI, Bonvicino CR, D'Andrea PS (2006) Reproductive isolation due to the genetic incompatibilities between Thrichomys pachyurus and two subspecies of Thrichomys apereoides (Rodentia, Echimyidae). Genome 49: 159–167. [DOI] [PubMed] [Google Scholar]
- 38.Woods CA, Kilpatrick W (2005) Infraorder Hytricognathi. In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. 3rd ed.Baltimore: Johns Hopkins University Press. [Google Scholar]
- 39.Paglia AP, Fonseca GAB, Rylands AB, Herrmann G, Aguiar LMS, et al.. (2012) Annotated checklist of Brazilian mammals. Arlington, VA: Conservation International.
- 40. Harrison RG (1989) Animal mitochondrial DNA as a genetic marker in population and evolutionary biology. Trends Ecol Evol 4: 6–11. [DOI] [PubMed] [Google Scholar]
- 41. Brown WM, George M Jr, Wilson AC (1979) Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA 76: 1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sunnucks P (2000) Efficient genetic markers for population biology. Trends Ecol Evol 15: 199–203. [DOI] [PubMed] [Google Scholar]
- 43. Hare MP (2001) Prospects for nuclear gene phylogeography. Trends Ecol Evol 16: 700–706. [Google Scholar]
- 44. Prychitko TM, Moore WS (2000) Comparative evolution of the mitochondrial cytochrome b gene and nuclear beta-fibrinogen intron 7 in woodpeckers. Mol Biol Evol 17: 1101–1111. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 46. Irwin DM, Kocher TD, Wilson AC (1991) Evolution of the cytochrome b gene of mammals. J Mol Evol 32: 128–144. [DOI] [PubMed] [Google Scholar]
- 47. Silva MN, Patton JL (1993) Amazonian phylogeography: mtDNA sequences variation in arboreal echimyid rodents (Caviomorpha). Mol Phylogenet Evol 2: 243–255. [DOI] [PubMed] [Google Scholar]
- 48. Matocq MD, Shurtliff QR, Feldman CR (2007) Phylogenetics of the woodrat genus Neotoma (Rodentia: Muridae): implications for the evolution of phenotypic variation in male external genitalia. Mol Phylogenet Evol 42: 637–652. [DOI] [PubMed] [Google Scholar]
- 49. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 50. Vilela RV, Machado T, Ventura K, Fagundes V, Silva MJJ, et al. (2009) The taxonomic status of the endangered thin-spined porcupine, Chaetomys subspinosus (Olfers, 1818), based on molecular and karyologic data. BMC Evol Biol 9: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rocha RG, Ferreira E, Costa BMA, Martins ICM, Leite YLR, et al. (2011) Small mammals of the mid-Araguaia River in central Brazil, with the description of a new species of climbing rat. Zootaxa 2789: 1–34. [Google Scholar]
- 52. Tamura K, Kumar S (2002) Evolutionary distance estimation under heterogeneous substitution pattern among lineages. Mol Biol Evol 19: 1727–1736. [DOI] [PubMed] [Google Scholar]
- 53. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 54. Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McLnerney JO (2006) Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 56. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- 58. Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol 55: 539–552. [DOI] [PubMed] [Google Scholar]
- 59. Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O (2011) Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol 60: 685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rambaut A, Drummond AJ (2007) Tracer v1.4. Available: http://beast.bio.ed.ac.uk/Tracer. Acessed 30 Jul 2012.
- 61. Heled J, Drummond AJ (2010) Bayesian inference of species trees from multilocus data. Mol Biol Evol 27: 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Drummond AJ, Ho SY, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gernhard T (2008) The conditioned reconstructed process. J Theor Biol 253: 769–778. [DOI] [PubMed] [Google Scholar]
- 65. Olivares AI, Verzi DH, Vucetich MG, Montalvo CI (2012) Phylogenetic affinities of the Late Miocene echimyid Pampamys and the age of Thrichomys (Rodentia, Hystricognathi). J Mammal 93: 76–86. [Google Scholar]
- 66. Verzi DH, Montalvo CI, Deschamps CM (2008) Biostratigraphy and biochronology of the Late Miocene of central Argentina: Evidence from rodents and taphonomy. Geobios 41: 145–155. [Google Scholar]
- 67. Verzi DH, Vucetich MG, Montalvo CI (1995) Un nuevo Eumysopinae (Rodentia, Echimyidae) del Mioceno tardío de la Provincia de La Pampa y consideraciones sobre la historia de la subfamilia. Ameghiniana 32: 191–195. [Google Scholar]
- 68. Ho SY, Phillips MJ (2009) Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst Biol 58: 367–380. [DOI] [PubMed] [Google Scholar]
- 69. Lemey P, Rambaut A, Welch JJ, Suchard MA (2010) Phylogeography takes a relaxed random walk in continuous space and time. Mol Biol Evol 27: 1877–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bielejec F, Rambaut A, Suchard MA, Lemey P (2011) SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 27: 2910–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ronquist F (1997) Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Syst Biol 46: 195–203. [Google Scholar]
- 72. Yu Y, Harris AJ, He X (2010) S-DIVA (Statistical Dispersal-Vicariance Analysis): A tool for inferring biogeographic histories. Mol Phylogenet Evol 56: 848–850. [DOI] [PubMed] [Google Scholar]
- 73. Nylander JA, Olsson U, Alström P, Sanmartín I (2008) Accounting for phylogenetic uncertainty in biogeography: a Bayesian approach to dispersal-vicariance analysis of the thrushes (Aves: Turdus). Syst Biol 57: 257–268. [DOI] [PubMed] [Google Scholar]
- 74. Kodandaramaiah U (2010) Use of dispersal-vicariance analysis in biogeography-a critique. J Biogeogr 37: 3–11. [Google Scholar]
- 75. Hey J (2010) Isolation with migration models for more than two populations. Mol Biol Evol 27: 905–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hey J, Nielsen R (2007) Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc Natl Acad Sci USA 104: 2785–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Beerli P (2006) Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22: 341–345. [DOI] [PubMed] [Google Scholar]
- 78. Beerli P, Felsenstein J (2001) Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci USA 98: 4563–4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beerli P (2010) Tutorial: Comparison of gene flow models using Bayes factors. Available: http://popgen.sc.fsu.edu/Migrate/Tutorials/Entries/2010/7/12_Day_of_longboarding.html. Acessed 2012 Dec 18.
- 80. Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22: 160–174. [DOI] [PubMed] [Google Scholar]
- 81. Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 82. Toews DP, Brelsford A (2012) The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol 21: 3907–3930. [DOI] [PubMed] [Google Scholar]
- 83. Moore WS (1995) Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene trees. Evolution 49: 718–726. [DOI] [PubMed] [Google Scholar]
- 84. Moore WS (1997) Mitochondrial-gene trees versus nuclear-gene trees, a reply to Hoelzer. Evolution 51: 627–629. [DOI] [PubMed] [Google Scholar]
- 85. Hoelzer GA (1997) Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene trees revisited. Evolution 51: 622–626. [DOI] [PubMed] [Google Scholar]
- 86. Leite YL, Patton JL (2002) Evolution of South American spiny rats (Rodentia, Echimyidae): the star-phylogeny hypothesis revisited. Mol Phylogenet Evol 25: 455–464. [DOI] [PubMed] [Google Scholar]
- 87. Galewski T, Mauffrey JF, Leite YL, Patton JL, Douzery EJ (2005) Ecomorphological diversification among South American spiny rats (Rodentia; Echimyidae): a phylogenetic and chronological approach. Mol Phylogenet Evol 34: 601–615. [DOI] [PubMed] [Google Scholar]
- 88. Upham NS, Patterson BD (2012) Diversification and biogeography of the Neotropical caviomorph lineage Octodontoidea (Rodentia: Hystricognathi). Mol Phylogenet Evol 63: 417–429. [DOI] [PubMed] [Google Scholar]
- 89. Wyss AR, Flynn JJ, Norell MA, Swisher CC, Charrier R, et al. (1993) South America's earliest rodent and recognition of a new interval of mammalian evolution. Nature 365: 434–437. [Google Scholar]
- 90. Flynn JJ, Wyss AR, Croft DA, Charrier R (2003) The Tinguiririca Fauna, Chile: biochronology, paleoecology, biogeography, and a new earliest Oligocene South American Land Mammal ‘Age’. Palaeogeogr Palaeoclimat Palaeoecol 195: 229–259. [Google Scholar]
- 91. Candela AM, Rasia LL (2012) Tooth morphology of Echimyidae (Rodentia, Caviomorpha): homology assessments, fossils, and evolution. Zool J Linn Soc 164: 451–480. [Google Scholar]
- 92. Potter PE (1997) The Mesozoic and Cenozoic paleodrainage of South America: a natural history. J S Am Earth Sci 10: 331–344. [Google Scholar]
- 93. Silva LA, Scariot A (2003) Composição florística e estrutura da comunidade arbórea em uma floresta estacional decidual em afloramento calcário (fazenda São José, São Domingos, GO, bacia do rio Paranã). Acta Bot Bras 17: 305–313. [Google Scholar]
- 94.Werneck FP (2006) Biogeografia e estrutura da comunidade de lagartos de enclaves de Floresta Estacional Decidual de São Domingos-GO (Vale do Paranã) [MSc]: Universidade de Brasília.
- 95. Carvalho-Júnior OA, Hermuche PM, Guimarães RF (2006) Identificação regional da Floresta Estacional Decidual na bacia do rio Paranã a partir da análise multitemporal de imagens MODIS. Rev Bras Geof 24: 319–332. [Google Scholar]
- 96. Pennington RT, Prado DE, Pendry CA (2000) Neotropical seasonally dry forests and Quaternary vegetation changes. J Biogeogr 27: 261–273. [Google Scholar]
- 97. Prado DE, Gibbs PE (1993) Patterns of species distributions in the dry seasonal forests of South America. Ann Mo Bot Gard 80: 902–927. [Google Scholar]
- 98. Werneck FP, Colli GR (2006) The lizard assemblage from Seasonally Dry Tropical Forest enclaves in the Cerrado biome, Brazil, and its association with the Pleistocene Arc. J Biogeogr 33: 1983–1992. [Google Scholar]
- 99. Moojen J, Locks M, Langguth A (1997) A new species of Kerodon Cuvier, 1825 from the State of Goiás, Brazil (Mammalia, Rodentia, Caviidae). Bol Mus Nac 377: 1–10. [Google Scholar]
- 100. Bezerra AMR, Bonvicino CR, Menezes AAN, Marinho-Filho J (2010) Endemic climbing cavy Kerodon acrobata (Rodentia: Caviidae: Hydrochoerinae) from dry forest patches in the Cerrado domain: new data on distribution, natural history, and morphology. Zootaxa 2724: 29–36. [Google Scholar]
- 101.Rodrigues MT (2003) Herpetofauna da Caatinga. In: Leal IR, Tabarelli M, Silva JMC, editors. Ecologia e conservação da Caatinga. Recife: Editora Universitária da Universidade Federal de Pernambuco. pp. 181–236. [Google Scholar]
- 102. Passoni JC, Benozzati ML, Rodrigues MT (2008) Phylogeny, species limits, and biogeography of the Brazilian lizards of the genus Eurolophosaurus (Squamata: Tropiduridae) as inferred from mitochondrial DNA sequences. Mol Phylogenet Evol 46: 403–414. [DOI] [PubMed] [Google Scholar]
- 103. Siedchlag AC, Benozzati ML, Passoni JC, Rodrigues MT (2010) Genetic structure, phylogeny, and biogeography of Brazilian eyelid-less lizards of genera Calyptommatus and Nothobachia (Squamata, Gymnophthalmidae) as inferred from mitochondrial DNA sequences. Mol Phylogenet Evol 56: 622–630. [DOI] [PubMed] [Google Scholar]
- 104. Nascimento FF, Pereira LG, Geise L, Bezerra AM, D'Andrea PS, et al. (2011) Colonization process of the Brazilian common vesper mouse, Calomys expulsus (Cricetidae, Sigmodontinae): a biogeographic hypothesis. J Hered 102: 260–268. [DOI] [PubMed] [Google Scholar]
- 105. Coutinho-Abreu IV, Sonoda IV, Fonseca JA, Melo MA, Balbino VQ, et al. (2008) Lutzomyia longipalpis s.l. in Brazil and the impact of the São Francisco River in the speciation of this sand fly vector. Parasit Vectors 1: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Rocha PLB (1995) Proechimys yonenagae, a new species of spiny rat (Rodentia: Echimyidae) from fossil sand dunes in the Brazilian Caatinga. Mammalia 59: 537–549. [Google Scholar]
- 107. Rodrigues MT, Juncá FA (2002) Herpetofauna of the Quaternary sand dunes of the middle Rio São Francisco: Bahia: Brazil. VII. Typhlops amoipira sp. nov., a possible relative of Typhlops yonenagae (Serpentes, Typhlopidae). Pap Avulsos Zool 42: 325–333. [Google Scholar]
- 108. Lencioni-Neto F (1994) Une nouvelle espèce de Chordeiles (Aves, Caprimulgidae) de Bahia (Brésil). Alauda 62: 241–245. [Google Scholar]
- 109. Pessôa LM, Oliveira JA, Lopes MOG (2004) Karyological and morphometric variation in the genus Thrichomys (Rodentia, Echimyidae). Mamm Biol 69: 258–269. [Google Scholar]
- 110.Bonvicino CR, Oliveira JA, D'Andrea PS (2008) Guia dos roedores do Brasil, com chaves para gêneros basedas em caracteres externos. Rio de Janeiro: Centro Pan-Americano de febre aftosa-OPAS/OMS. 120 p.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum likelihood phylogeny for cytb of Thrichomys. Similar topology was observed for Bayesian analysis. Numbers close to branches are SH-aLRT followed by posterior probability (pp) values. When identical values are observed, only one value is shown.
(PDF)
Maximum likelihood phylogeny for FGB of Thrichomys. Similar topology was observed for Bayesian analysis. Numbers close to branches are SH-aLRT followed by pp values. When identical values are observed, only one value is shown.
(PDF)
Summary of Bayes-DIVA analysis of Thrichomys. The tree is a MCC phylogeny generated with Beast for cytb. Circles at nodes show probabilities of ancestral ranges. Only the higher probabilities are shown. When two biogeographic regions are underlined in a node, it represents that ancestral range was at both regions. Biogeographic regions: A: southern Caatinga; B: northern Caatinga; C: central Caatinga; D: southern Cerrado; E: northern Cerrado; F: central Cerrado (see map in top left and Table S1).
(PDF)
List of Thrichomys specimens included in this study, their haplotype number (H), GenBank accession number, field or museum identification number (ID), localities, karyotypes and Biogeographic regions (Bio Regions).
(PDF)