Abstract
Recent findings provide evidence that tDNAs function as chromatin insulators from yeast to humans. TFIIIC, a transcription factor that interacts with the B-box in tDNAs as well as thousands of ETC sites in the genome, is responsible for insulator function. Though tDNAs are capable of enhancer-blocking and barrier activities for which insulators are defined, new insights into the relationship between insulators and chromatin structure suggest that TFIIIC serves a complex role in genome organization. We review the role of tRNA genes and TFIIIC as chromatin insulators, and highlight recent findings that have broadened our understanding of insulators in genome biology.
Keywords: chromatin, Epigenetics, CTCF, tRNA genes, ETC loci
Introduction
Insulators are a class of DNA regulatory elements defined by their ability to protect genes from position effects in transgene assays. Though numerous examples of insulator-mediated enhancer-blocking and heterochromatin barrier activities have been well studied, in depth interrogation of endogenous insulators and their relationship with the physical and functional organization of eukaryotic genomes paints a far more complex picture. Insulators are enriched at the borders of physical domains in both Drosophila melanogaster and mammals, consistent with a role in chromatin domain organization.1-4 However, neither mutations in or RNAi depletion of insulator proteins in D. melanogaster lead to substantial alterations in chromatin structure or gene activity,5,6 suggesting barrier activity is not a general feature of most endogenous insulator sites. Instead, insulator proteins appear to be involved in mediating long-range inter- and intra-chromosomal arrangements that can direct the nuclear co-localization of specific sequences. For example, insulator proteins localize to both repressive Polycomb (Pc) bodies7 and active transcription factories,8 and have been shown to underlie interactions between Pc target sites9 and the maintenance of H3K27me3 within repressive Pc domains.6 Mapping of interactions facilitated by insulator protein CTCF in mouse embryonic stem cells suggest insulators also contribute to genome organization by forming chromatin loops in which active or repressed genes are harnessed for coregulation, and by facilitating enhancer-promoter interactions.10 Supporting evidence comes from recent analyses of the HOXA locus, wherein developmental regulation of gene expression is accomplished in part by CTCF, which facilitates selective gene activation through chromatin loop formation.11
Though chromatin insulators continue to outgrow the classical barrier and enhancer-blocking roles that operationally defined these elements, these criteria have allowed for identification of the DNA elements and associated proteins required for insulator activity, including the recent demonstration that tRNA genes and TFIIIC act as insulators from yeast to humans.12 tDNA-mediated insulator activity depends on recruitment of RNA polymerase III (RNAP III) transcription factor TFIIIC,13,14 which also targets numerous RNAP III-independent sites that are equally capable of insulator activity when multimerized15,16. The parallel between TFIIIC recruitment to conserved DNA elements and other well characterized insulators, such as CTCF, suggests an exciting and novel role for TFIIIC in genome biology as the most highly conserved insulator complex. Here we review the role of tRNA genes and TFIIIC as chromatin insulators, including their discovery as heterochromatin barriers in yeast, and progress to our current understanding of insulators and their role in genome organization. We end by providing predictions for how tDNA insulators might contribute to chromatin organization and the mechanisms that likely underlie specialization and regulation of TFIIIC insulator function based on our rapidly evolving understanding of insulator proteins in other model systems.
The Most Highly Conserved Insulator
Discovery In Yeast
tRNA genes were first identified as insulators in Saccharomyces cerevisiae,17 wherein deletion of a tRNAThr gene at the transcriptionally silent HMR locus results in the spread of silencing and partial repression of a downstream gene.13 tDNA mediated insulator activity was subsequently demonstrated in Schizosaccharomyces pombe, wherein deletion of a centromeric tRNAAla gene leads to a spread of pericentromeric heterochromatin and gene silencing similar to studies in S. cerevisiae,18 though the mechanisms and components underlying silenced chromatin are distinct between the two species.19 However, not all tRNA or RNAP III transcribed genes are competent insulators, and further analyses revealed an important role for RNAP III transcription factors and tDNA promoter occupancy.13 Promoter organization within tRNA genes and most RNAP III transcribed units includes the A-box and B-box promoter elements, to which RNAP III transcription factor TFIIIC binds and recruits TFIIIB for transcription initiation.20 Point mutations in either A-box or B-box in the HMR tRNAThr boundary results in loss of insulator function, and S. cerevisiae strains mutant in components of TFIIIC or TFIIIB show similar loss of activity,13 suggesting an important role for TFIIIC and TFIIIB in tDNA mediated insulator function. TFIIIC is also essential for tDNA-mediated insulator activity in S. pombe,14 and the recruitment to highly conserved promoter elements is strikingly similar to the recruitment of insulator proteins to cognate regulatory elements in both Drosophila and mammals.
tDNA Insulators In Mammals
Identification and characterization of insulator elements and proteins capable of functional gene insulation began in D. melanogaster thanks to the many advantages of the robust fruit fly model system. Early studies demonstrated the ability of regions flanking the 87A7 heat shock locus, characterized by their specialized chromatin structures and labeled scs and scs’ accordingly,21 to protect reporter genes from chromosomal position effects.22,23 Insulator studies have since identified several proteins required for insulator function in Drosophila, including Zeste-white 5 and Boundary Element Associated Factor of 32 kDa, which are recruited to the scs and scs’ elements respectively,24,25 GAGA factor,26 Suppressor of Hairy-wing,27 and a Drosophila homolog that shares similar domain structure and insulator function with mammalian CTCF28 (Fig. 1). Despite numerous insulator proteins in D. melanogaster, studies in mammals have long been limited to examples of CTCF-mediated insulator activity,29 suggesting, until recently, that CTCF represents the most highly conserved insulator protein. However, two novel studies independently present evidence that tDNAs may also serve as chromatin insulators in mammals, supporting a highly conserved role for tRNA genes and TFIIIC in genome biology.
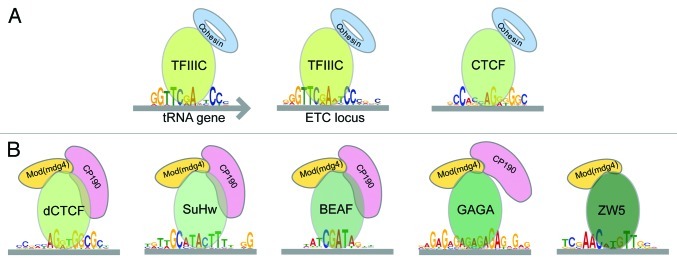
Figure 1. Structure and organization of insulators in eukaryotes. (A) From yeast to mammals, in organisms in which it has been studied, the TFIIIC protein interacts with the B-box sequence in tRNA genes or sites in the genome named ETC sites. TFIIIC interacts with cohesin, which is required for its function. The CTCF insulator, which is present in metazoans but not found in yeast or C. elegans, is composed of the CTCF protein that binds a specific sequence in the DNA and also interacts with cohesin. (B) Drosophila contains several different insulators with similar organization. Each insulator has a different DNA binding protein that recognizes distinct DNA sequences in the genome. dCTCF, Su(Hw) and BEAF interact with two accessory proteins, Mod(mdg4) and CP190, which are required for insulator function. GAGA and ZW5 interact with Mod(mdg4). Many GAGA sites in the genome also contain CP190, suggesting that this two proteins may also interact.
Transgenic reporter assays in murine erythroid leukemia (MEL) cells were used to demonstrate that clusters of tRNA genes are capable of protecting a reporter gene from silencing activity.30 Chromatin immunoprecipitation (ChIP) against RNAP III machinery confirms the enrichment of RNAP III, TFIIIB and TFIIIC, and deletion of A-box promoter elements leads to the spread of silencing chromatin, suggesting the recruitment of TFIIIC is also essential for insulator activity in mammalian cells. In humans, tRNA genes also cluster throughout the genome,31 and share a high degree of syntenic conservation, implying the locations of tDNAs are functionally significant.12 Comparison of tDNAs with transition zones that demarcate repressive chromatin domains, which are characterized by the presence of histone H3 K27 trimethylation (H3K27me3), reveals an enrichment of occupied tDNAs over inactive tDNAs, and analyses of individual loci demonstrate TFIIIC occupancy at these putative tDNA insulators.12 Transgenic reporter assays confirm the ability of human tDNAs to block repression, both in S. pombe and human embryonic kidney cells, and Raab et al. further show that human tDNAs posses enhancer-blocking activities that are dependent on intact B-box promoter elements.
Though intriguing, the correlation of tDNAs at transition zones and ability to function as enhancer-blockers or heterochromatin barriers in transgenic reporter assays shed little insight into the true nature of what roles tDNAs play in chromatin structure and genome organization in mammals. For one, CTCF is also enriched at H3K27me3 domain borders, both in Drosophila and mammals,32,33 yet is not essential for barrier activity at the well characterized β-globin locus,34-36 or at domain borders in D. melanogaster,6 suggesting most insulators do not function as heterochromatin barriers in their natural genomic context. Similarly, many insulators in Drosophila show little enhancer-blocking activity in reporter assays when compared with the gypsy transposon,4 suggesting that most insulators do not function as enhancer-blockers in vivo, or that insulators are finely tuned to function on the enhancers and promoters in their endogenous context, which are likely to vary in strength. Thus, reporter assays can be deceiving if they do not accurately represent the genomic environment of a given chromatin insulator. Meanwhile, short interspersed nuclear element (SINE) retrotransposons and promoters with paused polymerases also possess enhancer-blocking and barrier activities in transgenic reporter assays,37-39 and transition zones involving active domains show strong enrichments for transcriptionally active histone marks,1,2 together suggesting additional factors, including genomic context and recruitment of transcription factors, may play heavily into chromatin boundary formation in an endogenous context. Nevertheless, findings by Kamakaka and colleagues present an exciting possibility, in which tRNA genes and TFIIIC serve conserved and highly important roles both in protein biosynthesis and genome biology.
The TFIIIC Insulator Complex
The TFIIIC transcription factor is a multisubunit complex ultimately composed of six individual proteins, and this subunit composition is conserved from S. cerevisiae to humans.40 TFIIIC conservation parallels that of the promoter elements to which it binds, and several studies suggest TFIIIC localizes to many transcription-independent sites, called ETC (extra TFIIIC) sites, throughout both yeast and human genomes.41,42 In S. pombe, TFIIIC associates with B-box sequences flanking the mating-type (mat) heterochromatin domain independently of RNAP III, and is required for functional insulator activity,43 suggesting TFIIIC may function alone as a competent insulator (Fig. 1). TFIIIC bound ETC sites also possess insulator activity, particularly when multimerized in S. cerevisiae.16,44 However, multiple orphan B-box elements were unable to exhibit barrier activity in transgenic reporter assays conducted in MEL cells,30 suggesting additional factors may be necessary for competent insulator function in mammals, or that barrier assays do not accurately reflect the role of endogenous TFIIIC sites.
Insulators in D. melanogaster and mammals have been extensively characterized by their ability to mediate intra- and inter-chromosomal interactions.10,45-48 In each case, DNA-bound insulator proteins require the recruitment of additional proteins for insulator function, suggesting active insulator complexes are regulated both by the recruitment of insulator proteins to DNA, and the recruitment of essential co-factors.49 Mammalian CTCF specifically recruits the cohesin complex,50,51 a ring-shaped structure that mediates cohesion between sister chromatids from S-phase until mitosis, which may stabilize CTCF-mediated physical interactions through a similar mechanism. Remarkably, S. cerevisiae strains mutant for smc1 and smc3, which are conserved subunits of the cohesin complex, significantly disrupt tDNA mediated insulator function.17 RNAP III independent TFIIIC sites in mouse embryonic stem (ES) cells also associate with cohesins,52 suggesting the TFIIIC insulator complex may function similarly to CTCF through the recruitment of structural proteins. Interestingly, B-box elements in S. cerevisiae also represent loading sites for condensin,53 a cohesin related complex that is also essential for centromeric localization of dispersed RNAP III genes in S. pombe,54 together suggesting TFIIIC insulators likely also participate in three-dimensional genome structure.
In addition to recruiting essential co-factors, insulator proteins can also associate with other, distinct insulators to presumably establish a more robust chromatin insulator complex. For example, in D. melanogaster, CTCF clusters at many sites with BEAF-32 and/or Su(Hw), and these sites commonly flank physical chromatin domains, including repressive H3K27me3 domains.6 However, combinatorial knockdown of insulator proteins disrupts the level of H3K27me3 within rather than outside of these domains, suggesting insulators may align to strengthen long-range interactions important for Polycomb (Pc) mediated gene silencing.6 Though insulator alignment has not yet been observed in mammals, there is preliminary evidence to suggest a similar relationship may exist between TFIIIC and vertebrate CTCF. Mapping of TFIIIC sites in both human and mouse ES cells demonstrate that ETC sites bound by TFIIIC are often located close to CTCF-binding sites.42,52 Meanwhile, prediction of boundaries between topological domains in human cells based on chromatin and transcriptional states provides evidence for CTCF and tDNA enrichment at the boundaries between these domains,55 suggesting CTCF may cluster with TFIIIC, analogous to Drosophila proteins, at the borders of topological chromatin domains. The recruitment of cohesins and co-localization with CTCF at physical domain borders ultimately suggest TFIIIC may play an equally important role in eukaryotic genome organization.
Roles In Genome Organization
Microscopy-based interrogation of insulator proteins and genome-wide mapping of physical interactions provide mounting evidence that insulators are critical players in three-dimensional genome organization. In both D. melanogaster and mammals, insulator proteins interact with and localize to nuclear substructures, including the nuclear and nucleolar peripheries,56 and coalesce into distinct nuclear foci termed insulator bodies,7,57 together suggesting insulators interact and direct the localization of associated chromatin to defined nuclear compartments. Supporting evidence comes from recent demonstration that insulators underlie long-range Pc interactions and are important for the maintenance of H3K27me3 levels within Pc domains in Drosophila.6,9,58 Meanwhile, insulator proteins are distributed across the genome at thousands of sites,32,59-63 and are significantly enriched at the borders of lamina-associated domains,64,65 consistent with microscopy-based staining of perinuclear insulator bodies. Numerous studies have also characterized the ability of CTCF and Drosophila insulator proteins to mediate locus specific interactions,45-48 and recent genome-wide profiling of CTCF interactions in mouse ES cells suggests CTCF facilitates coregulation of related genes by establishing chromatin loops enriched for active or repressive epigenetic signatures, and by bridging interactions between enhancers and promoters.10
The recruitment of cohesin complexes to TFIIIC sites and the putative relationship between TFIIIC and CTCF would suggest a similar role for tDNAs in genome organization. Indeed, characterization of tDNA insulators has drawn many parallels to features implicating CTCF and Drosophila insulator proteins in nuclear architecture. Beyond having a similar genome-wide distribution, dispersed tRNA genes cluster in the nucleolus66 in a condensin-dependent manner,67 and immunofluorescent staining of TFIIIC in S. pombe reveals perinuclear and nucleolar proximal bodies, suggesting TFIIIC sites also cluster into insulator bodies.43 Perinuclear co-localization of two alleles depends on intact B-box elements, suggesting TFIIIC is essential for tethering target loci, termed Chromosome-Organizing Clamps (COC), to the nuclear periphery.43 Perinuclear positioning may also rely on nuclear pore proteins (NUPs), which commonly influence gene activity by regulating the intranuclear position of a given locus.68 For example, the silent HMR domain localizes to the nuclear periphery in S. cerevisiae, a feature that depends on NUPs, which localize to the tDNA bordering the HMR locus.69 TFIIIC has also been shown to direct RNAP III independent ETC sites to the nuclear periphery in S. cerevisiae, and induced degradation of subunit Tfc3 using an auxin-based degron system causes release, confirming that TFIIIC is directly involved in tethering.70 Perinuclear recruitment of ETC sites also relies on Mps3, an inner nuclear membrane domain protein, which effectively competes away peripheral tethering when overexpressed,70 suggesting a direct or indirect interaction with TFIIIC at the nuclear periphery. Interestingly, localization of ETC sites to the nuclear periphery is not essential for insulator activity, implying that the roles of TFIIIC in genome organization may be separable from the observed boundary function in yeast.
Beyond microscopy-based observations of TFIIIC mediated positioning, recent developments in genomic strategies for assaying chromosomal interactions have allowed an unprecedented look into the principles governing three-dimensional folding principles of interphase chromosomes,71,72 allowing for an unbiased query of the relationship between insulators and chromatin organization. In one study, Noble and colleagues recently devised a chromosome conformation capture (3C) based high-throughput derivative to map cis- and trans- interactions across the entire genome in S. cerevisiae. Interactions between tRNA genes are significantly enriched, and tDNAs generally co-localize into two clusters associated with the nucleolus or centromeres.73 Analogous determination of genome organization in both D. melanogaster and humans has revealed chromosomal organization in the form of discrete physical domains that can be epigenetically defined by chromatin and transcriptional signatures.1-3 Drosophila insulator proteins, mammalian CTCF, and tRNA genes are all enriched at the borders of physical domains genome-wide,1,2 providing evidence that in addition to classical insulator activities, tDNAs are distributed similarly to CTCF. Targeted mapping of interactions to a specific tDNA cluster in humans further demonstrated that tDNAs preferentially interact with other tDNAs, as well as ETC loci, in mammals.12 These findings provide compelling evidence that, like CTCF and other insulator proteins, TFIIIC establishes insulator-insulator interactions and thereby influences genome structure on a global scale.
Perspectives
New strategies for assaying genome structure and the improved accessibility of high resolution chromatin profiling have allowed for rapid growth in our understanding of how chromosomes are physically and functionally arranged, and to what degree chromatin insulators play a role in facilitating genome organization. Recent studies provide compounding evidence that insulators indeed play a large-scale role beyond the scope of simple enhancer-blocking and barrier activities that operationally defined these elements, but also raise questions concerning how individual insulator complexes are regulated and specialized. In particular, though insulator proteins are enriched at sites bordering discrete physical domains, they are also found dispersed within domains, leading one to ask what makes individual insulators different. Insulator studies in Drosophila suggest that the answer may involve cooperative binding between different classes of insulators, and through regulation of the recruitment of DNA-binding insulator proteins and additional co-factors. These findings lead us to speculate that future studies may uncover similar mechanisms underlying tDNA insulator activity, including potential collaboration with insulator protein CTCF.
Insulator Collaboration
CTCF aligns with Drosophila insulator proteins BEAF-32 and Su(Hw) at the borders of physical chromatin domains in D. melanogaster, where CTCF then becomes enriched for co-factors essential for insulator activity.6 The alignment of discrete insulators may provide advantages with respect to DNA accessibility and recruitment of DNA-binding insulator proteins. In D. melanogaster, CTCF, Su(Hw), and BEAF-32 all commonly function through the recruitment of additional proteins, Centrosomal Protein 190 (CP190) and Modifier of mdg4 (Mod(mdg4).6,74,75 Therefore, clustering may also allow for efficient recruitment of essential co-factors, thereby ensuring a functional and robust multi-insulator complex, perhaps to strengthen long-range chromosomal interactions. Genome-wide mapping of TFIIIC-bound ETC sites in mammals reveals a similar association with CTCF sites,42,52 and both are also enriched at the borders of physical domains in humans,2,3,55 supporting the possibility that insulator collaboration may be a conserved phenomenon. Whether TFIIIC insulator function depends on the cohesin complex in mammals, which is required for CTCF-mediated insulator activity, remains undetermined. However, preliminary studies have identified an association between TFIIIC and cohesins in MES cells,52 and SMC1 and SMC3 cohesin subunits are important for tDNA insulator activity in S. cerevisiae.17 Therefore, we speculate that, like insulators in D. melanogaster, TFIIIC and CTCF may similarly collaborate to efficiently recruit the cohesin complex, and thereby establish a robust multi-insulator complex capable of facilitating stable chromosomal interactions (Fig. 2A). In contrast to sites where CTCF aligns with other Drosophila insulators, independent CTCF sites are dispersed within physical domains, likely contributing to local interactions important for gene regulation. Independent insulator sites are also more susceptible to regulatory stimuli (Chintong Ong and V.G.C, unpublished data), suggesting aligned insulator complexes may have evolved to resist regulatory mechanisms that might otherwise destroy the physical organization of the genome.
Figure 2. Regulation of insulator function. (A) The genomic localization of insulator proteins is cell type-specific, suggesting that insulator function can be controlled by regulated binding of the DNA binding components and accessory proteins. (B) At some sites, which are presumably not functional, the DNA binding protein is present but the accessory proteins are absent. The cell may be able to regulate insulator function at these sites by controlling the recruitment of accessory proteins via covalent modification. (C) In Drosophila, clusters of insulator sites are enriched at the borders of topological chromosome domains and the borders of repressed domains containing H3K27me3. These clusters may represent very strong insulators that play a special role in genome organization. (D) Proposed model for how the CTCF and TFIIIC insulators in vertebrates could partner to create a stronger insulator
Regulation Of Insulator Proteins
The dependence of DNA-binding insulator proteins on additional proteins, such as the cohesin complex, presents an additional regulatory step and form of specialization among insulator sites. For example, chromatin architecture and gene expression at the HOXA locus is developmentally regulated by pluripotency factor OCT4, specifically by controlling cohesin recruitment to CTCF sites.11 Insulator proteins are similarly developmentally coordinated during the ecdysone hormone response in D. melanogaster,49 suggesting TFIIIC might also be regulated through recruitment of cohesins and condensins in both yeast and humans (Fig. 2B). Meanwhile, CpG methylation in mammals can regulate the occupancy of CTCF binding sites, a feature that has been well studied in the context of genomic imprinting.76 Occupancy of DNA-binding insulator proteins appears to also be regulated in D. melanogaster,49 though the mechanisms coordinating DNA-binding remain uncharacterized. ETC sites and tDNA insulators are therefore likely regulated similarly through the occupancy of TFIIIC. Nevertheless, to what degree tDNAs and TFIIIC-bound ETC chromatin insulators are regulated and specialized remain intriguing questions.
Conclusions
The discovery of tDNA insulator function in humans is significant and establishes tRNA genes as serving a highly conserved role in genome biology that parallels its fundamental role in protein biosynthesis. The level of TFIIIC conservation and its apparent role in genome organization perhaps reflects the importance of appropriate genome structure, and the need for chromatin insulators from yeast to humans. Recent advances have greatly extended our understanding of insulators and their role in genome organization, and provide a valuable framework for querying the importance of TFIIIC as a conserved insulator complex in future studies.
Acknowledgments
Research in the authors’ laboratory is supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM035463. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/21579
References
- 1.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz YB, Linder-Basso D, Kharchenko PV, Tolstorukov MY, Kim M, Li HB, et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 2012 doi: 10.1101/gr.138156.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soshnev AA, He B, Baxley RM, Jiang N, Hart CM, Tan K, et al. Genome-wide studies of the multi-zinc finger Drosophila Suppressor of Hairy-wing protein in the ovary. Nucleic Acids Res. 2012;40:5415–31. doi: 10.1093/nar/gks225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Bortle K, Ramos E, Takenaka N, Yang J, Wahi J, Corces V. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res. 2012 doi: 10.1101/gr.136788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacPherson MJ, Beatty LG, Zhou W, Du M, Sadowski PD. The CTCF insulator protein is posttranslationally modified by SUMO. Mol Cell Biol. 2009;29:714–25. doi: 10.1128/MCB.00825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melnik S, Deng B, Papantonis A, Baboo S, Carr IM, Cook PR. The proteomes of transcription factories containing RNA polymerases I, II or III. Nat Methods. 2011;8:963–8. doi: 10.1038/nmeth.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li HB, Müller M, Bahechar IA, Kyrchanova O, Ohno K, Georgiev P, et al. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol Cell Biol. 2011;31:616–25. doi: 10.1128/MCB.00849-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–8. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim YJ, Cecchini KR, Kim TH. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc Natl Acad Sci U S A. 2011;108:7391–6. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raab JR, Chiu J, Zhu J, Katzman S, Kurukuti S, Wade PA, et al. Human tRNA genes function as chromatin insulators. EMBO J. 2011 doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 2001;20:520–31. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott KC, White CV, Willard HF. An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1. PLoS One. 2007;2:e1099. doi: 10.1371/journal.pone.0001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallrath LL, Geyer PK. TFIIIC boxes in the genome. Cell. 2006;125:829–31. doi: 10.1016/j.cell.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 16.Valenzuela L, Dhillon N, Kamakaka RT. Transcription independent insulation at TFIIIC-dependent insulators. Genetics. 2009;183:131–48. doi: 10.1534/genetics.109.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol. 2006;16:119–29. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 19.Haldar D, Kamakaka RT. tRNA genes as chromatin barriers. Nat Struct Mol Biol. 2006;13:192–3. doi: 10.1038/nsmb0306-192. [DOI] [PubMed] [Google Scholar]
- 20.Orioli A, Pascali C, Pagano A, Teichmann M, Dieci G. RNA polymerase III transcription control elements: themes and variations. Gene. 2012;493:185–94. doi: 10.1016/j.gene.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 21.Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985;185:341–58. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 22.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–50. doi: 10.1016/0092-8674(91)90318-S. [DOI] [PubMed] [Google Scholar]
- 23.Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol. 1992;12:2424–31. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–89. doi: 10.1016/0092-8674(95)90008-X. [DOI] [PubMed] [Google Scholar]
- 25.Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohtsuki S, Levine M. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev. 1998;12:3325–30. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–97. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 28.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–70. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebersole T, Kim JH, Samoshkin A, Kouprina N, Pavlicek A, White RJ, et al. tRNA genes protect a reporter gene from epigenetic silencing in mouse cells. Cell Cycle. 2011;10:2779–91. doi: 10.4161/cc.10.16.17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17:620–8. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, et al. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–88. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, et al. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc Natl Acad Sci U S A. 2002;99:6883–8. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao S, Osborne CS, Bharadwaj RR, Pasceri P, Sukonnik T, Pannell D, et al. Retrovirus silencer blocking by the cHS4 insulator is CTCF independent. Nucleic Acids Res. 2003;31:5317–23. doi: 10.1093/nar/gkg742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barkess G, West AG. Chromatin insulator elements: establishing barriers to set heterochromatin boundaries. Epigenomics. 2012;4:67–80. doi: 10.2217/epi.11.112. [DOI] [PubMed] [Google Scholar]
- 37.Raab JR, Kamakaka RT. Insulators and promoters: closer than we think. Nat Rev Genet. 2010;11:439–46. doi: 10.1038/nrg2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chopra VS, Cande J, Hong JW, Levine M. Stalled Hox promoters as chromosomal boundaries. Genes Dev. 2009;23:1505–9. doi: 10.1101/gad.1807309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lunyak VV, Prefontaine GG, Núñez E, Cramer T, Ju BG, Ohgi KA, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–51. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 40.Dumay-Odelot H, Marck C, Durrieu-Gaillard S, Lefebvre O, Jourdain S, Prochazkova M, et al. Identification, molecular cloning, and characterization of the sixth subunit of human transcription factor TFIIIC. J Biol Chem. 2007;282:17179–89. doi: 10.1074/jbc.M611542200. [DOI] [PubMed] [Google Scholar]
- 41.Moqtaderi Z, Struhl K. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol Cell Biol. 2004;24:4118–27. doi: 10.1128/MCB.24.10.4118-4127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–40. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–72. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 44.Simms TA, Dugas SL, Gremillion JC, Ibos ME, Dandurand MN, Toliver TT, et al. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:2078–86. doi: 10.1128/EC.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003;17:664–75. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krivega M, Savitskaya E, Krivega I, Karakozova M, Parshikov A, Golovnin A, et al. Interaction between a pair of gypsy insulators or between heterologous gypsy and Wari insulators modulates Flp site-specific recombination in Drosophila melanogaster. Chromosoma. 2010;119:425–34. doi: 10.1007/s00412-010-0268-7. [DOI] [PubMed] [Google Scholar]
- 47.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–54. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103:10684–9. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, et al. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol Cell. 2011;44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, et al. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 52.Carrière L, Graziani S, Alibert O, Ghavi-Helm Y, Boussouar F, Humbertclaude H, et al. Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Res. 2012;40:270–83. doi: 10.1093/nar/gkr737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–27. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K. Centromeric localization of dispersed Pol III genes in fission yeast. Mol Biol Cell. 2010;21:254–65. doi: 10.1091/mbc.E09-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Lunyak VV, Jordan IK. Genome-wide prediction and analysis of human chromatin boundary elements. Nucleic Acids Res. 2012;40:511–29. doi: 10.1093/nar/gkr750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–8. doi: 10.1016/S1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 57.Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol Cell. 2000;6:1025–35. doi: 10.1016/S1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 58.Comet I, Schuettengruber B, Sexton T, Cavalli G. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc Natl Acad Sci U S A. 2011;108:2294–9. doi: 10.1073/pnas.1002059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–50. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, et al. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nègre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 63.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–45. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 65.van Bemmel JG, Pagie L, Braunschweig U, Brugman W, Meuleman W, Kerkhoven RM, et al. The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS One. 2010;5:e15013. doi: 10.1371/journal.pone.0015013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–14. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–17. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 69.Ruben GJ, Kirkland JG, MacDonough T, Chen M, Dubey RN, Gartenberg MR, et al. Nucleoporin mediated nuclear positioning and silencing of HMR. PLoS One. 2011;6:e21923. doi: 10.1371/journal.pone.0021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hiraga SI, Botsios S, Donze D, Donaldson AD. TFIIIC localizes budding yeast ETC sites to the nuclear periphery. Mol Biol Cell. 2012;23:2741–54. doi: 10.1091/mbc.E11-04-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rajapakse I, Groudine M. On emerging nuclear order. J Cell Biol. 2011;192:711–21. doi: 10.1083/jcb.201010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–7. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell. 2007;28:761–72. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ghosh D, Gerasimova TI, Corces VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 2001;20:2518–27. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ohlsson R, Bartkuhn M, Renkawitz R. CTCF shapes chromatin by multiple mechanisms: the impact of 20 years of CTCF research on understanding the workings of chromatin. Chromosoma. 2010;119:351–60. doi: 10.1007/s00412-010-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]



