Abstract
The proto-oncogene c-Maf has been shown to be an important transcriptional regulator in the differentiation of a number of cellular contexts, like the eye and hematopoietic system. Here we discuss the recent progress made in understanding c-Maf function in the nervous system.
Keywords: c-Maf, Ret, touch, mechanoreceptors, development, skin innervation, pacinian corpuscles, sensory neuron
Introduction
Maf was first identified in the late 1980s as a retroviral oncogene encoded by the avian musculoaponeurotic fibrosarcoma virus AS42.1 The corresponding proto-oncogene c-Maf has cell transforming activity when overexpressed in fibroblasts, and chromosomal translocations of the c-Maf locus that result in overexpression occur in human tumors, particularly in multiple myeloma.2,3 c-Maf encodes a transcription factor that binds DNA directly and recognizes a sequence motif that is known as Maf Recognition Element (MARE). c-Maf contains a basic-leucine-zipper (bZIP) domain, an evolutionary conserved sequence located N-terminal to the basic domain, called extended homology region/ancillary DNA binding domain, and an acidic transactivation domain. The basic domain and the evolutionary conserved ancillary DNA binding domain located N-terminal to the basic domain participate in DNA binding.2,4 The leucine-zipper, a part of the bZIP domain, is responsible for dimerization of c-Maf and is present also in other transcription factors like Jun and Fos. c-Maf forms homo- and heterodimers with other bZIP factors, expanding its regulatory repertoire. In mammals, further paralogs of c-Maf exist; the large Maf proteins MafA, MafB (mutated in the kreisler mouse) and Nrl have a similar domain structure as c-Maf, whereas the small Maf proteins MafF, MafG and MafK lack the N-terminal transactivation domain and act as dimerization partners of other bZIP factors.
Maf transcription factors have been shown to be important transcriptional regulators in a number of cellular contexts, like the eye and lens (c-Maf), hindbrain (MafB), bone (c-Maf) and the hematopoietic system (c-Maf and MafB). Their role in hematopoietic cells has received considerable attention, for instance, the fact that MafB/c-Maf co-operate to suppress self-renewal in terminally differentiated, mature monocytes and macrophages.5,6 Thus, in the absence of MafB/c-Maf, the typical link between cell cycle exit and terminal differentiation is revoked. c-Maf is also expressed in several neuronal cells (Fig. 1) and we recently identified the first function of c-Maf in the development of sensory neurons that we will discuss in depth below.
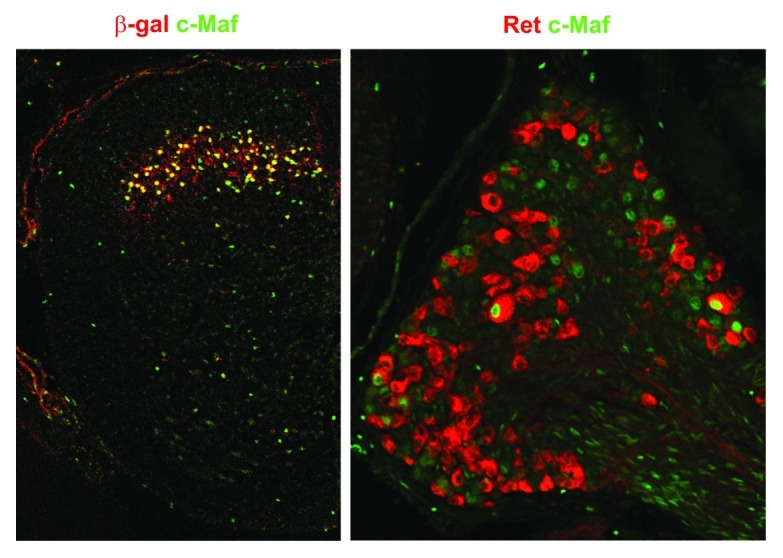
Figure 1. c-Maf expression in the spinal cord and sensory neurons. Analysis of c-Maf expression in the dorsal spinal cord (left) and dorsal root ganglia (right). In the dorsal spinal cord, c-Maf (green) is expressed in layer III neurons that receive mechanosensory information from the periphery. In dorsal root ganglia, c-Maf (green) is expressed in large diameter neurons that co-express Ret (red). For the analysis shown on the left, mice that carry a heterozygous c-MaflacZ allele were used. In c-MaflacZ mice, LacZ is expressed under the control of the c-Maf locus, and expression of the LacZ gene product, β-galactosidase, is useful to follow the expression of the allele.
Development of sensory neurons
Sensory neurons are part of the peripheral nervous system and are located in dorsal root ganglia (DRG). Many functionally distinct sensory neuron subclasses exist that detect a remarkable variety of physical and chemical stimuli, like force, temperature or acid. Sensory neurons are classified into (1) low-threshold mechanoreceptors that end in specialized anatomical structures (e.g., Pacinian and Meissner corpuscles, Merkel cells, laceolate and circumferential endings) and detect innocuous touch, (2) nociceptors that terminate as free nerve endings and sense noxious (i.e., potentially harmful physical, chemical or thermal stimuli) and (3) proprioceptors that innervate muscles and tendons to provide information about their movement and position. According to their electrophysiological properties, mechanoreceptors are classified as rapidly adapting mechanoreceptors that sense for instance vibration, and slowly adapting mechanoreceptors that detect stimuli like indentation and stretch of the skin. Neurotrophin receptors provide classical markers that distinguish sensory neuron subtypes7: Mechanoreceptors express the neurotrophin receptor TrkB and/or Ret (early Ret+ neurons), proprioceptive neurons express TrkC, and nociceptive neurons express TrkA. TrkA is expressed in all nociceptive neurons during early development, but during maturation subpopulations of nociceptors extinguish TrkA and begin to express Ret.
The developmental mechanisms that generate the functional and anatomical diversity of sensory neurons are incompletely understood. Sensory neurons derive from neural crest cells that delaminate from the dorsal neural tube and migrate to condense into dorsal root ganglia. It is now clear that sensory neurons are generated in two major neurogenic waves. During the first wave (E9.5) neural crest cells express the transcription factor neurogenin 2 (Ngn2), which drives formation of mechanoreceptors and proprioceptors. A second wave of neurogenesis (E11-E13) depends on neurogenin 1 (Ngn1) and mostly generates nociceptors.8 Most or all sensory neurons express the homeobox transcription factors Islet1 and brain-specific homeobox/POU domain protein 3A (Brn3a) after they exit the cell cycle and begin to differentiate. Islet1 and Brn3a are essential for the correct expression of many markers of the sensory neuron lineages.9,10 Interestingly, sensory ganglia of Islet1 mutant mice ectopically express transcription factors that are normally found in the central nervous system, indicating that Islet1 suppresses inappropriate differentiation programs.10
The diversification into specialized mechanoreceptor and nociceptor subtypes, however, begins once neurogenesis is completed. For instance, diversification and maturation of nociceptors start around E15.5 and are only completed after birth and depend on the neurotrophin NGF, the transcription factor Runx1 and the tyrosine kinase receptor Met.11-13 Compared with nociceptors, mechanoreceptor mature faster.14 Mechanoreceptors represent a small population of sensory neurons and, until recently, few markers for this lineage were known. The scarcity of molecular data had made it difficult to analyze the development of mechanoreceptors and to define the basis of their diversity and function.
Molecular mechanisms of mechanoreceptor development
The first genes found to drive development and differentiation of mechanoreceptive and proprioceptive neurons were the tyrosine kinase receptor Ret, and the transcription factors Shox2 and Runx3. Ret expression appears early during development of mechanoreceptors, and in the absence of Ret, cutaneous mechanoreceptive end-organs are underdeveloped and Pacinian corpuscles are absent.15,16 The transcription factor short stature homeobox 2 (Shox2) promotes TrkB expression in mechanoreceptors, and Shox2 is required for formation of most TrkB+ neurons. Consistent with this, Shox2 mutants have a deficit in a subset of mechanoreceptors that terminate in Meissner corpuscles and Merkel cells, and light touch sensation is strongly impaired.17,18 Conversely, Runx3 blocks inappropriate gene expression programs in prospective proprioceptive neurons. Runx3 is expressed in TrkC+ proprioceptive neurons, promoting TrkC and suppressing Shox2 in these cells.17,19 The molecular identity of signals controlling Ret, Shox2 and Runx3 expression in early-born sensory neurons is unclear, leaving it open how the diversification of mechanoreceptive and proprioceptive neurons is initiated. Similarly, little is known about the genes that execute the differentiation program of mechanoreceptive neurons.
c-Maf controls development and function of cutaneous mechanoreceptors
We identified c-Maf as a transcription factor expressed in mechanoreceptive neurons shortly after their birth (E11). c-Maf expression in sensory neurons is conserved in evolution and is detected in rodents, birds and humans.20 c-Maf expression defines two main subgroups of sensory neurons: (1) neurons that co-express c-Maf, Ret and MafA (about 7% of all sensory neurons; these correspond to rapidly adapting mechanoreceptors) and (2) c-Maf-positive neurons that do not express Ret and MafA (about 15% of all sensory neurons; these correspond to slowly adapting mechanoreceptors, proprioceptors and a small subpopulation of nociceptors). Mice lacking c-Maf die in the perinatal period. To study the role of c-Maf in sensory neurons, conditional mutant mice were generated. These Isl1Cre c-Mafflox/- mice are hereafter called c-Maf mutants.20
Anatomical analysis showed that a lack of c-Maf primarily affects the morphology of rapidly adapting mechanoreceptors, whereas slowly adapting mechanoreceptors and D-hair mechanoreceptor morphologies are left intact. Several types of rapidly adapting mechanoreceptors that innervate the skin are disrupted in the mutants. Meissner corpuscles—the end organs of rapidly adapting mechanoreceptors in the non-hairy skin—and a substantial proportion of lanceolate endings that associate with hair follicles showed an aberrant morphology. The fact that not all lanceolate endings were altered indicated that these endings are heterogeneous, which was verified by the use of a battery of neurochemical markers. This analysis revealed that neurochemically distinct neuron types, some of which are dependent on c-Maf, innervate the same hair follicle.20 Consistent with recent observations by others,21 three types of lanceolate endings could be distinguished. Lanceolate endings that express Calbindin and NF200 correspond to rapidly adapting mechanoreceptors and require c-Maf for development.20 Those that express high levels of TrkB do not depend on c-Maf and most likely represent D-hair mechanoreceptors.20,21 A third subpopulation is also not c-Maf-dependent and does not expresses Calbindin, NF200 or TrkB, and appears to correspond to C-fiber low threshold receptors.20,21 Finally, circumferential endings are altered in morphology in c-Maf mutant mice. Interestingly, despite the dramatic effects on the morphology of the peripheral nerve terminals in the skin, cell death of the corresponding neurons was not observed. However, the changed morphology was accompanied by sensory dysfunction.
Sensory function was assessed by electrophysiology using the in vitro skin-nerve preparation. This technique allows qualitative and quantitative characterizations of subtypes of mechanoreceptors, such as rapidly adapting, slowly adapting, and D-hair mechanoreceptors, which are distinguished by firing patterns, force sensitivities, and action potential conduction velocities.22 Rapidly adapting mechanoreceptors of c-Maf mutant mice had a reduced conduction velocity and aberrant firing properties, i.e., action potentials were increased and fired in a prolonged manner. At first glance, it is counterintuitive that a morphological disruption of rapidly adapting mechanoreceptors results in increased and prolonged firing. However, c-Maf controls the expression of many genes, including several voltage-gated ion channels that control neuronal excitability. Thus, it is unlikely that morphological changes solely account for the altered firing properties. Slowly adapting mechanoreceptors were only mildly affected in c-Maf mutants and showed a reduction of thresholds to mechanical stimulation and reduced conduction velocities, but their firing pattern was not significantly changed. D-hair mechanoreceptors remained fully functional.
The c-Maf mutation affects the expression of many downstream genes. Interestingly, c-Maf is essential for appropriate expression of Ret, and the morphology of rapidly adapting mechanoreceptors in Ret mutant mice resemble those observed in c-Maf mutants.15,16,20 Several deregulated genes encode potassium channels; among these Kcnq4 is already known to affect firing properties of rapidly adapting mechanoreceptors and contributes to the changes observed in c-Maf mutants.20,23 MafA is co-expressed with c-Maf in rapidly adapting mechanoreceptors and the two closely related factors might act redundantly. The fact that c-Maf regulates MafA expression explains the dominating c-Maf function in these neurons.16,20 Interestingly, genes like Netrin G1 and G2, whose ligand (Lrrc4) is expressed in spinal cord lamina III/IV were also deregulated; these receptors/ligands might participate in the establishment of the mechanoreceptive sensory circuitry and the formation of synapses.
c-Maf function and the detection of high frequency vibration
The electrophysiological characterization described above was restricted to sensory neurons innervating the mouse hairy skin and thus did not assess Pacinian corpuscles. Pacinian corpuscles are found at high densities in rodent periostea, the membranes covering bones, but they are abundant in human skin. These structures are particularly sensitive to high frequency vibration.24 Ablation of c-Maf in mice leads to a pronounced loss of Pacinian corpuscles, and the few remaining had an abnormal morphology. Furthermore, in contrast to the axons of cutaneous mechanoreceptors, axons innervating Pacinian corpuscles were lost. Thus, among all mechanoreceptive neuron types, those that end in Pacinian corpuscles react particularly sensitive to a loss of c-Maf function.
In addition to its important role in sensory neurons, c-Maf controls eye and lens development in mice and humans.25,26 c-Maf is highly expressed in the lens where it controls the transcription of crystallin genes encoding structural lens proteins. The lack of c-Maf in mice abrogates crystallin gene expression and causes microphthalmia due to a failure in differentiation of lens fibers.27 Ocular abnormalities and cataracts have been reported in families that carry dominant mutations in the c-MAF gene. These patients suffer from juvenile cataract, microcornea and a malformation of the iris (coloboma). All known mutations that cause such dysfunctions locate to sequences encoding the c-MAF ancillary DNA binding domain, and all act dominantly and thus affect heterozygous carriers.25,28,29 The evolutionary conservation suggested a function in sensory neuron development of humans, and therefore a family comprising four carriers of the dominant Arg288→Pro288 (R288P) mutation was tested for touch sensitivity. The function of Meissner and Pacinian corpuscles was assessed by applying a vibrational stimulus of increasing amplitude to the nail bed of the little finger, and the subjects were asked to signal the detection of the stimulus by pressing a button. A wide range of frequencies (5–240 Hz) covering the detection range of Pacinian corpuscles and other mechanoreceptors was tested. All four carriers were strongly impaired in their ability to detect high frequency vibrations, but not low frequencies, indicating that Pacinian corpuscles are affected by the dominant c-MAF mutation.20 However, it cannot be ruled out that other mechanisms might contribute, for instance abberant processing of sensory information in the central nervous system. For instance, c-Maf is also present in neurons of dorsal horn layers III/IV, which are known to receive mechanosensory input, and was recently found to regulate gene expression in these neurons.30 We conclude that c-MAF/c-Maf is critical for mechanoreception in humans and in mice.
Conclusion
The richness of our touch percepts requires integration of signals from various types of mechanically sensitive end-organs. Depending on the stimulus quality and strength, distinct combinations of end-organs respond to tactile stimuli. The diversity of mechanoreceptors and the scarcity of molecular data had made it difficult to analyze the development of mechanoreceptors and to define the basis of their functional heterogeneity. c-Maf is a useful marker for mechanoreceptors and the analysis of c-Maf functions provided a wealth of information about ion channels and other genes expressed by mechanosensory neurons. Rapidly adapting mechanoreceptors are particularly dependent on c-Maf function, but it is also clear that other cutaneous Aβ-fibers are affected, albeit to a lesser extent, in c-Maf mutant mice (summarized in Fig. 2), indicating that c-Maf functions extends beyond rapidly adapting mechanoreceptors.
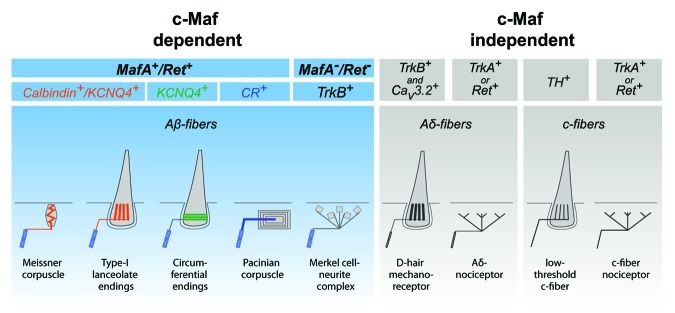
Figure 2. Schematic representation of low threshold mechanoreceptor endings and their dependence on c-Maf. Aβ-fiber depends on c-Maf. The c-Maf mutation affects strongly the development and function of rapidly adapting mechanoreceptors like Meissner corpuscles, type-I lanceolate endings and Pacinian corpuscles.
Interestingly, in the central nervous system, c-Maf is expressed by neurons thought to integrate mechanoreceptive information, indicating that c-Maf marks a neuronal circuit that participates in touch sensation. To unravel c-Maf function in the processing of the peripheral signals will be one of the intriguing challenges lying ahead.
Glossary
Abbreviations:
- bZIP
basic-leucine-zipper
- MARE
Maf Recognition Element
- c-Maf
musculoaponeurotic fibrosarcoma oncogene homolog
- MafA
musculoaponeurotic fibrosarcoma oncogene homolog A
- MafB
musculoaponeurotic fibrosarcoma oncogene homolog B
- Nrl
neural retina leucine zipper
- Ngn1
neurogenin 1
- Ngn2
neurogenin 2
- TrkA
tropomyosin receptor kinase A
- TrkB
tropomyosin receptor kinase B
- TrkC
tropomyosin receptor kinase C
- Ret
ret proto-oncogene
- Brn3a
Brain-specific homeobox/POU domain protein 3A
- DRG
dorsal root ganglion
- Isl1
islet-1
- Cre
Cre recombinase
- Kcnq4
potassium voltage-gated channel, KQT-like subfamily, member 4
- NF200
Neurofilament 200
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/21809
References
- 1.Nishizawa M, Kataoka K, Goto N, Fujiwara KT, Kawai S. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc Natl Acad Sci U S A. 1989;86:7711–5. doi: 10.1073/pnas.86.20.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kataoka K, Nishizawa M, Kawai S. Structure-function analysis of the maf oncogene product, a member of the b-Zip protein family. J Virol. 1993;67:2133–41. doi: 10.1128/jvi.67.4.2133-2141.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurt EM, Wiestner A, Rosenwald A, Shaffer AL, Campo E, Grogan T, et al. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell. 2004;5:191–9. doi: 10.1016/S1535-6108(04)00019-4. [DOI] [PubMed] [Google Scholar]
- 4.Swaroop A, Xu JZ, Pawar H, Jackson A, Skolnick C, Agarwal N. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci U S A. 1992;89:266–70. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho I-C, Glimcher LH. Transcription: tantalizing times for T cells. Cell. 2002;109(Suppl):S109–20. doi: 10.1016/S0092-8674(02)00705-5. [DOI] [PubMed] [Google Scholar]
- 6.Aziz A, Soucie E, Sarrazin S, Sieweke MH. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science. 2009;326:867–71. doi: 10.1126/science.1176056. [DOI] [PubMed] [Google Scholar]
- 7.Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marmigère F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci. 2007;8:114–27. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- 9.McEvilly RJ, Erkman L, Luo L, Sawchenko PE, Ryan AF, Rosenfeld MG. Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature. 1996;384:574–7. doi: 10.1038/384574a0. [DOI] [PubMed] [Google Scholar]
- 10.Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 2008;11:1283–93. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smeyne RJ, Klein R, Schnapp A, Long LK, Bryant S, Lewin A, et al. Severe sensory and sympathetic neuropathies in mice carrying a disrupted Trk/NGF receptor gene. Nature. 1994;368:246–9. doi: 10.1038/368246a0. [DOI] [PubMed] [Google Scholar]
- 12.Chen C-L, Broom DC, Liu Y, de Nooij JC, Li Z, Cen C, et al. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron. 2006;49:365–77. doi: 10.1016/j.neuron.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 13.Gascon E, Gaillard S, Malapert P, Liu Y, Rodat-Despoix L, Samokhvalov IM, et al. Hepatocyte growth factor-Met signaling is required for Runx1 extinction and peptidergic differentiation in primary nociceptive neurons. J Neurosci. 2010;30:12414–23. doi: 10.1523/JNEUROSCI.3135-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lechner SG, Frenzel H, Wang R, Lewin GR. Developmental waves of mechanosensitivity acquisition in sensory neuron subtypes during embryonic development. EMBO J. 2009;28:1479–91. doi: 10.1038/emboj.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo W, Enomoto H, Rice FL, Milbrandt J, Ginty DD. Molecular identification of rapidly adapting mechanoreceptors and their developmental dependence on ret signaling. Neuron. 2009;64:841–56. doi: 10.1016/j.neuron.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourane S, Garces A, Venteo S, Pattyn A, Hubert T, Fichard A, et al. Low-threshold mechanoreceptor subtypes selectively express MafA and are specified by Ret signaling. Neuron. 2009;64:857–70. doi: 10.1016/j.neuron.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Abdo H, Li L, Lallemend F, Bachy I, Xu X-J, Rice FL, et al. Dependence on the transcription factor Shox2 for specification of sensory neurons conveying discriminative touch. Eur J Neurosci. 2011;34:1529–41. doi: 10.1111/j.1460-9568.2011.07883.x. [DOI] [PubMed] [Google Scholar]
- 18.Scott A, Hasegawa H, Sakurai K, Yaron A, Cobb J, Wang F. Transcription factor short stature homeobox 2 is required for proper development of tropomyosin-related kinase B-expressing mechanosensory neurons. J Neurosci. 2011;31:6741–9. doi: 10.1523/JNEUROSCI.5883-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kramer I, Sigrist M, de Nooij JC, Taniuchi I, Jessell TM, Arber S. A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron. 2006;49:379–93. doi: 10.1016/j.neuron.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Wende H, Lechner SG, Cheret C, Bourane S, Kolanczyk ME, Pattyn A, et al. The transcription factor c-Maf controls touch receptor development and function. Science. 2012;335:1373–6. doi: 10.1126/science.1214314. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Rutlin M, Abraira VE, Cassidy C, Kus L, Gong S, et al. The functional organization of cutaneous low-threshold mechanosensory neurons. Cell. 2011;147:1615–27. doi: 10.1016/j.cell.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewin GR, Moshourab R. Mechanosensation and pain. J Neurobiol. 2004;61:30–44. doi: 10.1002/neu.20078. [DOI] [PubMed] [Google Scholar]
- 23.Heidenreich M, Lechner SG, Vardanyan V, Wetzel C, Cremers CW, De Leenheer EM, et al. KCNQ4 K(+) channels tune mechanoreceptors for normal touch sensation in mouse and man. Nat Neurosci. 2012;15:138–45. doi: 10.1038/nn.2985. [DOI] [PubMed] [Google Scholar]
- 24.Johnson KO, Yoshioka T, Vega-Bermudez F. Tactile functions of mechanoreceptive afferents innervating the hand. J Clin Neurophysiol. 2000;17:539–58. doi: 10.1097/00004691-200011000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Jamieson RV, Perveen R, Kerr B, Carette M, Yardley J, Heon E, et al. Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum Mol Genet. 2002;11:33–42. doi: 10.1093/hmg/11.1.33. [DOI] [PubMed] [Google Scholar]
- 26.Mihelec M, St Heaps L, Flaherty M, Billson F, Rudduck C, Tam PPL, et al. Chromosomal rearrangements and novel genes in disorders of eye development, cataract and glaucoma. Twin Res Hum Genet. 2008;11:412–21. doi: 10.1375/twin.11.4.412. [DOI] [PubMed] [Google Scholar]
- 27.Reza HM, Yasuda K. Roles of Maf family proteins in lens development. Dev Dyn. 2004;229:440–8. doi: 10.1002/dvdy.10467. [DOI] [PubMed] [Google Scholar]
- 28.Vanita V, Singh D, Robinson PN, Sperling K, Singh JR. A novel mutation in the DNA-binding domain of MAF at 16q23.1 associated with autosomal dominant “cerulean cataract” in an Indian family. Am J Med Genet A. 2006;140:558–66. doi: 10.1002/ajmg.a.31126. [DOI] [PubMed] [Google Scholar]
- 29.Hansen L, Eiberg H, Rosenberg T. Novel MAF mutation in a family with congenital cataract-microcornea syndrome. Mol Vis. 2007;13:2019–22. [PubMed] [Google Scholar]
- 30.Hu J, Huang T, Li T, Guo Z, Cheng L. c-Maf is required for the development of dorsal horn laminae III/IV neurons and mechanoreceptive DRG axon projections. J Neurosci. 2012;32:5362–73. doi: 10.1523/JNEUROSCI.6239-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]


