Abstract
A recent study provides evidence that RNA polymerase uses 2- to ~4-nt RNAs, species termed “nanoRNAs,” to prime transcription initiation in Escherichia coli. Priming of transcription initiation with nanoRNAs represents a previously undocumented component of transcription start site selection and gene expression.
Keywords: RNA polymerase, RNA-seq, gene expression, small RNA, start site selection, stationary phase
Identification of growth phase-dependent nanoRNA-mediated priming in E. coli
Working in collaboration with the lab of Simon Dove we identified growth phase-dependent nanoRNA-mediated priming in E. coli by analyzing the effect of decreasing the concentration of nanoRNAs on transcription start site selection.1 This strategy was based upon prior evidence indicating that priming of transcription initiation with 2- to 4-nt RNAs can alter the position of transcription initiation to template positions upstream of the start site observed during initiation with nucleoside triphosphates (NTPs) only, i.e., de novo initiation. In particular, studies with bacterial RNA polymerase (RNAP) indicate that 2- to 4-nt RNAs can effectively compete with NTPs for use as primers during transcription initiation in vitro provided the 5′ end of the RNA is complementary to sequences between positions –3 and +1 and the 3′ end is complementary to position +1, +2 or +3, where position +1 is the site of de novo initiation.2-8 Thus, we developed a strategy that would enable the identification of promoters where nanoRNA-mediated priming resulted in the production of transcripts initiating at template positions upstream of the start site observed during de novo initiation. Specifically, we reasoned that such promoters could be identified on the basis that they possessed transcription start sites that were shifted to downstream template positions when the concentrations of nanoRNAs were decreased via overproduction of an exonuclease that degrades nanoRNAs.
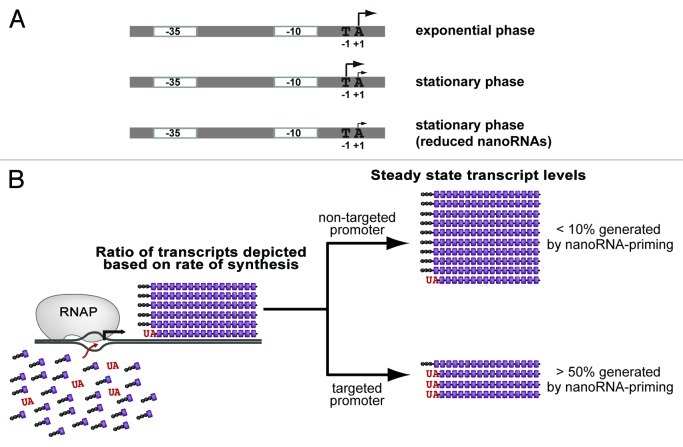
We found that decreasing the concentration of nanoRNAs had no effect on transcription start site selection during the exponential phase of growth. In contrast, we identified seven promoters with start sites that shifted to downstream template positions when the concentrations of nanoRNAs were decreased during the stationary phase of growth. The transcription start sites associated with each of these seven promoters exhibited similar characteristics (Fig. 1A). In particular, transcription initiation at these promoters during stationary phase occurred at two consecutive template positions: a T (designated position –1) and an A (designated position +1). In contrast, transcription initiation during exponential phase only occurred at +1 A. Furthermore, decreasing the concentrations of nanoRNAs during stationary phase eliminated transcription initiation from –1 T, resulting in initiation only from +1 A, as observed during exponential phase. We also found that the transcripts initiating from –1 T observed during stationary phase carry a 5′ hydroxyl while the transcripts initiating from position +1 A carry a 5′ triphosphate. Thus, we concluded that the transcripts initiating from position –1 T were produced by priming of transcription initiation with nanoRNAs carrying a 5′ hydroxyl while transcripts initiating from position +1 A were produced by de novo initiation. These studies reveal that nanoRNA-mediated priming occurs in E. coli and leads to growth phase-dependent alterations in both transcription start site selection (shifting the transcription start site from position +1 A to position –1 T) and in the phosphorylation status of the transcript 5′ ends (shifting them from a 5′ triphosphate to a 5′ hydroxyl).
Figure 1. NanoRNA-mediated priming alters transcription start site selection in Escherichia coli. (A) Transcription start sites observed at promoters targeted by nanoRNA-mediated priming. (B) Model to account for the differential susceptibility of –1/+1 TA promoters to nanoRNA-mediated priming. Depicted is the competition between NTPs (purple) and the dinucleotide UA (red) for use by RNAP during initiation at a –1/+1 TA promoter. The abundance of transcripts generated by de novo initiation is greater than the abundance of transcripts that are generated by priming with UA. The steady-state levels of transcripts reflect the relative stability of those transcripts generated by nanoRNA-mediated priming compared with those generated by de novo initiation. At the majority of –1/+1 TA promoters (non-targeted) the stability of transcripts generated by nanoRNA-mediated priming is less than or equal to the stability of transcripts generated by de novo initiation. At the targeted promoters the stability of transcripts generated by nanoRNA-mediated priming is significantly greater than the stability of transcripts generated by de novo initiation.
NanoRNA-mediated priming: a means of circumventing the “rules” governing de novo transcription initiation
In vitro and in vivo analysis have established a general set of rules governing how transcription start site selection occurs with bacterial RNAP during de novo transcription initiation. In particular, RNAP initiates transcription at template positions located within a 5 base pair window centered 7 base pairs downstream of the promoter –10 element (i.e., positions +5 to +9 with respect to the last base pair of the –10 element).9-14 Within this window, RNAP prefers to initiate transcription at position +7 using a purine nucleotide (A or G). If the sequence of the template strand is such that a purine nucleotide cannot be incorporated at position +7, RNAP will initiate transcription at the template position nearest to +7 where it can incorporate a purine nucleotide. However, if a purine nucleotide cannot be incorporated at template positions located within the +5 to +9 window, RNAP will initiate transcription at position +7 using a pyrimidine nucleotide (U or C). (It is important to note that the precise position at which RNAP initiates de novo RNA synthesis is strongly influenced by the concentrations of NTPs.)
Our findings reveal that nanoRNA-mediated priming can lead to apparent deviations from the “rules” governing de novo transcription start site selection. For example, at several of the promoters targeted by nanoRNA-mediated priming, RNAP initiates transcription using a pyrimidine nucleotide (U) at template position +6 instead of using a purine nucleotide (A) corresponding to position +7. NanoRNA-mediated priming also leads to the incorporation of a hydroxyl group onto the 5′ end of RNA transcripts during transcription initiation. Incorporation of 5′ hydroxyl is highly disfavored during de novo initiation because the affinity of an NTP for an initiation complex is significantly greater than the affinity of a nucleoside for an initiation complex.15 In contrast, incorporation of a hydroxyl group onto the 5′ end of an RNA transcript is not disfavored during nanoRNA-mediated priming because the affinity of a nanoRNA carrying a 5′ hydroxyl for an initiation complex can be significantly greater than the affinity of an NTP for an initiation complex.7,16 Thus, nanoRNA-mediated priming serves as a mechanism to circumvent the “rules” imposed by de novo transcription initiation regarding the position that RNAP starts and the requirement that the first nucleotide incorporated into the transcript carry a triphosphate.
How is nanoRNA-mediated priming manifest in a growth phase-dependent manner?
Our findings indicate that nanoRNA-mediated priming is manifest during stationary phase but not during exponential phase. Perhaps the simplest explanation to account for why nanoRNA-mediated priming is manifest in a growth phase-dependent manner is that nanoRNAs accumulate specifically during stationary phase, possibly in response to limiting resources and/or changes in the cell density of the culture. Alternatively, nanoRNAs might be present throughout all phases of growth, but nanoRNA-mediated priming may only be manifest during stationary phase. For example, as nanoRNAs will compete with NTPs for use during transcription initiation, the concentrations of nanoRNAs relative to the concentrations of NTPs during exponential phase might be too low to enable the detection of transcripts produced by nanoRNA-mediated priming. Accordingly, the reduction in NTP concentrations that accompanies the transition from exponential to stationary phase17 would increase the percentage of transcription initiation events that occurred via nanoRNA-mediated priming. Another possibility is that transcripts produced by nanoRNA-mediated priming might be highly unstable during exponential phase but become stable (and thus detectable) during stationary phase. To address these various models it will be important to define the precise point during the transition from exponential phase to stationary phase that nanoRNA-mediated priming is first manifest and to develop methods for directly detecting and quantifying nanoRNAs in cells.
How is nanoRNA-mediated priming targeted to a specific set of promoters?
Each of the seven promoters identified as targets of nanoRNA-mediated priming carried a T at position –1 and an A at position +1. Thus, the targeting of growth phase-dependent nanoRNA-mediated priming to specific promoters is facilitated, in part, by the presence of the sequence “–1/+1 TA.” In the context of these –1/+1 TA promoters, nanoRNA-mediated priming produces transcripts beginning with the sequence “UA” that carry a 5′ hydroxyl. These transcripts could only be generated by priming with 5′ hydroxyl nanoRNAs beginning with the sequence UA. Thus, the simplest explanation to account for why nanoRNA-mediated priming is targeted to –1/+1 TA promoters is that 5′ hydroxyl nanoRNAs beginning with the sequence UA preferentially accumulate in cells.
How might nanoRNAs beginning with the sequence UA carrying a 5′ hydroxyl preferentially accumulate? One possibility we have proposed1 is that they are produced as terminal products of RNA degradation. According to this proposal during stationary phase full-length RNA transcripts are degraded through a multistep process. The first step in this process is the endonucleolytic cleavage of full length transcripts specifically targeted to the phosphodiester bonds located at the 5′ end of the sequence UA. This cleavage would generate a 2′, 3′ cyclic phosphate on one of the cleavage products and a 5′ hydroxyl on the other. Because E. coli does not contain any 5′ to 3′ exonucleases, further processing of the initial cleavage fragments would be performed by the four major 3′ to 5′ exonucleases: polynucleotide phosphorylase, RNase II, RNase R, and Oligoribonuclease. Among these four exonucleases, Oligoribonuclease is the only one that can efficiently degrade RNAs less than ~5-nt in length.18 However, Oligoribonuclease is not thought to contribute to the degradation of longer RNA transcripts.19-21 Thus, processing of the cleavage fragments by polynucleotide phosphorylase, RNase II, or RNase R would generate 5′ hydroxyl-carrying nanoRNAs beginning with UA. These nanoRNAs would either be used to prime transcription initiation or degraded to nucleotides by Oligoribonuclease. We note that none of the characterized endonucleases of E. coli show a preference for cleaving the phosphodiester bond 5′ to the sequence UA. Thus, if the model described above is correct, the putative endonuclease responsible for the initial cleavage event awaits identification.
It is important to mention that in vitro studies indicate that 2-nt RNAs are, in general, more effective than longer RNAs at competing with NTPs for use during transcription initiation.8 Therefore, we consider it likely that the dinucleotide UA is fully responsible for the observed nanoRNA-mediated priming. In principle, UA could be generated through the proposed RNA degradation pathway described above or, perhaps, by an enzymatic activity that fuses uridine with an adenosine nucleotide.
Another unresolved aspect of how nanoRNA-mediated priming is targeted to specific promoters stems from the observation that not all “–1/+1 TA” promoters are targeted by nanoRNA-mediated priming. In fact, only ~15% of the “–1/+1 TA” promoters we analyzed were significantly impacted by nanoRNA-mediated priming.1 Thus, features other than “–1/+1 TA” must determine whether or not a promoter is targeted by nanoRNA-mediated priming. Although several possibilities could account for the differential susceptibility of “–1/+1 TA” promoters to nanoRNA-mediated priming, our preferred model is as follows (Fig. 1B): First, we propose that the concentrations of nanoRNAs during stationary phase are relatively low compared with the concentrations of NTPs. Second, we propose that nanoRNA-mediated priming comprises only a small fraction of the total number of initiation events at all “–1/+1 TA” promoters. Thus, in the context of the majority of “–1/+1 TA” promoters transcripts produced by nanoRNA-mediated priming are undetectable because they exhibit similar or lower stability than transcripts generated by de novo initiation. In contrast, in the context of a few “–1/+1 TA” promoters transcripts produced by nanoRNA-mediated priming are significantly more stable than transcripts generated by de novo initiation. When considering the steady-state levels of transcripts produced from these ‘targeted’ promoters, transcripts produced by nanoRNA-mediated priming will account for a significant fraction. According to this model the transcripts that are generated by nanoRNA-mediated priming at the targeted promoters are stabilized by an unknown mechanism, perhaps involving cis-acting sequences in the RNA transcript itself and/or the presence of a 5′ hydroxyl (see below). We note that this model, which proposes that transcripts generated by nanoRNA-mediated priming are differentially stable compared with those produced by de novo initiation, could also account for the effects of nanoRNA-mediated priming on gene expression as described below.
How does nanoRNA-mediated priming impact gene expression?
NanoRNA-mediated priming not only serves as a mechanism to influence transcription start site selection, but also serves as a mechanism to influence gene expression. In particular we found that nanoRNA-mediated priming increased the expression of at least two genes: tomB and bhsA. As mentioned above, we consider it likely that nanoRNA-mediated priming activates the expression of these genes because the transcripts generated by nanoRNA-mediated priming are significantly more stable than transcripts generated by de novo initiation. One possibility is that the presence of a 5′ hydroxyl instead of a 5′ triphosphate on the tomB and bhsA transcripts leads to increased transcript stability. In support of this notion, it has been shown that the endonuclease that initiates the decay of most mRNAs in E. coli, RNase E, does not work efficiently on transcripts carrying a 5′ hydroxyl.22-24 Furthermore, it has been demonstrated in vivo that transcripts carrying a 5′ hydroxyl can be more stable than transcripts of identical sequence carrying a 5′ triphosphate.25 However, as mentioned above, transcripts that are generated by nanoRNA-mediated priming in the context of different promoters may not necessarily exhibit an increase in stability compared with transcripts generated by de novo initiation. Thus, perhaps in certain instances the presence of a 5′ hydroxyl protects against degradation while in other cases the presence of a 5′ hydroxyl has no effect or leads to a decrease in transcript stability [see, for example, ref. 26].
Although we have identified two genes whose expression is impacted by nanoRNA-mediated priming, only 225 promoters were included in our initial analysis. Thus, the full extent of the “nanoRNA regulon” (i.e., the complete set of genes whose expression is directly affected by nanoRNA-mediated priming) remains to be elucidated. In addition, we currently do not know what impact nanoRNA-mediated priming has on cellular physiology. In this regard, the two genes we have identified whose expression is regulated by nanoRNA-mediated priming (tomB and bhsA) have previously been implicated in biofilm formation and response to stress.27-29 Thus, nanoRNA-mediated priming may contribute to biofilm formation and/or the survival of cells that have been exposed to various stresses (e.g., acid stress, oxidative stress, heat shock, etc.).
Future Prospects
The most important question to address in future studies is whether or not nanoRNA-mediated priming impacts gene expression in organisms other than E. coli. The strategy employed to identify nanoRNA-mediated priming in E. coli, which relied upon determining the effect of overproducing an exonuclease that degrades nanoRNAs on transcription start site selection, should be broadly applicable to the study of nanoRNA-mediated priming in other organisms.
NanoRNA-mediated priming represents a previously undocumented mechanism by which the 5′ ends of RNA transcripts can be generated in cells. Furthermore, as indicated by our studies in E. coli, nanoRNA-mediated priming provides a means of altering the phosphorylation status of the 5′ ends of RNA transcripts in a manner that does not require post transcriptional modification by an RNA decapping enzyme or an RNA endonuclease. Therefore, the potential contribution of nanoRNA-mediated priming should be considered when the phosphorylation status of the 5′ ends of RNAs are used as a means of differentiating those 5′ ends that were generated by transcription initiation vs. those ends that were generated post transcriptionally. In this regard, recent analysis of mRNAs isolated from Saccharomyces cerevisiae uncovered transcripts carrying a 5′ monophosphate that were associated with transcription start sites and insensitive to the presence or absence of any known decapping enzymes.30 Perhaps these species of unknown origin were generated by priming with 5′ monophosphate nanoRNAs.
Acknowledgments
This work is supported by NIH grants GM096454 and GM088343.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/21903
References
- 1.Vvedenskaya IO, Sharp JS, Goldman SR, Kanabar PN, Livny J, Dove SL, et al. Growth phase-dependent control of transcription start site selection and gene expression by nanoRNAs. Genes Dev. 2012;26:1498–507. doi: 10.1101/gad.192732.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman DJ, Niyogi SK. RNA initiation with dinucleoside monophosphates during transcription of bacteriophage T4 DNA with RNA polymerase of Escherichia coli. Proc Natl Acad Sci U S A. 1973;70:574–8. doi: 10.1073/pnas.70.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minkley EG, Pribnow D. Transcription of the early region of bacteriophage T7: selective initiation with dinucleotides. J Mol Biol. 1973;77:255–77. doi: 10.1016/0022-2836(73)90335-5. [DOI] [PubMed] [Google Scholar]
- 4.Di Nocera PP, Avitabile A, Blasi F. In vitro transcription of the Escherichia coli histidine operon primed by dinucleotides. Effect of the first histidine biosynthetic enzyme. J Biol Chem. 1975;250:8376–81. [PubMed] [Google Scholar]
- 5.Smagowicz WJ, Scheit KH. Primed abortive initiation of RNA synthesis by E. coli RNA polymerase on T7 DNA. Steady state kinetic studies. Nucleic Acids Res. 1978;5:1919–32. doi: 10.1093/nar/5.6.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grachev MA, Zaychikov EF, Ivanova EM, Komarova NI, Kutyavin IV, Sidelnikova NP, et al. Oligonucleotides complementary to a promoter over the region -8...+2 as transcription primers for E. coli RNA polymerase. Nucleic Acids Res. 1984;12:8509–24. doi: 10.1093/nar/12.22.8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruetsch N, Dennis D. RNA polymerase. Limit cognate primer for initiation and stable ternary complex formation. J Biol Chem. 1987;262:1674–9. [PubMed] [Google Scholar]
- 8.Goldman SR, Sharp JS, Vvedenskaya IO, Livny J, Dove SL, Nickels BE. NanoRNAs prime transcription initiation in vivo. Mol Cell. 2011;42:817–25. doi: 10.1016/j.molcel.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoyama T, Takanami M. Essential structure of E. coli promoter II. Effect of the sequences around the RNA start point on promoter function. Nucleic Acids Res. 1985;13:4085–96. doi: 10.1093/nar/13.11.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong W, Kang C. Start site selection at lacUV5 promoter affected by the sequence context around the initiation sites. Nucleic Acids Res. 1994;22:4667–72. doi: 10.1093/nar/22.22.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis DE, Adhya S. Axiom of determining transcription start points by RNA polymerase in Escherichia coli. Mol Microbiol. 2004;54:692–701. doi: 10.1111/j.1365-2958.2004.04318.x. [DOI] [PubMed] [Google Scholar]
- 12.Liu C, Heath LS, Turnbough CL., Jr. Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes Dev. 1994;8:2904–12. doi: 10.1101/gad.8.23.2904. [DOI] [PubMed] [Google Scholar]
- 13.Sørensen KI, Baker KE, Kelln RA, Neuhard J. Nucleotide pool-sensitive selection of the transcriptional start site in vivo at the Salmonella typhimurium pyrC and pyrD promoters. J Bacteriol. 1993;175:4137–44. doi: 10.1128/jb.175.13.4137-4144.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker KA, Osuna R. Factors affecting start site selection at the Escherichia coli fis promoter. J Bacteriol. 2002;184:4783–91. doi: 10.1128/JB.184.17.4783-4791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClure WR, Cech CL, Johnston DE. A steady state assay for the RNA polymerase initiation reaction. J Biol Chem. 1978;253:8941–8. [PubMed] [Google Scholar]
- 16.Smagowicz W, Scheit KH. A minimal mechanism for abortive initiation of transcription of T7 DNA. Nucleic Acids Res. 1981;9:5845–54. doi: 10.1093/nar/9.21.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol. 2008;190:718–26. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci U S A. 1999;96:4372–7. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–66. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belasco JG. All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nat Rev Mol Cell Biol. 2010;11:467–78. doi: 10.1038/nrm2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Condon C. Molecular Biology of RNA Processing and Decay in Prokaryotes. London, Uk: Academic Press, 2009. [Google Scholar]
- 22.Jiang X, Belasco JG. Catalytic activation of multimeric RNase E and RNase G by 5′-monophosphorylated RNA. Proc Natl Acad Sci U S A. 2004;101:9211–6. doi: 10.1073/pnas.0401382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koslover DJ, Callaghan AJ, Marcaida MJ, Garman EF, Martick M, Scott WG, et al. The crystal structure of the Escherichia coli RNase E apoprotein and a mechanism for RNA degradation. Structure. 2008;16:1238–44. doi: 10.1016/j.str.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tock MR, Walsh AP, Carroll G, McDowall KJ. The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′-end-dependent ribonuclease that has context-dependent broad sequence specificity. J Biol Chem. 2000;275:8726–32. doi: 10.1074/jbc.275.12.8726. [DOI] [PubMed] [Google Scholar]
- 25.Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giliberti J, O’Donnell S, Etten WJ, Janssen GR. A 5′-terminal phosphate is required for stable ternary complex formation and translation of leaderless mRNA in Escherichia coli. RNA. 2012;18:508–18. doi: 10.1261/rna.027698.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.García-Contreras R, Zhang XS, Kim Y, Wood TK. Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage killer genes. PLoS One. 2008;3:e2394. doi: 10.1371/journal.pone.0002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mermod M, Magnani D, Solioz M, Stoyanov JV. The copper-inducible ComR (YcfQ) repressor regulates expression of ComC (YcfR), which affects copper permeability of the outer membrane of Escherichia coli. Biometals. 2012;25:33–43. doi: 10.1007/s10534-011-9510-x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XS, García-Contreras R, Wood TK. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J Bacteriol. 2007;189:3051–62. doi: 10.1128/JB.01832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harigaya Y, Parker R. Global analysis of mRNA decay intermediates in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2012;109:11764–9. doi: 10.1073/pnas.1119741109. [DOI] [PMC free article] [PubMed] [Google Scholar]