Abstract
Mature pepper (Capsicum sp.) fruits come in a variety of colors, including red, orange, yellow, brown, and white. To better understand the genetic and regulatory relationships between the yellow fruit phenotype and the capsanthin-capsorubin synthase gene (Ccs), we examined 156 Capsicum varieties, most of which were collected from Northwest Chinese landraces. A new ccs variant was identified in the yellow fruit cultivar CK7. Cluster analysis revealed that CK7, which belongs to the C. annuum species, has low genetic similarity to other yellow C. annuum varieties. In the coding sequence of this ccs allele, we detected a premature stop codon derived from a C to G change, as well as a downstream frame-shift caused by a 1-bp nucleotide deletion. In addition, the expression of the gene was detected in mature CK7 fruit. Furthermore, the promoter sequences of Ccs from some pepper varieties were examined, and we detected a 176-bp tandem repeat sequence in the promoter region. In all C. annuum varieties examined in this study, the repeat number was three, compared with four in two C. chinense accessions. The sequence similarity ranged from 84.8% to 97.7% among the four types of repeats, and some putative cis-elements were also found in every repeat. This suggests that the transcriptional regulation of Ccs expression is complex. Based on the analysis of the novel C. annuum mutation reported here, along with the studies of three mutation types in yellow C. annuum and C. chinense accessions, we suggest that the mechanism leading to the production of yellow color fruit may be not as complex as that leading to orange fruit production.
Introduction
Plants have colored flowers and fruits to attract insects or other animals acting as pollinators and seed dispersers [1]. The Capsicum genus, which is generally accepted to originate in South America, comprises 25–30 species [2]. Five of these species have been domesticated, including C. annuum, C. baccatum, C. chinense, C. frutescens and C. pubescens. The species with the greatest number of cultivated varieties worldwide is C. annuum. The wild progenitor of C. annuum is thought to be the bird pepper, whose fruit is small and red when ripe [2]. The ripe color of pepper has undergone selection during domestication, which has resulted in some new phenotypes, including yellow, orange, brown and even white fruits, which are found in all cultivated pepper species [3]. Since diverse colors (from white to red) can found across Capsicum varieties, pepper has become a good model system for studying the mechanism of fruit color change [3].
The color found in ripe pepper fruit (Capsicum sp.) mainly depends on the variable accumulation of capsanthin and capsorubin, both of which are involved in the carotenoid biosynthetic pathway. Red peppers accumulate higher levels of total carotenoid during ripening [4]. Some studies have indicated that the expression levels of carotenoid biosynthesis-related genes are directly linked to high total carotenoid accumulation in Capsicum. The expression levels of some carotenoid biosynthetic gene, including phytoene synthase (Psy), phytoene desaturase (Pds) and capsanthin-capsorubin synthase (Ccs), are relatively high in red peppers, whereas some of these genes are not expressed in peppers with lower levels of total carotenoid [4].
Test crosses between red and white-fruited varieties indicated that the inheritance of mature fruit color is controlled by three loci—c1, c2 and y [5], [6]. The presence of dominant alleles at all three loci leads to the production of red ripe-color fruit [7]. The c2 locus is firstly considered as a major locus for orange fruit color and is associated with the Psy gene [8], [9]. The deletion of the upstream region of the Ccs gene causes a second orange color phenotype [10]. Additionally, in the orange C. annuum cultivar Fogo, a mutation in Ccs produces premature termination [11]. Recently, Rodriguez-Uribe et al. [12] suggested that the control of color development in orange C. annuum involves a more complex process than the presence of a deletion in a structural gene for a step in pigment biosyntheses; orange ripe color may involve transcriptional regulation of Psy and/or Ccs expression.
The yellow pepper fruit color phenotype, which is recessive to red, is controlled by the y locus. Linkage analysis indicated that the candidate gene of the y locus is also the Ccs gene [4], [13]. To the best of our knowledge, three types of mutations for the Ccs gene have been found in yellow fruit pepper, including the following: a deletion that lacks the distal 220 bp of the 3′ end of the gene in C. annuum cultivars [7], [13], a premature stop-codon and a frame-shift in the coding sequence in two C. chinense accessions [4].
Since diverse modes of regulation have been found to occur between Ccs and the orange color phenotype, further studies about the relationship between Ccs and yellow color ripe-fruit are needed. Here, we identified a new mutant ccs variant in the yellow fruit cultivar CK7, which was derived from a local Northwest Chinese pepper population. Cluster analysis was used to examine the genetic relationship between members of the population. We found that CK7, which is a C. annuum species, shares low genetic similarity with other yellow C. annuum varieties. In the coding sequence of the ccs allele of CK7, we detected a premature stop codon derived from a C to G change, along with a downstream frame-shift caused by a 1-bp deletion. The second mutant position was the same as the type found in the Fogo cultivar [11]. In addition, we detected the expression of ccs in CK7, which was also detected in orange Fogo fruits [12]. Since the differential expression of Ccs can influence fruit color, we analyzed the promoter sequences of this gene in various pepper varieties. Unexpectedly, a 176-bp tandem repeat sequence was found in the promoter regions. In all C. annuum varieties studied, the repeat number was three, compared with four in two C. chinense accessions, thus leading to the presence of a relatively long promoter sequence in the C. chinense accessions. The level of similarity among the four repeat types ranged from 84.8% to 97.7%, and some putative cis-elements were also found in all of the repeats. These results suggest that there is complex transcriptional regulation of Ccs expression. The results of the novel mutation found in C. annuum and three other types of mutations in C. annuum and C. chinense accessions suggest that yellow fruit color is controlled by one structural gene in Capsicum.
Materials and Methods
Plant materials
All the 156 pepper cultivars were obtained from the College of Horticulture, Northwest Agriculture & Forestry University. Among these cultivars, 14 accessions were selected from the Asian Vegetable Research and Development Center (Table 1). All cultivars were maintained by inbreeding at the Horticulture Farm in Yangling, China. Twelve plants per line were grown in greenhouses from March to July, under natural light, at a maximum and minimum temperature of 35 and 16°C, respectively. The fruits were harvested both at the immature (nearly 30 days after anthesis and before the breaking period) and fully mature (more than 40 days after anthesis, with the final fruit color fixed) stages. For each stage, more than five fruits were collected from the same pepper lines and frozen in liquid nitrogen for subsequent RNA extractions. The fruit colors were observed and photographed at the mature stage. Young leaves were used for DNA extractions.
Table 1. Descriptive characteristics of pepper cultivars, developed by the Asian Vegetable Research and Development Center (AVRDC-The World Vegetable Center).
| Name in this study | Vegetable introduction number in AVRDC | Genus and species | Country of collection | Color at mature stage |
| R11 | VI059346 | C. annuum | USA | Red |
| R12 | VI012493 | C. annuum | Canada | Yellow |
| R13 | VI012712 | C. annuum | Bulgaria | Red |
| R14 | VI012755 | C. sp | Korea | Red |
| R15 | VI047076 | C. annuum | Canada | Red |
| R19 | VI012852 | C. annuum | USA | Yellow |
| R25 | VI047102 | C. annuum | USA | Red |
| R26 | VI046805 | C. baccatum | Peru | Red |
| R28 | VI044315 | C. chinense | Unknown | Red |
| R29 | VI046935 | C. frutescens | Mexico | Red |
| R30 | VI047018 | C. chinense | USA | Red |
| R34 | VI027861 | C. annuum | Turkey | Red |
| R37 | VI012716 | C. annuum | Bulgaria | Red |
| R38 | VI012496 | C. baccatum | Chile | Red |
SSR analysis
DNA from young leaves was extracted as described by Li et al. [14]. A total of 25 genomic SSR loci [15] and seven other SSR markers (suggested by Prof. Alain Palloix, INRA, France) were used to analyze the phylogenetic relationship between the cultivars (Table S1). PCR was performed as follows: 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The PCR products were analyzed in 6% denatured polyacrylamide gels in 1 × TBE [16].
Data analysis
Each fingerprint profile was scored for the presence or absence of bands in a gel. Pairwise genetic similarities among individuals were computed with NTSYSpc 2.02j [17]. Pairwise genetic distances were averaged among individuals within each accession to generate an accession pairwise genetic distance matrix. Dendrograms were constructed using the UPGMA method (Unweighted Pair Group Method with Arithmetic mean).
Cloning of Ccs gene and promoter sequences
Sixteen genetically distinct pepper cultivars were selected based on the color types in fully ripe fruit (eight red and eight yellow), including CK6 (C. annuum, red), CK8 (C. annuum, red), R29 (C. frutescens, red), R37 (C. annuum, red), R28 (C. chinense, red), R30 (C. chinense, red), R26 (C. baccatum, red), R38 (C. baccatum, red), P123-1-1 (C. annuum, yellow), CK4 (C. annuum, yellow), CK4-1 (C. annuum, yellow), CK18 (C. annuum, yellow), CK7 (C. annuum, yellow), CK26-1 (C. annuum, yellow), R12 (C. annuum, yellow), and R19 (C. annuum, yellow). The CDS (coding sequence) and promoter fragments of the Ccs gene from all sixteen cultivars were amplified using a Long-Distance PCR Kit (Takara, China) with gene-specific primers (Table S2). The resulting PCR products were subcloned and sequenced as described previously [14].
Multiple sequence alignment of full-length CDS was performed using ClustalX software (http://www.ch.embnet.org/software/clustal_X.html) and displayed using the DNAMAN 4.0 program (Lynnon Biosoft. Co.). Phylogenetic trees were constructed using MEGA3.1 software (http://www.megasoftware.net/) based on the neighbor-joining method. Bootstrap values were calculated from 1,000 trials.
The promoter fragments from pepper lines CK8 (C. annuum), R28 (C. chinense) and R30 (C. chinense) were compared to identify the homologous tandem repeat regions and cis-acting elements. Putative cis-acting elements were identified by searching the PLACE database (http://www.dna.affrc.go.jp/PLACE/) [18] and PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Expression of Ccs
The expression of Ccs gene was analyzed using quantitative RT-PCR. Total RNA was extracted from immature and ripe red/yellow pericarps using a previously described method [16], [19]. PCR was performed in a 96-well plate using an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, USA), with SYBR Green Realtime PCR Master Mix (TaKaRa, China). The amplification was initiated by heating to 94°C for 10 min followed by 40 cycles at 94°C for 5 s and 62°C for 30 s. The amplification specificity was tested by producing a dissociation curve (65°C to 90°C). To compare results from different reactions and samples, Ccs amplification was normalized to that of the ubiquitin gene using the CT values [20]. The expression results represent the average of three independent biological replicates from different fruits of a single plant.
Results
Color phenotypes and cluster analysis of pepper cultivars
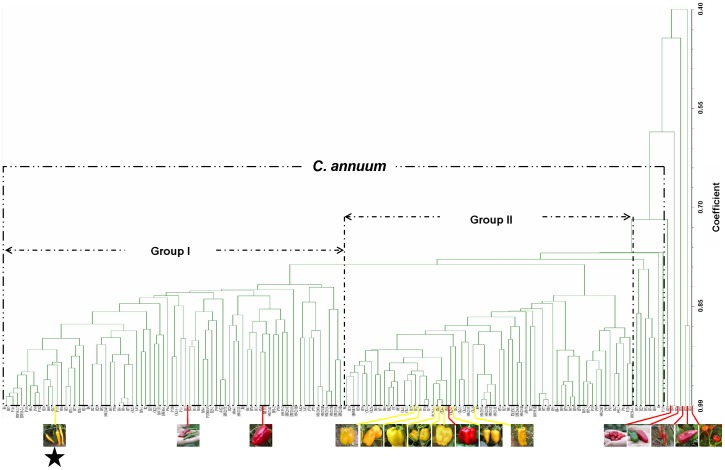
All 156 pepper cultivars were grown in a field to examine the mature fruit colors. Most of the cultivars showed a diverse degree of red coloring in fruit, which included dark red, red and light red (Figure 1). In addition, some lines producing orange or brown ripe fruit, and eight cultivars producing yellow ripe fruit, were also observed (Figure 1). Since six lines with yellow ripe fruit were local peppers, cluster analysis was necessary to identify the genetic relationship between these lines and to determine which Capsicum species these lines belong to.
Figure 1. Mature red and yellow pepper lines used in this study.
Top two lines (red cultivars): (A) CK6, (B) CK8, (C) R29, (D) R37, (E) R28, (F) R30, (G) R26, (H) R38; bottom two lines (yellow cultivars): (I) P123-1-1, (J) CK4, (K) CK4-1, (L) CK18, (M) CK7, (N) CK26-1, (O) R12, (P) R19.
Across all the 156 cultivars evaluated, 32 SSR markers were analyzed, and 154 scorable polymorphic bands were detected. An UPGMA dendrogram (Figure 2) revealed one main cluster (C. annuum) and four genetically distinct species. The majority of the cultivars evaluated fell into the main cluster, which could be subdivided into two large (I, II) and one small group. All of the unknown local yellow pepper lines were grouped in the main cluster, revealing that these lines are all C. annuum cultivars. Among the eight yellow cultivars, seven are in group II, which mainly includes sweet or bell pepper. One exception is the yellow fruit line CK7, which is found in group I. Most chili peppers in this study are found in this group, and CK7 is also a hot pepper. Therefore, the genetically distinct background between CK7 and other yellow cultivars suggests that the yellow color fruit genotype may have an independent mutational origin.
Figure 2. The clustering pattern obtained for all 156 pepper cultivars examined in this study using SSR analysis.
The novel yellow ripe-color pepper line, CK7, is marked with an asterisk.
Sequence diversity in the coding sequence of the Ccs allele
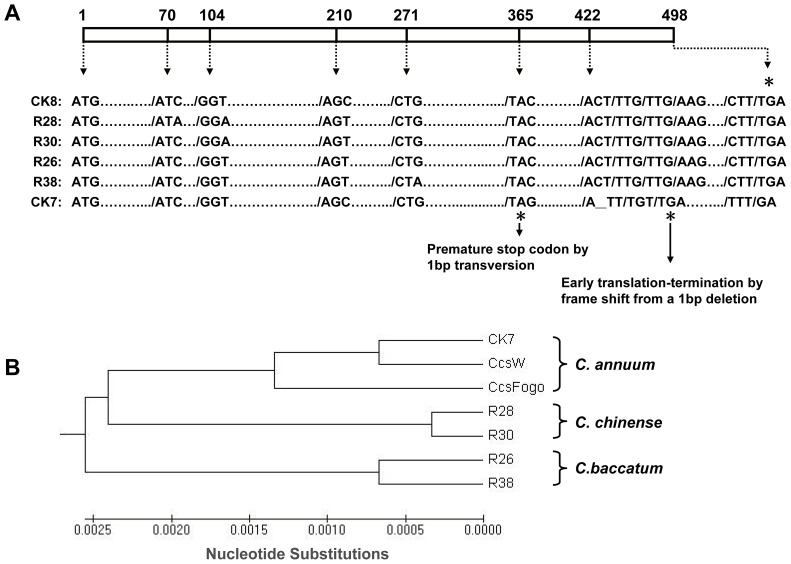
PCR was performed to analyze the genomic coding sequences of Ccs in the 156 pepper varieties. All non-yellow lines produced positive PCR results after electrophoresis (Figure 3 showed a part of the results). As expected, we did not detect the Ccs gene in seven yellow color cultivars, which is agreement with the results of Lefebvre et al. [13]. However, one yellow line, CK7, produced the same banding pattern as that of the red cultivars (Figure 3). The PCR band from CK7 was extracted from the gel and sequenced. Sequence analysis revealed that CK7 contains a new variant of Ccs, with two types of early translation termination of the Ccs gene (Figure 4A). One mutation is a frame shift derived from a 1-bp deletion (1,265 bp downstream of the start codon), which is the same as the mutation detected in the C. annuum cultivar Fogo [11]. The second mutation, a novel premature stop codon produced by a point mutation (1,095 bp downstream of the start codon, cytosine to guanine) was found upstream of the deletion described above.
Figure 3. PCR amplification of the Ccs gene and its promoters from the genomes of the indicated Capsicum varieties.
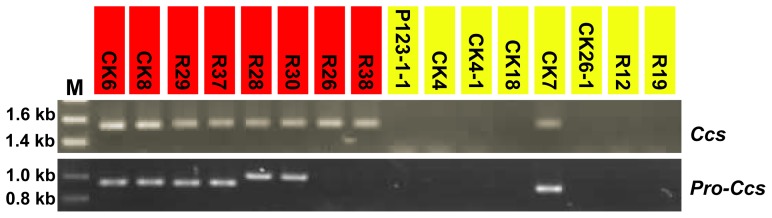
The order of the pepper lines is the same as in Figure 1, and the mature fruit colors are indicated with red and yellow highlighting.
Figure 4. Comparisons of the Ccs coding sequences in the indicated Capsicum varieties.
(A) Schematic representation of the mutations in Ccs among different cultivars. The nucleotide sequences were aligned, and the resulting missense mutations in the amino acid sequence are marked with asterisks. The 1-bp deletion in the coding sequence, which leads to early translation-termination, is underlined in the CK7 sequence. ATG and TGA indicate the start and stop codons, respectively. (B) Phylogenetic tree of the Ccs gene generated by multiple alignments of the coding sequence. The sequence of CcsW, which served as the positive control, was obtained from the NCBI (Accession: ×76165), and the sequence of CcsFogo was obtained from Guzman et al. [11] (GenBank GU122933).
The premature stop codon in the ccs gene in CK7 encodes a putative 364 amino-acid truncated protein that lacks the 134 amino-acid region found in the C-terminus of the wild-type protein. The estimate molecular weight and isoelectric point (pI) of the mutant CCS protein are 41.4 kDa and 8.47, respectively, in contrast to 56.6 kDa and 8.77, respectively, in the wild type. Ccs belongs to the lycopene cyclase gene family (LYC), which encodes lycopene beta and epsilon cyclase proteins. Alignment of putative proteins encoded by several genes from this family, which share high homology with CCS, revealed that the amino acids at the C-termini are more conservative than those at the N-termini (Figure S1). The last deduced domain, which is located near amino acids 430–460, is totally lacking in the ccs variant of CK7.
The Ccs CDS regions from five other cultivars were also sequenced to analyze the structural polymorphism in this gene (Figure 4A). Among the cultivars, CK8, which belongs to C. annuum species and exhibits typical wild red fruit phenotype, was used as a positive check; four other lines (R28, R30, R26 and R38) were chosen due to their different promoter profiles (as described below). Some polymorphic nucleotides were found, whereas no changes in the amino acid sequence were detected in the four lines examined (Figure 4A). Together with the ccs sequence from CK7 and Fogo [11], a phylogenetic tree examining the Ccs gene was generated by multiple alignments of the nucleotide sequences in the CDS region (Figure 4B). Despite the presence of several mutations, the gene sequences from CK7, Fogo and wild C. annuum were still grouped into a same cluster, in contrast to the C. chinense and C. baccatum species. This suggests that an independent evolution and mutation may have occurred in the Ccs gene after Capsicum interspecies differentiation.
Cultivar-specific Ccs expression
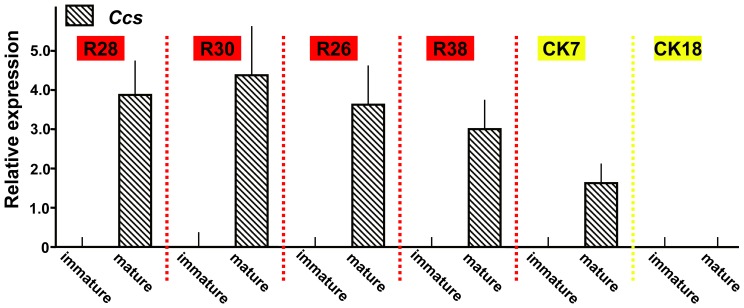
Given the variability in gene structures and mature color phenotypes, we used quantitative RT-PCR to test the expression of Ccs in the lines that produced positive PCR results for the Ccs CDS (Figure 5). Transcripts for Ccs were not detectable in the immature fruits of any of the pepper lines examined (Figure 5). In the four red fruit lines, including two C. chinense (R28 and R30) and two C. baccatum lines (R26 and R38), Ccs expression was detected in fully mature fruits. We also detected some accumulation of Ccs transcript in CK7, which has a mutant ccs gene and yellow fruit, although the expression level was lower than that detected in the red fruit lines; a similar pattern was revealed in the Fogo accession [12]. As expected, none of the other yellow lines exhibited Ccs transcript accumulation.
Figure 5. Expression pattern of Ccs.
Total RNA obtained from immature and mature pericarps of R28, R30, R26, R38, CK7 and CK18 was used for quantitative RT-PCR analysis. Each result represents the average of three independent biological replicates ± SE (n = 3), with a significance level of P≤0.05.
Structural variations in the Ccs promoters of different species
As the expression patterns for Ccs gene were variable across the pepper cultivars [4], [12], we examined the promoter regions of this gene in all of the varieties using PCR (Figure 3). All red mature C. annuum lines produced a similar 920-bp band as the result submitted in GenBank (Y14165). Two larger bands were detected in two C. chinense species (R28 and R30), and no PCR product was detected in two C. baccatum accessions (R26 and R38). The yellow mutant cultivar evaluated in this study, CK7, produced the similar PCR band as in red-fruited C. annuum lines, while none of the other yellow peppers produced a positive PCR result. The promoters from CK8 (used as a positive check for red ripe-fruit lines in C. annuum), CK7, R28 and R30 were sequenced, and no sequence differences in the two C. annuum lines (CK8 and CK7) were likely to be associated with the differences in Ccs gene transcription.
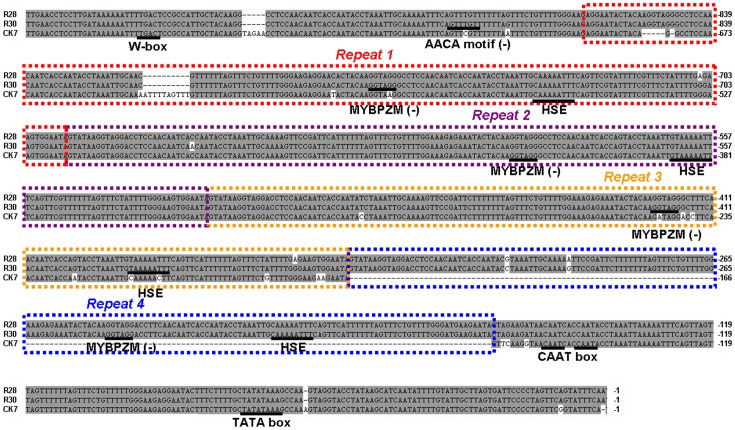
After multiple alignments of the nucleotide sequences, some similar insertions/deletions in the promoter sections from different species reported by Ha et al. [4] were confirmed, including a 11/10-bp deletion and 176-bp insertion in the C. chinense lines (Figure 6). Unexpectedly, we found a tandem repeat structure in the promoter region, with the 176-bp insertion/deletion comprising just one unit of the repeat. The start position of this repeat structure is 165-bp upstream of the transcriptional start point. In addition, three continuous repeats (Repeat 1–Repeat 3; Figure 6) were revealed in the C. annuum lines, whereas four units (Repeat 1–Repeat 4) were detected in the C. chinense accessions. In the C. chinense accession R28, the similarity of the four repeat units ranged from 84.8% (Repeat 1 and Repeat 4) to 97.7% (Repeat 3 and Repeat 4) (Figure S2).
Figure 6. Sequence comparisons in the Ccs promoter region among the Capsicum varieties examined in this study.
The boxes (red, purple, yellow and blue) indicate the four repeat units found in this region. Major cis–elements as predicted by PLACE and PlantCARE softeare are underlined.
Various potential cis-regulatory elements were identified; the most notable ones are listed in Table 2 and underlined in Figure 6. Analysis of the 176 bp repeat unit sequence indicated that this sequence contains a heat stress-related cis-element (HSE, A/TAAAAATTTC) and an Myb homolog binding site (MYBPZM, CCT/AACC). In maize, the Myb binding site is a target recognized by the product of the maize P gene, which is associated with red pigmentation of the kernel pericarp and flavonoid biosynthesis [21].
Table 2. Specific putative cis-acting elements in the promoter of Ccs from C. chinense accession.
| Position | |||||||
| Response/Binding Site | Sequence (5′ to 3′) | Region Ia (−1∼−165) | Repeat 4b (−166∼−341) | Repeat 3 (−342∼−517) | Repeat 2 (−518∼−693) | Repeat 1 (−694∼−869) | Region II (−870∼−977) |
| Heat stress response | CCAATBOX1[CCAAT] | −223, −311 | −399, −487 | −663 | −751, −838 | −909 | |
| Heat stress response | YAAAAATTTC | −204 | −380 | −556 | −732 | ||
| Light response | IBOXCORE[GATAA] | −961 | |||||
| Light response | INRNTPSADB[YTCANTYY] | −193, −198 | −369, −374 | −550, −633 | −726 | ||
| Light response | SORLIP2AT[GGGCC] | −591 | |||||
| Low-temperature response | LTRE1HVBLT49[CCGAAA] | −265c | |||||
| MYB binding site | MYBZM[CCWACC] | −330 c | −506 c | −682 c | |||
| SEF4 binding site | SEF4MOTIFGM7S[RTTTTTR] | −129 c | −206 c | −382 c | −558 c | −734 c | |
| SPBF binding site | SP8BFIBSP8BIB[TACTATT] | −339 c | −515 c | −691 c | |||
| TATA box | TATATAA | −70 | |||||
| CAAT box | CAAT | −144, −151 | |||||
| W box | TTTGACY | −949 | |||||
Note: negative values indicate the first nucleotide position of the cis-element with respect to the transcription initiation site located at position +1.
aThe promoter of Ccs from C. chinense accession is divided into six parts, which consists of four tandem repeat (Repeat 1 - 4) flanked with proximal (Region I) and 5′-distal (Region II) sequence.
bRepeat 4 is lacking in C. annuum accession.
cSequence of the complementary strand.
Discussion
Since Capsicum exhibits variations in morphology and genetic composition, many studies of this plant involving cluster analysis with different molecular markers systems have been reported [22]–[26]. In these studies, AFLP analysis was useful for revealing high levels of polymorphism, while SSR method was useful for detecting specific genetic relationships between pepper cultivars [23], [25]. Here, we selected 32 SSR loci, including seven markers used by INRA (France), to perform cluster analysis of all of our local pepper cultivars. In this study, 32 SSR markers produced 154 polymorphic bands, with an average allelic variation of 4.8. In addition to Capsicum species, we could also distinguish the major chili and bell pepper lines among C. annuum cultivars. In fact, we failed to detect genetic difference between only five pairs of pepper lines (Figure 2). Among these pairs, B3 and B35, B6 and B27, B8 and B10, CK11 and CK13 were all selected from a same locality in Northwest China, with similar morphologies, which suggests that both lines in each pair are closely related. The remaining pair, HW201A and HW201B, is a male sterile line and its maintainer. This study revealed that, in addition to the AFLP method, SSR analysis is a powerful method for analyzing the phylogenetic relationship between C. annuum species.
The complex taxonomic relationship among Capsicum species implies that diverse genetic variation can lead to similar phenotype. To date, the heredity of mature pepper fruit color is still not fully understood. In the orange-fruited cultivars, ripe fruit color involves not only one or two structural changes in the genes of the carotenoid biosynthetic pathway, but also the transcriptional and/or post-transcriptional regulation of these genes [12]. However, in the yellow ripe-color pepper lines, fruit color is determined by variations in the Ccs gene [4], [7], [13]. Three structural mutations in the Ccs gene have thus far been identified in yellow fruit pepper lines. The first mutation involves a deletion in the gene region, which maybe didn’t contain the distal 220 bp of the 3′ end of this gene in yellow C. annuum cultivars [7], [13]. The second and third mutations, which involve a premature stop codon and a frame-shift, respectively, were detected in two C. chinense accessions [4]. Similar mutations in the Ccs gene in C. annuum and C. chinense have not previously been reported. Here, using cluster analysis, we determined that a local yellow-colored ripe fruit cultivar, CK7, is a C. annuum line that harbors a new ccs variant. In CK7, the gene has the same frame-shift mutation in CDS as identified in Fogo, an orange-colored mature C. annuum accession [11]. The mutant ccs variant in Fogo could result into a truncated production, and prevented the synthesis of capsanthin and capsorubin [12]. In addition, the ccs gene shares a similar expression pattern in both lines, which differs from the expression pattern detected in the yellow-colored mature C. chinense accessions [4]. These results suggest that there is a close genetic relationship between Fogo and CK7. Considering the different fruit colors exhibited in these two lines, the mutation in the Ccs gene may have occurred before the modified regulation of the gene, at least in the Fogo and CK7 accessions. Furthermore, the mutation that produced the yellow fruit phenotype may have occurred prior to the post-regulation detected in orange-colored mature fruit pepper. Additionally, a special upstream premature codon was found in CK7, whose appearance can be explained by the theory of high rate random mutation in a nonfunctioning gene.
Since the regulation of gene expression is another important area of investigation in the analysis of pepper fruit color, we thought it was important to examine the promoters of the Ccs gene. Ha et al. [4] identified three classes of promoter sequences in three Capsicum species and suggested that differences in this region reflect species-specific variations in Capsicum. We repeated their experiment, and PCR amplifications in two C. baccatum accessions (R26 and R38) yielded negative results (Figure 3). This may have been due to the different pepper varieties examined between both studies, as well as the complex genetic background of pepper. We detected the longer amplicons in the C. chinense lines (R28 and R30) that were also found by Ha et al. [4]. Sequence analysis revealed a unique tandem repeat structure in the Ccs promoter region. The tandem repeats comprise a major part of the promoter (65%, 528/812 bp in the amplicon from C. annuum) and are located near the transcriptional start point (165-bp upstream). Therefore, the presence of putative cis-elements, and the evolutionary significance of the repeats, cannot be ignored. Using bioinformatics, we detected some light and temperature sensitive elements in every repeat unit (Table 2), which may explain why the accumulation of capsanthin/capsorubin increases in response to abundant sunshine and high temperatures.
The proximal 176-bp tandem repeat structure in promoters is very rare in plants. The presence of this structure may have led to the deletion of the Ccs gene in most yellow-colored fruit C. annuum. Since the Ccs promoter is functional in tomato [27], it would be both valuable and feasible to study the effects of the proximal 165-bp promoter region and repeat unit on the transcriptional regulation of Ccs.
Supporting Information
Alignment of the deduced amino acid sequence of CCS and its homologous proteins.
(DOC)
Sequence comparisons of the four repeat units in the Ccs promoter region.
(DOC)
Simple sequence repeat (SSR) loci used in this study.
(DOC)
Gene-specific primers used in this study.
(DOC)
Acknowledgments
The authors wish to thank Prof. Alain Palloix (INRA PACA Avignon, France) for SSR primer sequences and the germplasm genotyping results.
Funding Statement
This work was supported by National Natural Science Foundation of China (No. 31101540 and No. 31272163), Research Fund for the Doctoral Program of Higher Education of China (No. 20110204120004), and Basic Scientific Research Fund from Northwest A&F University (No. QN2012010). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bartley GE, Scolnik PA (1995) Plant carotenoid: pigments for photoprotection, visual attraction, and human health. Plant Cell 7: 1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eshbaugh WH (1993) History and exploitation of a serendipitous new crop discovery. In: Janick J, Simon JE, editors.New crops.New York:Wiley. pp. 132–139. [Google Scholar]
- 3. Paran I, van der Knaap E (2007) Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J Exp Bot 58: 3841–3852. [DOI] [PubMed] [Google Scholar]
- 4. Ha SH, Kim JB, Park JS, Lee SW, Cho KJ (2007) A comparison of the carotenoid accumulation in Capsicum varieties that show different ripening colours: deletion of the capsanthin-capsorubin sythase gene is not a prerequisite for the formation of a yellow pepper. J Exp Bot 58: 3135–3144. [DOI] [PubMed] [Google Scholar]
- 5. Kormos J, Kormos K (1960) Die genetischen typen der carotenoid-systeme der paprikafrucht. Acta Bot Acad Sci Hun 6: 305–319. [Google Scholar]
- 6. Hurtado-Hernandez H, Smith PG (1985) Inheritance of mature fruit colour in Capsicum annuum L. . J Hered 76: 211–213. [Google Scholar]
- 7. Popovsky S, Paran I (2000) Molecular genetics of the y locus in pepper: its relation to capsanthin-capsorubin synthase and to fruit color. Theor Appl Genet 101: 86–89. [Google Scholar]
- 8. Thorup TA, Tanyolac B, Livingstone KD, Popovsky S, Paran I, et al. (2000) Candidate gene analysis of organ pigmentation loci in the Solanaceae. Proc Natl Acad Sci, USA 97: 11192–11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huh JH, Kang BC, Nahm SH, Kim S, Ha KS, et al. (2001) A candidate gene approach identified phytoene synthase as the locus for mature fruit color in red pepper (Capsicum spp). Theor Appl Genet 102: 524–530. [Google Scholar]
- 10. Lang Y, Yanagawa S, Sasanuma T, Sasakuma T (2004) Orange fruit colour in Capsicum due to deletion of capsanthin-capsorubin synthesis gene. Breeding Sci 54: 33–39. [Google Scholar]
- 11. Guzman I, Hamby S, Romero J, Bosland PW, O′Connell MA (2010) Variability of carotenoid biosynthesis in orange colored Capsicum spp. Plant Sci 179: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rodriguez-Uribe L, Guzman I, Rajapakse W, Richins RD, O′Connell MA (2012) Carotenoid accumulation in orange-pigmented Capsicum annuum fruit, regulated at multiple levels. J Exp Bot 63: 517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lefebvre V, Kuntz M, Camara B, Palloix A (1998) The capsanthin-capsorubin synthase gene: a candidate gene for the y locus controlling the red fruit colour in pepper. Plant Mol Biol 36: 785–789. [DOI] [PubMed] [Google Scholar]
- 14. Li Z, Pan JS, Guan Y, Tao QY, He HL, et al. (2008) Development and fine mapping of three co-dominant SCAR markers linked to the M/m gene in the cucumber plant (Cucumis sativus L.). Theor Appl Genet 117: 1253–1260. [DOI] [PubMed] [Google Scholar]
- 15. Minamiyama Y, Tsuro M, Hirai M (2006) An SSR-based linkage map of Capsicum annuum . Mol Breeding 18: 157–169. [Google Scholar]
- 16. Li Z, Huang SW, Liu SQ, Pan JS, Zhang ZH, et al. (2009) Molecular isolation of the M gene suggests that a conserved-residue conversion induces the formation of bisexual flowers in cucumber plants. Genetics 182: 1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohlf FJ (2000) NTSYS-PC, Numerical Taxonomy System for the PC, Exeter Software, Ver. 2.1. Available: http://www.exetersoftware.com/cat/ntsyspc/ntsyspc.html.Accessed 2013 Mar 26.
- 18. Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Z, Wang S, Tao QY, Pan JS, Si LT, et al. (2012) A putative positive feedback regulation mechanism in CsACS2 expression suggests a modified model for sex determination in cucumber (Cucumis sativus L.). J Exp Bot 63: 4475–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Borovsky Y, Paran I (2008) Chlorophyll breakdown during pepper fruit ripening in the chlorophyll retainer mutation is impaired at the homolog of the senescence-inducible stay-green gene. Theor Appl Genet 117: 235–240. [DOI] [PubMed] [Google Scholar]
- 21. Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene. Cell 76: 543–553. [DOI] [PubMed] [Google Scholar]
- 22. Toquica SP, Rodriguez F, Martinez E, Duque MC, Tohme J (2003) Molecular characterization by AFLPs of Capsicum germplasm from the amazon department in Colombia. Genet Res Crop Evol 50: 639–647. [Google Scholar]
- 23. Tam SM, Mhiri C, Vogelaar A, Kerkveld M, Pearce SR, et al. (2005) Comparative analyses of genetic diversities within tomato and pepper collections detected by retrotransposon-based SSAP, AFLP and SSR. Theor Appl Genet 110: 819–831. [DOI] [PubMed] [Google Scholar]
- 24. Portis E, Nervo G, Cavallanti F, Barchi L, Lanteri S (2006) Multivariate analysis of genetic relationships between Italian pepper landraces. Crop Sci 46: 2517–2525. [Google Scholar]
- 25. Joy N, Abraham Z, Soniya EV (2007) A preliminary assessment of genetic relationships among agronomically important cultivars of black pepper. BMC Genet 8: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thul ST, Darokar MP, Shasany AK, Khanuja SPS (2012) Molecular profiling for genetic variability in Capsicum species based on ISSR and RAPD markers. Mol Biotechnol 51: 137–147. [DOI] [PubMed] [Google Scholar]
- 27. Kuntz M, Chen HC, Simkin AJ, Romer S, Shipton CA, et al. (1998) Upregulation of two ripening-related genes from a non-climacteric plant (pepper) in a transgenic climacteric plant (tomato). Plant J 13: 351–361. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the deduced amino acid sequence of CCS and its homologous proteins.
(DOC)
Sequence comparisons of the four repeat units in the Ccs promoter region.
(DOC)
Simple sequence repeat (SSR) loci used in this study.
(DOC)
Gene-specific primers used in this study.
(DOC)