Abstract
The metastatic cascade is a complex and extremely inefficient process with many potential barriers. Understanding this process is of critical importance because the majority of cancer mortality is associated with metastatic disease. Recently, it has become increasingly clear that microRNAs (miRNAs) play important roles in tumorigenesis and metastasis, yet few studies have examined how germline variations may dysregulate miRNAs in turn affecting metastatic potential. To explore this possibility, the highly metastatic MMTV-PyMT mice were crossed with 25 AKXD (AKR/J × DBA/2J) recombinant inbred strains to produce F1 progeny with varying metastatic indices. When mammary tumors from the F1 progeny were analyzed by miRNA microarray, miR-290 (containing miR-290-3p and miR-290-5p) was identified as a top candidate progression-associated miRNA. The microarray results were validated in vivo when miR-290 up-regulation in two independent breast cancer cell lines suppressed both primary tumor and metastatic growth. Computational analysis identified breast cancer progression gene Arid4b as a top target of miR-290-3p, which was confirmed by luciferase reporter assay. Surprisingly, pathway analysis identified estrogen receptor (ER) signaling as the top canonical pathway affected by miR-290 upregulation. Further analysis demonstrated ER levels were elevated in miR-290-expressing tumors and positively correlated with apoptosis. Taken together, our results suggest miR-290 targets Arid4b while simultaneously enhancing ER signaling and increasing apoptosis, thereby suppressing breast cancer progression. This, to the best of our knowledge, is the first example of inherited differences in miRNA expression playing a role in breast cancer progression.
Keywords: miR-290, Arid4b, estrogen receptor, metastasis, breast cancer
INTRODUCTION
The metastatic cascade is a complex and extremely inefficient process with many potential barriers. Because as much as 90% of cancer-associated mortality is associated with metastatic disease rather than primary tumor burden, understanding the molecular mechanisms and pathways exploited by cancer cells during the metastatic process is of critical importance. Decades of metastasis research have led to a variety of different models of the metastatic process (1). Although the metastatic process likely involves a combination of models, our laboratory has focused on inherited genetic variants that influence metastatic potential (2). These efforts have resulted in the identification of a number of genes associated with metastatic potential in both mouse and humans (2–5). Since these genes do not appear to be frequently somatically mutated in breast cancer, identification and characterization of inherited metastasis susceptibility genes provide novel insights into the mechanisms underlying the terminal stages of breast cancer progression.
More recently there has been significant interest in characterizing the role of microRNAs in tumor progression. The discovery of microRNAs (miRNAs -- small, non-coding RNAs) in 1993 sparked a paradigm shift in our understanding of gene regulation. Initially, miRNAs were classified as functionless stretches within ‘junk DNA’, but are currently viewed as powerful regulators of gene expression as it relates to cellular development, death and proliferation (6), as well as disease (7). Because miRNAs negatively regulate gene expression post-transcriptionally, they can act as either tumor suppressors or oncogenes (8) depending on their gene targets and cellular context (9). Recently, much attention has been focused on the role dysregulated microRNAs (miRNAs) have on tumorigenesis (10) and metastasis (11), and whether miRNA profiles can be used for diagnosis and prognosis in cancer (12). Most interesting, germline mutations in microRNAs have been associated with skeletal and growth defects (13), chronic lymphocytic leukemia (CLL) (14) and breast cancer (15), thereby supporting the notion that germline mutations in certain miRNAs may create a predisposition towards tumorigenesis.
In this study, we analyzed miRNA expression levels in the mammary tumors of the [PyMT × AKXD] F1 progeny described previously (16) to determine if a link exists between inherited differences in miRNA expression and mammary tumor progression. We provide evidence that miR-290-3p, a member of the pluripotency miR-290-295 cluster (17–19), an ortholog of the human miR-371-373 cluster (19), is differentially expressed between the highly metastatic AKR/J and poorly metastatic DBA2/J strains of mice. Increased expression of miR-290 suppresses tumor growth and progression in a tumor autonomous manner, in part by targeting the recently described tumor progression gene, Arid4b (20). To the best of our knowledge this is the first example of an inherited miRNA expression difference being associated with tumor progression.
MATERIALS & METHODS
Cell lines
All cells were cultured in DMEM media with 10% FBS and antibiotics. Puromycin was used for selection.
Mouse strains
The AKXD RI mice were generated as described (21). The PyMT mouse strain FVB/N-TgN(MMTV-PyVT)634Mul (22) was then bred to 18 of the AKXD RI strains to generate 18 [PyMT × AKXD]F1 sublines (16).
miRNA microarray analysis of AKXD RI tumors
The LMT_miRNA_v2 microarray was designed using the Sanger miR9.0 database (http://microrna.sanger.ac.uk) and manufactured as custom-synthesized 8 × 15K microarrays (Agilent Technologies, San Jose, CA). Each mature miRNA was represented by + and − (reverse complement) strand sequences. Each probe has 4 replicates within each microarray, providing technical replicates for consistency and performance of the microarray. Each unique mature miRNA was represented by 8 probes (4 + strand and 4 − strand). A total of 3,556 unique LMT seq IDs (miRNA, positive and negative controls, +/− strand) were on the microarray, each with 4 replicates.
Total RNA (1 ug) was labeled using the miRCURY™ LNA microRNA Array Power Labeling kit (Exiqon Inc, Woburn, MA). The 3′-end of the RNA was enzymatically labeled with Hy3 for the sample RNA and Hy5 fluorescent dye (Exiqon) for the reference RNA (Ambion reference RNA) and co-hybridized onto the microarrays. The washed and dried slides were scanned using the Agilent scanner. The Feature Extraction program extracted spot intensities. The Log2Ratio of all signals was used for data analysis.
mRNA microarray analysis of 6DT1 miRNA tumors
The Agilent 2100 Bioanalyzer (Agilent Technologies) verified each sample RNA had a high quality score (RIN >9). The RNA (100 ng) was reverse transcribed and amplified using the Ambion WT Expression Kit. Sense strand cDNA was fragmented and labeled using the GeneChip WT Terminal Labeling and Controls Kit. Four replicates of each sample were hybridized to GeneChip Mouse Gene 1.0 ST Array in the GeneChip Hybridization Oven 645 while shaking at 60 rpm at 45°C for 16 hrs. Washing and staining were performed on the GeneChip Fluidics Station 450 and scanned on the GeneChip Scanner 3000. Data were collected using the GeneChip Command Console Software (AGCC). All reagents, software and instruments used, except for the Agilent 2100 Bioanalyzer, were from Affymetrix.
RNA isolation
Tumors were snap-frozen upon harvesting and stored at −80°C. All tumors were homogenized on dry ice in an Rnase-free environment. The RNA was isolated using the mirVana miRNA Isolation Kit (Ambion). The RNA for all remaining samples, including cell lines, was isolated using the RNAeasy Kit (Qiagen).
Quantitative real-time PCR and Western Blot
Total RNA was reverse transcribed with iScript cDNA Synthesis Kit (Bio-Rad) and PCR amplified using QuantiTect SYBR Green PCR Kit (Qiagen) on the Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosytems). The standard curve method was used for quantitation and normalized to endogenous control Ppib levels. TaqMan MicroRNA Assays (Applied Biosystems) were used to measure miRNA expression. Expression of miRNA was defined from the threshold cycle, and relative expression levels were calculated using the 2-ΔΔCt method (23) after normalization with reference snoRNA135.
Primers for RT-PCR: Ppib and Arid4b.
Mouse PPIB: 5′-GGAGATGGCACAGGAGGAAAGAG-3′ (forward)
Mouse PPIB: 5′-TGTGAGCCATTGGTGTCTTTGC-3′ (reverse)
Mouse ARID4B: 5′-AACAAAGGTGCAGGTGAAGC-3′ (forward)
Mouse ARID4B: 5′-ACATCAGTGCCCACTGTCAA-3′ (reverse)
Mouse ESR1: 5′-TCTCTGGGCGACATTCTTCT-3′ (forward)
Mouse ESR1: 5′-CATGGTCATGGTAAGTGGCA-3′ (reverse)
Mouse EGFR: 5′-GGCGTTGGAGGAAAAGAAAG-3′ (forward)
Mouse EGFR: 5′-ATCCTCTGCAGGCTCAGAAA-3′ (reverse)
Mouse C3: 5′-GGCCTTCTCTCTAACAGCCA-3′ (forward)
Mouse C3: 5′-TGCAGGTGACTTTGCTTTTG-3′ (reverse)
Mouse DLC1: 5′-CCTGGCTGGAATAGCATCAT-3′ (forward)
Mouse DLC1: 5′-ATGCATGGGTCAAGGAAGAG-3′ (reverse)
Mouse IL6ST: 5′-CTGAGGGACCGGTGGTGT-3′ (forward)
Mouse IL6ST: 5′-TCATGTTCCTTCTATCGGGTC-3′ (reverse)
Mouse IL2RA: 5′-TTGCTGATGTTGGGGTTTCT-3′ (forward)
Mouse IL2RA: 5′-AGGAGAGGGCTTTGAATGTG-3′ (reverse)
Mouse CDH1: 5′-GAGGTCTACACCTTCCCGGT-3′ (forward)
Mouse CDH1: 5′-AAAAGAAGGCTGTCCTTGGC-3′ (reverse)
Western blot with Arid4b was performed as described in (20). For the remaining Western blots, a PARP antibody (#9544, Cell Signaling) and an ER-alpha antibody (ab2746, Abcam) were used. Protein lysate was generated from tumors by homogenizing in RIPA buffer without detergents. Detergents were subsequently added (1% NP-40, 0.5% Na Deoxycholate, and 0.1% SDS) after homogenizing. Western blot bands were quantified using ChemiDoc-It Imaging System with VisionWorks LS software.
Generation of stable cell lines
The pEZX-MR06 plasmids (GeneCopoeia) that contained either miR-290 or a scrambled insert were transformed into GCl-L3 Chemically Competent E. coli Cells (GeneCopoeia) and purified using Plasmid Midi Kit (Qiagen). Two days before transfection, 293ta packaging cells (GeneCopoeia) were plated in a 10-cm dish with DMEM and 10% heat-inactivated FBS. The cells were transfected at 70–80% confluency according to the manufacturer’s instructions in the Lenti-Pac FIV Expression Kit (GeneCopoeia). Mvt-1 and 6DT1 cells were transduced using 200 ul of purified lentivirus and Polybrene. Media containing puromycin was applied 5 days post-transduction. Stable clones were generated by plating 5 cells/ml of media in each well of a 96-well plate. Fresh media containing puromycin was added to each well 48 h later. When the cells were ~ 80% confluent, they were collected and transferred to a T25 flask.
In vivo analysis
FVB/N (24) female mice at 6 weeks of age were orthotopically injected with 1.0 × 105 cells. Mice were euthanized 30 days after injection of 6DT1 cells and 42 days after injection of Mvt-1. Mammary tumors were removed and weighed, and each lung was analyzed for surface metastases and internal metastasis after histological sectioning: calculated as the mean number of metastases detected from 3 separate lung sections having 20 sections between each section.
Luciferase reporter assays
HEK293T cells were seeded in a 24-well plate and a construct containing the full-length 3′-UTR of human ARID4B adjacent to the Renilla gene (SwitchGear) was cotransfected with 50 nM of miRIDAN miRNA mimic (Dharmacon) and a control vector pGL4.13 [luc2/SV40] (Promega) using DharmaFECT DUO (Dharmacon) when the cells were ≥ 80% confluent. After 24 hours, the plate was stored at −80°C overnight. The following day, the cells in each well were mixed by pipetting and then divided evenly. Half of the sample was treated with Steady-Glo reagent (Promega) and the remaining half was treated with LightSwitch reagent (Switchgear). All samples were quantified using the Lumat LB 9507 (EG&G Berthold) luminometer and the Renilla luciferase activity was normalized to the Firefly luciferase activity. miRIDAN miRNA mimic negative control was used as the control miRNA. Experiments were performed in triplicate.
Statistical and bioinformatic analysis
Error bars depict S.E. of the mean (SEM). GraphPad Prism was applied to calculate statistical significance. The in vivo data was analyzed using Mann-Whitney while remaining calculations were performed using the unpaired Student’s t-test. Microarray data was analyzed using Partek Genomics Suite. Differentially expressed genes were identified with ANOVA analysis; genes with a p < 0.05 were considered significant. Significant genes were analyzed for pathway enrichment using Ingenuity Pathway Analysis software. The MFold program was used to determine the structure of mmu-miR-290. TargetScan, miRDB, and microRNA.org were applied using default parameters. *p < 0.05, **p < 0.01, ***p < 0.001.
Site-directed mutagenesis
Site-directed mutagenesis was performed using high-fidelity DNA polymerase, ARID4B 3′-UTR vector (#S210100, SwitchGear Genomics) and two synthetic oligonucleotide primers containing the desired mutation in a temperature cycler. The mutagenesis reaction was transformed into DH5a supercompetent cells, and then the mutant plasmid DNA (isolated from a single colony) was verified by DNA sequencing.
RESULTS
miR-290 expression is significantly correlated with tumor latency and metastatic index
Previously, the highly metastatic FVB/N-TgN (MMTV-PyMT) mice were crossed with 18 AKXD RI (AKR/J × DBA/2J) strains to map modifiers associated with tumor latency, tumor burden and metastatic index (Supplementary Fig. S1) (16). To identify miRNAs that might contribute to metastatic progression we evaluated miRNA expression levels within all mammary tumors by miRNA microarray. The miRNA microarray data was analyzed using Partek Genomics Suite software and correlated with tumor burden, tumor latency and metastatic index ([log2(number of metastases/lung area (μm2))]) (Table 1). 1-way ANOVA analysis revealed a significant positive correlation between miR-290 and latency and a significant negative correlation with metastatic index. Further 2-way ANOVA analysis produced a significant negative correlation between miR-290 with metastatic index and tumor burden; and metastatic index and latency. Lastly, 3-way ANOVA analysis produced a significant negative correlation between miR-290 with metastatic index, latency, and tumor burden. Importantly, these ANOVA results suggested miR-290 may play a suppressor role in during breast cancer progression.
Table 1.
Correlation Between miR-290 and Phenotype
| Phenotype | P-value | Factor | Correlation |
|---|---|---|---|
| 1-way Metastatic Index (MI) | 0.014 | 7.62 | −0.56 |
| * 2-way MI and Tumor Burden (TB) | 0.006 | 10.3 | −0.63 |
| * 2-way MI and Latency | 0.021 | 6.66 | −0.54 |
Only the metastatic index statistics are listed.
Only parameters producing a mean f ratio much greater than 1.0 are listed.
This was not the case with 1-way ANOVA of tumor burden (F = 1.02) and 2-way ANOVA of tumor burden (F = 1.04) and latency (F = 1.48).
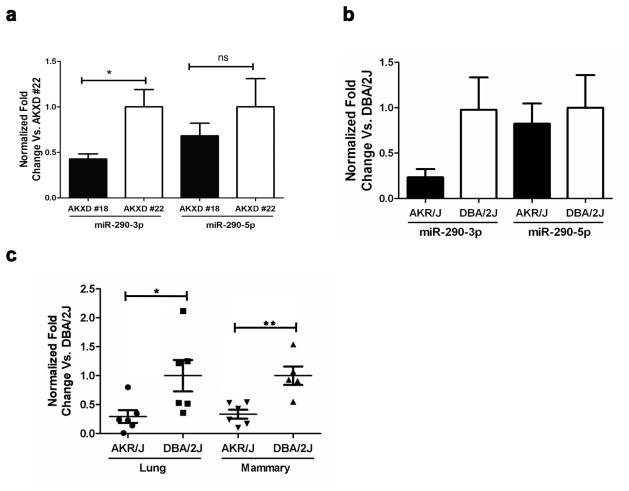
The structure of miR-290 includes two functionally active mature miRNAs: miR-290-3p and miR-290-5p (Supplementary Fig. S2). To validate the microarray findings and determine if only one or both of the mature miRNAs are correlated with tumor progression, the expression levels of miR-290-3p and miR-290-5p were compared in mammary tumors from AKXD strain #18 (the most metastatic) and AKXD strain #22 (the least metastatic) (Fig. 1a). The miR-290-3p expression was downregulated more than 50% in AKXD strain #18 compared to AKXD strain #22. In contrast, miR-290-5p displayed virtually no expression difference between the two strains. Likewise, the expression levels of miR-290-3p and miR-290-5p were evaluated in mammary tumors from the AKXD parental mouse strains, the highly metastatic AKR/J mice and poorly metastatic DBA/2J mice (Fig. 1b). Consistent with an anti-metastatic role, the expression of miR-290-3p was down-regulated 70% in the AKR/J tumors compared to the DBA/2J tumors, while miR-290-5p was only slightly down-regulated. Lastly, to explore whether certain genetic strains may contain a predisposition towards tumorigenesis and metastasis through an inherited up- or down-regulation of specific miRNAs, miR-290-3p expression was measured in normal mammary and normal lung tissue from AKR/J mice and DBA/2J mice (Fig. 1c). Interestingly, both tissues displayed more than a 50% down-regulation of miR-290-3p expression in the AKR/J mice, thereby supporting a potential predisposition towards breast carcinogenesis in the AKR/J strain. Taken together, these results further suggested miR-290-3p may act as a tumor and metastasis suppressor miRNA, and thereby warranted further investigation.
Fig. 1.
miR-290-3p and miR-290-5p expression analysis in mammary tumors and normal tissues. (a) miR-290-3p expression was measured in mammary tumors from the highly metastatic AKXD strain #18 versus the less metastatic AKXD strain #22 and in the (b) highly metastatic AKR/J strain (n = 5) versus the less metastatic DBA/2J strain (n = 4). (c) miR-290-3p expression was measured in normal lung and normal breast tissues from the AKR/J versus the DBA/J strain. RNA from the AKXD samples was analyzed five times via qRT-PCR. *p < 0.05, **p < 0.01, ***p < 0.001.
miR-290 suppresses mammary tumor progression
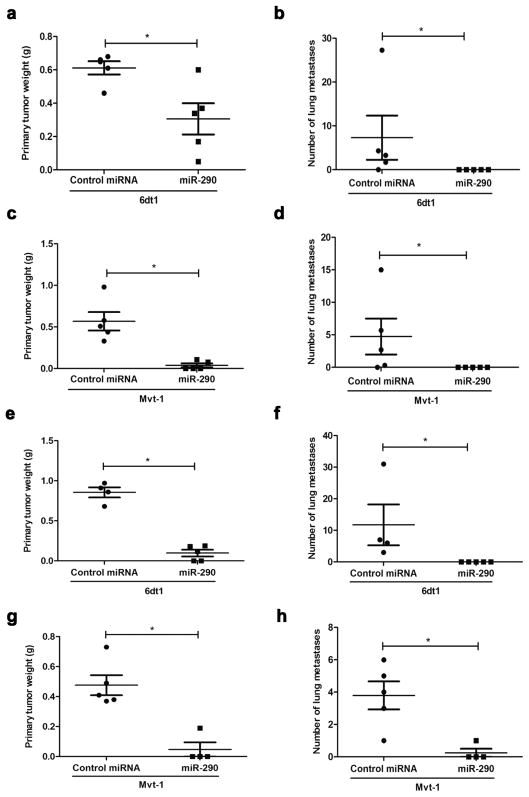
To determine the phenotypic effects of miR-290 expression on breast cancer progression in vivo, a lentivirus construct was used to up-regulate miR-290 expression in the highly metastatic mouse mammary tumor cell lines 6DT1 and Mvt-1 (Supplementary Table S1). Heterogeneous populations expressing the miR-290 construct were injected orthotopically into the mammary fat pad of FVB/N mice. In vivo analysis demonstrated miR-290 expression in the 6dt1 cell population significantly decreased the primary tumor weight approximately 50% (Fig. 2a) and the number of lung metastases from approximately 7 lesions per lung section to zero lesions (Fig. 2b). Likewise, miR-290 expression in the Mvt-1 cell population reduced primary tumor weight by approximately 50% (Fig. 2c) and the number of lung metastases from approximately 5 lesions to zero lesions (Fig. 2d).
Fig. 2.
miR-290 suppresses breast cancer tumorigenesis and lung metastasis. miR-290 expression in the 6dt1 and Mvt-1 heterogenous populations suppressed primary tumor weight (a, c) and the number of lung metastasis (b, d). Clones generated from the heterogenous populations also suppressed primary tumor weight (e, g) and the number of lung metastasis (f, h).
To confirm these results and eliminate any potential bias due to population heterogeneity, single cell clones were generated from both the Mvt1 and 6DT1 heterogeneous populations and injected as above. The results were even more dramatic with mammary tumor burden reduced 75% (Fig. 2e) and the average number of lung lesions reduced from 10 to zero (Fig. 2f) for the miR-290 6dt1 clone. Similarly, the miR-290 Mvt-1 clone displayed a 80% reduction in primary tumor size (Fig. 2g) and an average of 1 versus 4 lung lesions in the negative control (Fig. 2h). To be sure miRNA expression was maintained after orthotopic injection, the mammary tumors generated from the 6DT1 clones were analyzed. Expression analysis demonstrated all the miR-290 tumors displayed elevated miR-290-3p and miR-290-5p expression compared to the negative control tumors (Supplementary Fig. S3).
miR-290-3p targets breast cancer progression gene Arid4b
To identify interesting potential targets of miR-290-3p, bioinformatic analysis using TargetScan, miRDB and microRNA.org was applied. Targetscan and miRDB identified Arid4b as the top target, while microRNA.org identified Arid4b as the second-ranked target (Table 2). Interestingly, elevated levels of Arid4b have been previously linked to breast carcinogenesis (25, 26) and recent studies in our laboratory have shown Arid4b promotes breast cancer progression (20). Therefore, analysis was performed to determine whether miR-290-3p might inhibit mammary cancer progression in part by suppression of Arid4b.
Table 2.
Top Bioinformatic Targets of miR-290-3p
| Program | Gene | Gene Name | Scoreŧ | Rank |
|---|---|---|---|---|
| TargetScan | ARID4B | AT rich interactive domain 4B | −0.85 | 1 |
| miRDB | ARID4B | AT rich interactive domain 4B | 96 | 1 |
| microRNA.org | RBL2 | retinoblastoma-like 2 | −2.98 | 1 |
| ARID4B | AT rich interactive domain 4B | −2.22 | 2 |
Target scan = total context score; miRDB = score; microRNA.org = mirSVR score
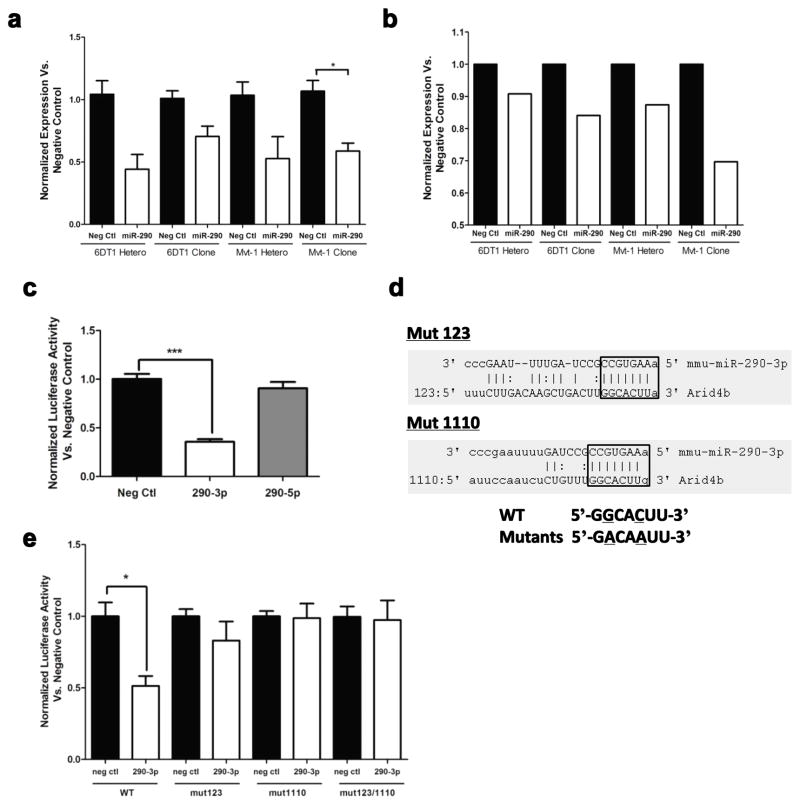
First, Arid4b RNA levels were compared in the clones and heterogeneous populations. Expression analysis detected an approximate 50% reduction in Arid4b mRNA levels (Fig. 3a) and a 10% to 30% reduction in protein levels (Fig. 3b) in the miR-290 up-regulated cells. Next, to determine if miR-290-3p directly targets the 3′-UTR of Arid4b, a luciferase construct containing the 3′-UTR of Arid4b was transfected with a miRNA negative control mimic, a miR-290-3p mimic or a miR-290-5p mimic; a 50% reduction in luciferase activity was observed only with the miR-290-3p mimic (Fig. 3c). To further demonstrate miR-290-3p interacts directly with two seed binding regions within the Arid4b 3′-UTR, two point mutations were generated in each seed binding region and denoted Mut123 and Mut1110 (Fig. 3d). Although a significant reduction in luciferase activity was observed for the WT construct, high luciferase activity was maintained in all of the mutants (Fig. 3e), thereby supporting the direct interaction between miR-290-3p and these two targeted regions within the Arid4b 3′-UTR.
Fig. 3.
miR-290-3p targets the 3′UTR of Arid4b. Arid4b expression was measured by (a) qRT-PCR and (b) Western blot in all four stable cell lines with upregulated miR-290. c) Luciferase activity was measured after transfection of a vector containing the 3′-UTR of ARID4B adjacent to a Renilla gene with miR-290-3p or miR-290-5p. d) Two miR-290-3p targeted regions within the 3′-UTR of ARID4B were identified and then mutated using 2 point mutations each. e) Luciferase activity was measured for the WT 3′UTR of ARID4B and the mutants after miR-290-3p transfection. *p < 0.05, **p < 0.01, ***p < 0.001.
miR-290 up-regulates ER expression, ER signaling and ER-associated apoptosis
Because miRNAs affect numerous signaling pathways and genes, rather than a single gene, and because the tumor microenvironment can dramatically influence gene expression, cDNA microarray analysis was conducted on the tumors generated from the 6DT1 clones. These samples were chosen because they displayed the greatest suppression of pulmonary metastasis (Fig. 2f) and because they were previously shown to maintain miR-290 expression within the individual tumors (Supplementary Fig. S3). A total of 4 negative control tumors and 3 miR-290 expressing tumors (the remaining 2 tumors were too small for analysis) were arrayed.
Bioinformatic analysis of the 6DT1 tumors generated a list of 7,015 differentially regulated genes (p < 0.05) with fold-changes between approximately −6-fold to 74-fold in the miR-290 tumors (Supplemental Table 2S). The top ten down-regulated genes included granzyme D and E precursors, and mast cell protease 1 precursor. The list of top ten up-regulated genes included the metabolism-associated genes Cyp2e1 and Ces1d. Interestingly, many genes associated with adipocytes were up-regulated in the miR-290 tumors: Retn (27) and Cfd (28) are both secreted by adipocytes; Adipoq (29) is expressed exclusively in adipose tissue and has anti-inflammatory activities (30); lgals12 is a primary regulator of the early stages of adipose tissue development (31); and lastly Cidec may mediate adipocyte apoptosis (32).
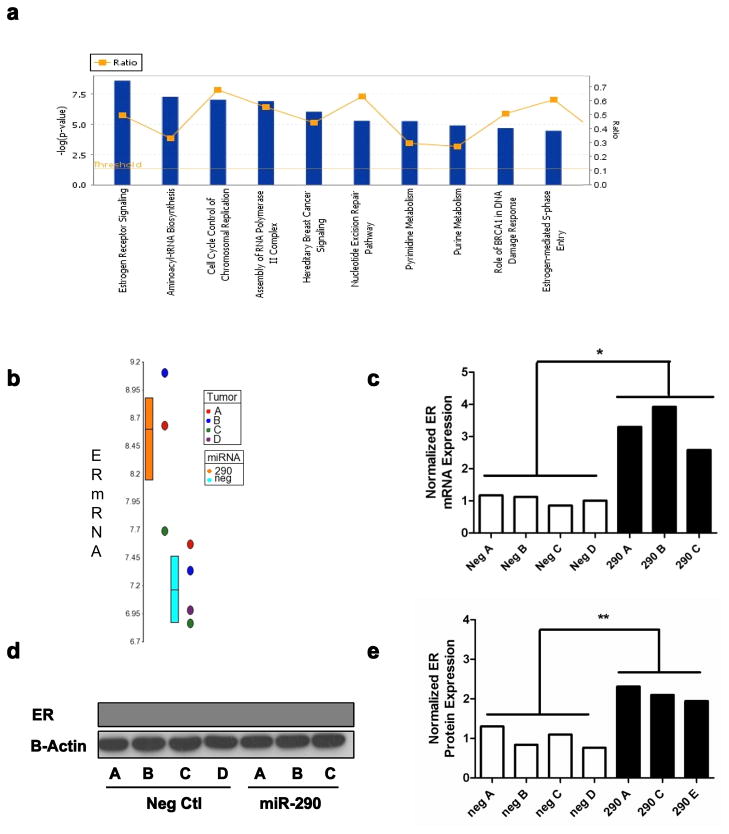
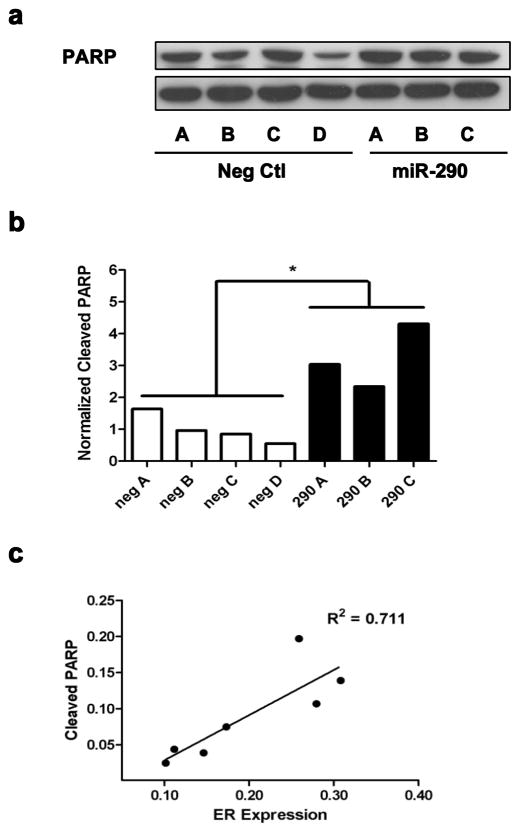
The expression data was then processed using Ingenuity Pathway Analysis (IPA) to identify biological functions associated with miR-290 expression; a total of 38 dysregulated canonical pathways (p < 0.01) were identified. The most significantly dysregulated pathways identified were pyrimidine and purine metabolism, aminoacyl-tRNA biosynthesis, cell cycle control of chromosomal replication, assembly of RNA polymerase II complex, nucleotide excision repair pathway, and a number of pathways associated with breast cancer and estrogen receptor (ER) signaling: hereditary breast cancer signaling, role of BRCA1 in DNA damage response, estrogen-mediated S-phase entry and estrogen receptor signaling (Fig. 4a). Dysregulation of the ER signaling pathway in the miR-290 tumors was further validated by measuring the expression level of a subset of genes within the ER signaling pathway (EGFR, C3, DLC1, IL6ST, IL2RA, CDH1)(33, 34) via microarray and qRT-PCR (Supplementary Fig. S4). Furthermore because estrogen receptor signaling was the most significantly dysregulated we sought to understand if the enhanced signaling was a consequence of upregulated ER expression. For this, we analyzed the mRNA levels observed in the microarray results (Fig. 4b) and validated those via qRT-PCR (Fig. 4c); both methods confirmed ER upregulation in the miR-290 tumors. Likewise, Western blot analysis (Fig. 4d) confirmed significantly elevated ER levels in the miR-290 tumors (Fig. 4e). Furthermore because previous publications have documented a link between elevated ER signaling and apoptosis (35, 36) this relationship was explored in vivo. Apoptosis was measured via Western blot in the 6DT1 tumors through detection of the 89 kDa PARP fragment resulting from caspase cleavage (Fig. 5a). Interestingly, a significantly elevated level of apoptosis was observed in the miR-290 tumors (Fig. 5b) that was positively correlated with ER expression (r2 = 0.711) (Fig. 5c).
Fig. 4.
miR-290 enhances ER-alpha signaling and expression levels. a) The top ten canonical pathways dysregulated by miR-290. The blue bar represents p-value and the yellow squares represent the ratio of genes dysregulated in the pathway. ER mRNA levels were measured in the miR-290 tumors by b) microarray analysis and c) qRT-PCR. ER protein levels were detected in the miR-290 tumors by d) Western blot and e) quantified.
Fig. 5.
Elevated ER expression is positively correlated with apoptosis. The level of apoptosis was measured in the 6dt1 tumors through a) Western blot detection of cleaved PARP and b) quantitation. c) A correlation between cleaved PARP and ER expression was observed in the 6dt1 tumors.
DISCUSSION
Metastasis is a complex phenotype that requires many molecular and cellular events. Recent evidence suggests that in addition to tumor cell autonomous events, such as somatic mutation, non-tumor cells and tissues also play a significant role in tumor progression. Cancer initiation and metastasis should therefore be considered a disease of the whole organism, rather than focused on the loss of proliferative control in an individual tissue. As a consequence, a complete understanding of the metastatic process requires better characterization of factors that influence the biology of the entire organism, not just a specific cell or tissue type.
In addition to key environmental factors that affect organismal biology, genetic background is an underappreciated factor that can dramatically influence tumor initiation and progression. By influencing gene expression and efficiency of gene function, inherited polymorphisms determine not only the morphological features that make individuals unique, but also establish the spectrum of sensitivity or resistance to particular disease states, such as cancer, heart disease, diabetes, etc. Along these lines, studies from our laboratory have previously established the strong relationship between genetic predisposition and the metastatic process (37). Therefore the identification and characterization of polymorphic factors that establish sensitivity or resistance to metastatic disease should in turn provide invaluable incite regarding the mechanistic basis of tumor dissemination and progression.
Our laboratory previously focused on how polymorphisms can influence the metastatic function of protein coding genes. Yet recent evidence has demonstrated that miRNA dysregulation can significantly alter signaling networks thereby affecting tumor progression and metastasis (38). We therefore extended our research focus to explore whether constitutional differences in miRNA expression might influence the metastatic potential of mammary tumors. This involved profiling miRNA expression across an AKXD recombinant inbred genetic reference panel to identify miRNAs that are associated with metastatic progression. The AKXD RI panel consisted of 20 substrains of mice derived from an original cross between the high metastatic AKR/J and low metastatic DBA/2J inbred strains of mice (21). By profiling miRNA expression across this panel, we observed a correlation between miR-290 expression and metastatic burden, thus suggesting miR-290 is a potential metastasis-associated miRNA. Subsequent in vivo analysis confirmed miR-290 suppresses breast cancer progression through suppression of both primary and metastatic tumor growth. In addition, analysis of non-neoplastic tissues from AKR/J and DBA/2J mice demonstrated that differential expression of miR-290 exists prior to oncogenesis, thereby suggesting miR-290-associated suppressive effects are at least in part an inherited rather than a purely somatic event, yet the exact nature of the expression polymorphism has not been established.
When bioinformatics analysis was performed to establish a mechanistic basis for the suppressive effect of miR-290, the recently described metastasis susceptibility gene Arid4b (20) was unexpectedly identified as a top target, and the direct targeting was subsequently validated by luciferase assay. Arid4b had previously been identified as a potential metastasis susceptibility gene in our laboratory based on the strong correlation observed between Arid4b expression with both tumor growth and metastatic burden in the AKXD RI panel. In brief, multiple amino acid substitutions in the Arid4b gene were identified in the AKR/J strain compared to the reference mouse genome. These non-synonymous substitutions increased binding of the AKR/J strain of Arid4b to members of the SIN3 HDAC complex and reduced metastatic burden compared to the DBA/2J allele. Similarly, over expression of either allele was found to increase tumor growth compared to the control. In contrast, knock down of endogenous Arid4b reduced metastatic burden, yet did not significantly affect primary tumor growth. Hence the suppression of tumor growth and metastasis associated with miR-290-3p targeting of Arid4b is consistent with previous findings in our lab, and suggests the tumor suppression properties of miR-290 are at least partially related to Arid4b targeting.
Although part of the suppressive effect of miR-290-3p may be accounted for by Arid4b targeting, additional pathways are likely affected because, unlike the Arid4b knock down, miR-290 expression significantly suppresses primary tumor burden. Along these lines, subsequent microarray analysis demonstrated miR-290 enhances estrogen receptor expression and signaling. These unexpected results are particularly noteworthy because they are contrary to the currently accepted and long held belief that stimulation of ER by estrogen fuels cell proliferation and breast cancer progression (39), thereby creating a vast industry focused on the development of antiestrogen therapies and aromatase inhibitors to block ERα activity (40, 41). Yet, as mentioned previously, some reports have documented the therapeutic benefits of high-dose estrogen therapy for treatment of breast cancer (42–44), arguing that estrogen can collapse the enhanced survival pathways (i.e. HER2/neu and NFκβ) supported by exhaustive antiestrogen treatment while simultaneously promoting apoptosis by activating caspase-8 and synthesis of Fas receptor (35, 36). Consequently, the activation of apoptosis as measured by PARP cleavage in miR-290-expressing cells is consistent with the latter scenario, which suggests this relationship may contribute to the tumor suppressive effects observed.
Another potential mechanism of tumor and metastasis suppression by miR-290 is suppression of differentiation. Support for this mechanism comes from reports demonstrating miR-290 is enriched in ES cells and reduced after differentiation (45), and members of the miR-290-295 cluster make up greater than 70% of the miRNAs in ES cells (46). In addition, current hypotheses suggest cells at the invading fronts of tumor masses, as well as disseminating cells, undergo an obligate epithelial-to-mesenchymal transition and acquire stem cell-like characteristics. Once established in a secondary site, cells are thought to reverse this process to re-establish a more epithelial-like state before proliferation resumes. Furthermore, recent work on induced pluripotent stem cells (iPSCs) suggests miR-290 may play an important role in reprogramming and maintenance of a stem cell-like state. In short, reprogramming has been achieved when the four reprogramming factors OSKM (Oct3/4, Sox2, Klf4, and c-Myc) are ectopically expressed in mouse embryonic fibroblasts and human fibroblasts (47). And reports show c-Myc induces the expression of miRNAs associated with pluripotency such as the miR-290-295 (48), miR-302 (49), and the miR-17-92 (50) clusters. Hence sustained expression of miR-290 within tumor cells might inhibit the subsequent mesenchymal-to-epithelial transition required for tumor growth and proliferation at a secondary site.
In summary, we have demonstrated that expression of miR-290 in highly metastatic breast cancer cell lines significantly decreases tumor progression. Moreover, our results suggest that inherited differences in expression of miRNAs, in addition to somatically acquired alterations in expression, may be important determinants of tumor progression. The precise mechanism for miR-290 tumor suppression is unclear but likely involves multiple factors, such as Arid4b targeting; enhanced ER signaling and apoptosis; and cellular reprogramming. Further research into each of these explanations is certainly required to determine whether any, or a combination of these is a contributing to the effect. Nonetheless, the results described herein certainly spark future investigations by providing additional molecular insights into distinct factors that regulate breast cancer progression.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Glenn Merlino for critical reading of the manuscript and helpful discussion. This work was supported by the National Institutes of Health (NIH), NCI, Center for Cancer Research, Intramural Research Program and the Howard Hughes Medical Institute-NIH Research Scholars Program.
Footnotes
Conflict of interest statement: The authors of this manuscript certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
References
- 1.Hunter KW, Crawford NP, Alsarraj J. Mechanisms of metastasis. Breast cancer research : BCR. 2008;10 (Suppl 1):S2. doi: 10.1186/bcr1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford NP, Ziogas A, Peel DJ, Hess J, Anton-Culver H, Hunter KW. Germline polymorphisms in SIPA1 are associated with metastasis and other indicators of poor prognosis in breast cancer. Breast Cancer Res. 2006;8:R16. doi: 10.1186/bcr1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford NP, Alsarraj J, Lukes L, Walker RC, Officewala JS, Yang HH, et al. Bromodomain 4 activation predicts breast cancer survival. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6380–5. doi: 10.1073/pnas.0710331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford NP, Qian X, Ziogas A, Papageorge AG, Boersma BJ, Walker RC, et al. Rrp1b, a new candidate susceptibility gene for breast cancer progression and metastasis. PLoS Genet. 2007;3:e214. doi: 10.1371/journal.pgen.0030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsieh SM, Lintell NA, Hunter KW. Germline polymorphisms are potential metastasis risk and prognosis markers in breast cancer. Breast Dis. 2006;26:157–62. doi: 10.3233/bd-2007-26114. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–62. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 8.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–96. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 9.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–78. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–4. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 11.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 12.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 13.de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S, et al. Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43:1026–30. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 15.Li W, Duan R, Kooy F, Sherman SL, Zhou W, Jin P. Germline mutation of microRNA-125a is associated with breast cancer. J Med Genet. 2009;46:358–60. doi: 10.1136/jmg.2008.063123. [DOI] [PubMed] [Google Scholar]
- 16.Hunter KW, Broman KW, Voyer TL, Lukes L, Cozma D, Debies MT, et al. Predisposition to efficient mammary tumor metastatic progression is linked to the breast cancer metastasis suppressor gene Brms1. Cancer Res. 2001;61:8866–72. [PubMed] [Google Scholar]
- 17.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–67. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 18.Houbaviy HB, Dennis L, Jaenisch R, Sharp PA. Characterization of a highly variable eutherian microRNA gene. RNA. 2005;11:1245–57. doi: 10.1261/rna.2890305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 20.Winter SF, Lukes L, Walker RC, Welch DR, Hunter KW. Allelic variation and differential expression of the mSIN3A histone deacetylase complex gene Arid4b promote mammary tumor growth and metastasis. PLoS Genetics. 2012 doi: 10.1371/journal.pgen.1002735. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mucenski ML, Taylor BA, Jenkins NA, Copeland NG. AKXD recombinant inbred strains: models for studying the molecular genetic basis of murine lymphomas. Mol Cell Biol. 1986;6:4236–43. doi: 10.1128/mcb.6.12.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. MCB. 1992;12:954–61. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Taketo M, Schroeder AC, Mobraaten LE, Gunning KB, Hanten G, Fox RR, et al. FVB/N: an inbred mouse strain preferable for transgenic analyses. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:2065–9. doi: 10.1073/pnas.88.6.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao J, Gao T, Stanbridge EJ, Irie R. RBP1L1, a retinoblastoma-binding protein-related gene encoding an antigenic epitope abundantly expressed in human carcinomas and normal testis. J Natl Cancer Inst. 2001;93:1159–65. doi: 10.1093/jnci/93.15.1159. [DOI] [PubMed] [Google Scholar]
- 26.Cui D, Jin G, Gao T, Sun T, Tian F, Estrada GG, et al. Characterization of BRCAA1 and its novel antigen epitope identification. Cancer Epidemiol Biomarkers Prev. 2004;13:1136–45. [PubMed] [Google Scholar]
- 27.Kim KH, Lee K, Moon YS, Sul HS. A cysteine-rich adipose tissue-specific secretory factor inhibits adipocyte differentiation. The Journal of biological chemistry. 2001;276:11252–6. doi: 10.1074/jbc.C100028200. [DOI] [PubMed] [Google Scholar]
- 28.White RT, Damm D, Hancock N, Rosen BS, Lowell BB, Usher P, et al. Human adipsin is identical to complement factor D and is expressed at high levels in adipose tissue. The Journal of biological chemistry. 1992;267:9210–3. [PubMed] [Google Scholar]
- 29.Hu E, Liang P, Spiegelman BM. AdipoQ is a novel adipose-specific gene dysregulated in obesity. The Journal of biological chemistry. 1996;271:10697–703. doi: 10.1074/jbc.271.18.10697. [DOI] [PubMed] [Google Scholar]
- 30.Peake PW, Shen Y, Campbell LV, Charlesworth JA. Human adiponectin binds to bacterial lipopolysaccharide. Biochemical and biophysical research communications. 2006;341:108–15. doi: 10.1016/j.bbrc.2005.12.162. [DOI] [PubMed] [Google Scholar]
- 31.Yang RY, Hsu DK, Yu L, Chen HY, Liu FT. Galectin-12 is required for adipogenic signaling and adipocyte differentiation. The Journal of biological chemistry. 2004;279:29761–6. doi: 10.1074/jbc.M401303200. [DOI] [PubMed] [Google Scholar]
- 32.Tang X, Xing Z, Tang H, Liang L, Zhao M. Human cell-death-inducing DFF45-like effector C induces apoptosis via caspase-8. Acta biochimica et biophysica Sinica. 2011;43:779–86. doi: 10.1093/abbs/gmr073. [DOI] [PubMed] [Google Scholar]
- 33.Cardamone MD, Bardella C, Gutierrez A, Di Croce L, Rosenfeld MG, Di Renzo MF, et al. ERalpha as ligand-independent activator of CDH-1 regulates determination and maintenance of epithelial morphology in breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7420–5. doi: 10.1073/pnas.0903033106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leunen K, Gevaert O, Daemen A, Vanspauwen V, Michils G, De Moor B, et al. Recurrent copy number alterations in BRCA1-mutated ovarian tumors alter biological pathways. Human mutation. 2009;30:1693–702. doi: 10.1002/humu.21135. [DOI] [PubMed] [Google Scholar]
- 35.Osipo C, Gajdos C, Liu H, Chen B, Jordan VC. Paradoxical action of fulvestrant in estradiol-induced regression of tamoxifen-stimulated breast cancer. J Natl Cancer Inst. 2003;95:1597–608. doi: 10.1093/jnci/djg079. [DOI] [PubMed] [Google Scholar]
- 36.Liu H, Lee ES, Gajdos C, Pearce ST, Chen B, Osipo C, et al. Apoptotic action of 17beta-estradiol in raloxifene-resistant MCF-7 cells in vitro and in vivo. J Natl Cancer Inst. 2003;95:1586–97. doi: 10.1093/jnci/djg080. [DOI] [PubMed] [Google Scholar]
- 37.Lifsted T, Le Voyer T, Williams M, Muller W, Klein-Szanto A, Buetow KH, et al. Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int J Cancer. 1998;77:640–4. doi: 10.1002/(sici)1097-0215(19980812)77:4<640::aid-ijc26>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 38.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 39.Jordan VC, Brodie AM. Development and evolution of therapies targeted to the estrogen receptor for the treatment and prevention of breast cancer. Steroids. 2007;72:7–25. doi: 10.1016/j.steroids.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osipo C, Liu H, Meeke K, Jordan VC. The consequences of exhaustive antiestrogen therapy in breast cancer: estrogen-induced tumor cell death. Exp Biol Med (Maywood) 2004;229:722–31. doi: 10.1177/153537020422900804. [DOI] [PubMed] [Google Scholar]
- 41.Jordan VC. Tamoxifen: a most unlikely pioneering medicine. Nat Rev Drug Discov. 2003;2:205–13. doi: 10.1038/nrd1031. [DOI] [PubMed] [Google Scholar]
- 42.Haddow A, Watkinson JM, Paterson E, Koller PC. Influence of Synthetic Oestrogens on Advanced Malignant Disease. Br Med J. 1944;2:393–8. doi: 10.1136/bmj.2.4368.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lonning PE, Taylor PD, Anker G, Iddon J, Wie L, Jorgensen LM, et al. High-dose estrogen treatment in postmenopausal breast cancer patients heavily exposed to endocrine therapy. Breast Cancer Res Treat. 2001;67:111–6. doi: 10.1023/a:1010619225209. [DOI] [PubMed] [Google Scholar]
- 44.Ingle JN. Estrogen as therapy for breast cancer. Breast Cancer Res. 2002;4:133–6. doi: 10.1186/bcr436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu P, Reid JG, Gao X, Shaw CA, Creighton C, Tran PL, et al. Novel microRNA candidates and miRNA-mRNA pairs in embryonic stem (ES) cells. PLoS One. 2008;3:e2548. doi: 10.1371/journal.pone.0002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–33. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–6. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 48.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–9. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.